The Role of Phosphatidylinositol 3-Kinase Catalytic Subunit Type 3 in the Pathogenesis of Human Cancer
Abstract
1. Introduction
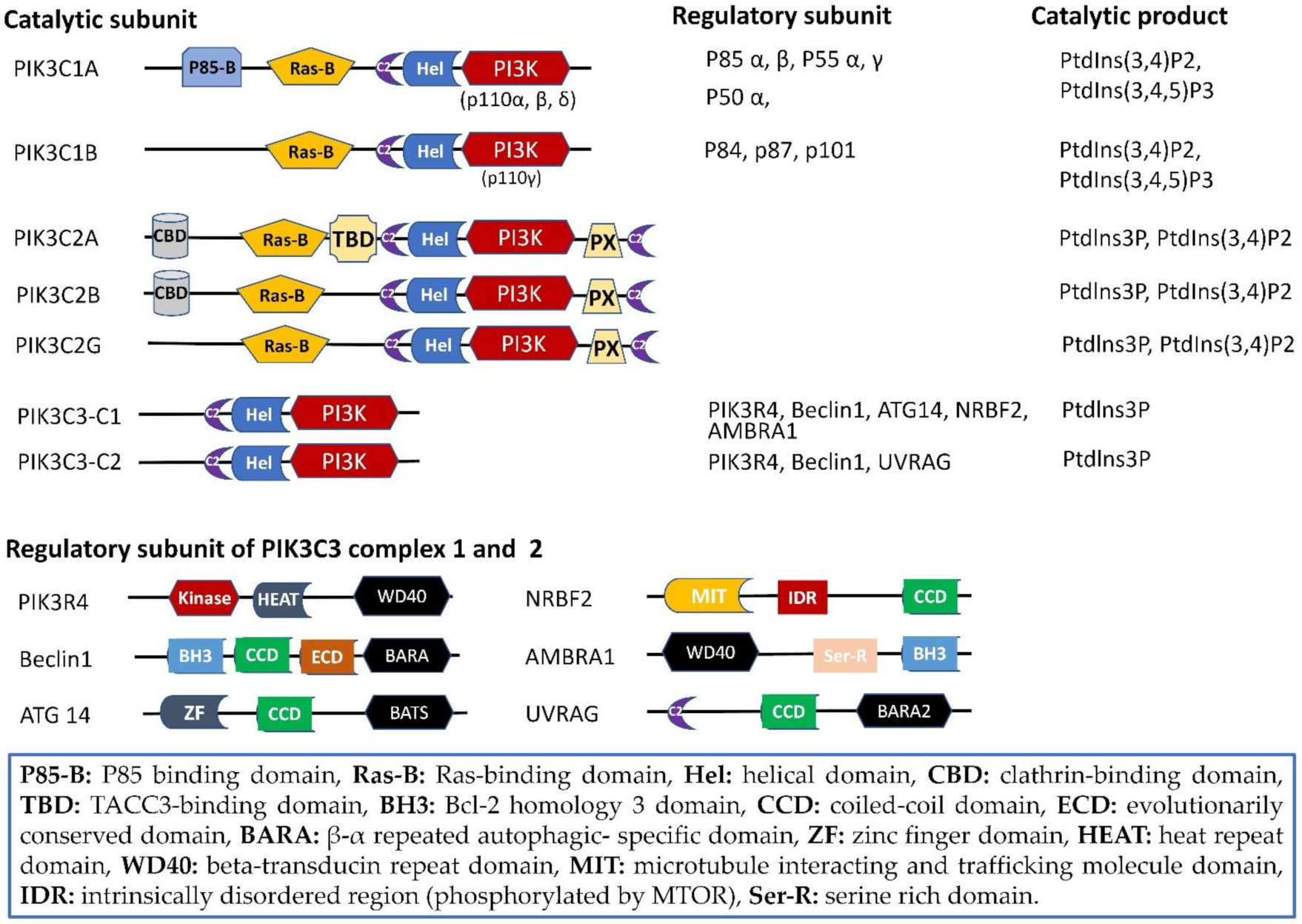
2. Phosphoinositide 3-Kinases (PI3Ks): Gene, Protein, and Structure
2.1. Structure Domain
2.2. PIK3C2
2.3. PIK3C3
2.3.1. PIK3C3-C1
2.3.2. PIK3C3-C2
3. Expression of PIK3C3 in Human Tissues
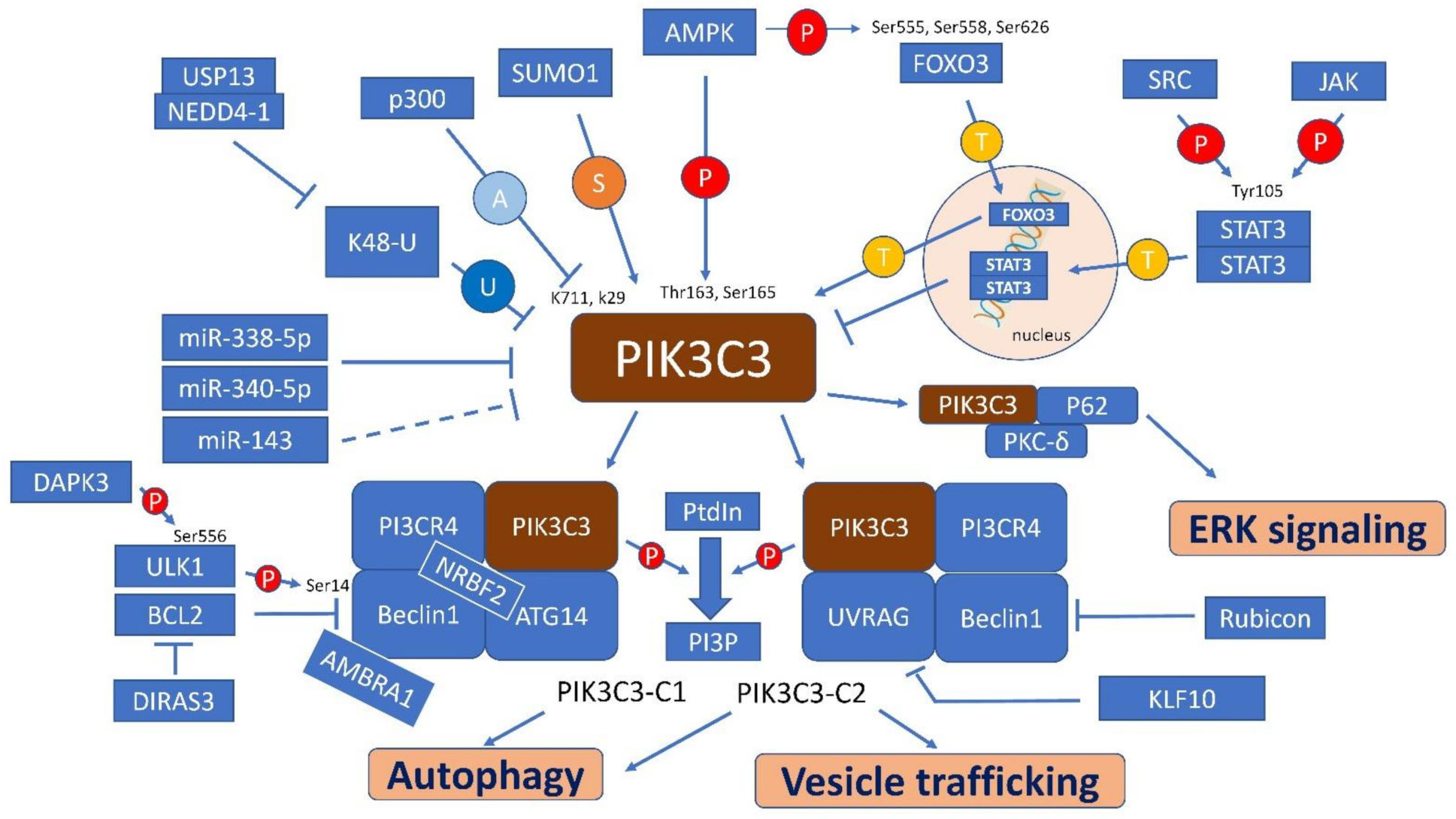
4. Regulatory Factors, Pathways and Interaction Molecules for PIK3C3
5. Biological Functions of PIK3C3
5.1. Proliferation, Apoptosis, and Tumor Growth
5.2. Migration, Invasion, and Metastasis
6. Significance of PIK3C3 in Human Cancer
6.1. Cancer Stem Cells (CSCs)
6.2. The Role of PIK3C3 in Cancer Immunity
6.3. Expression of PIK3C3 in Human Cancer
6.4. Prognostic Significance of PIK3C3 in Human Cancer
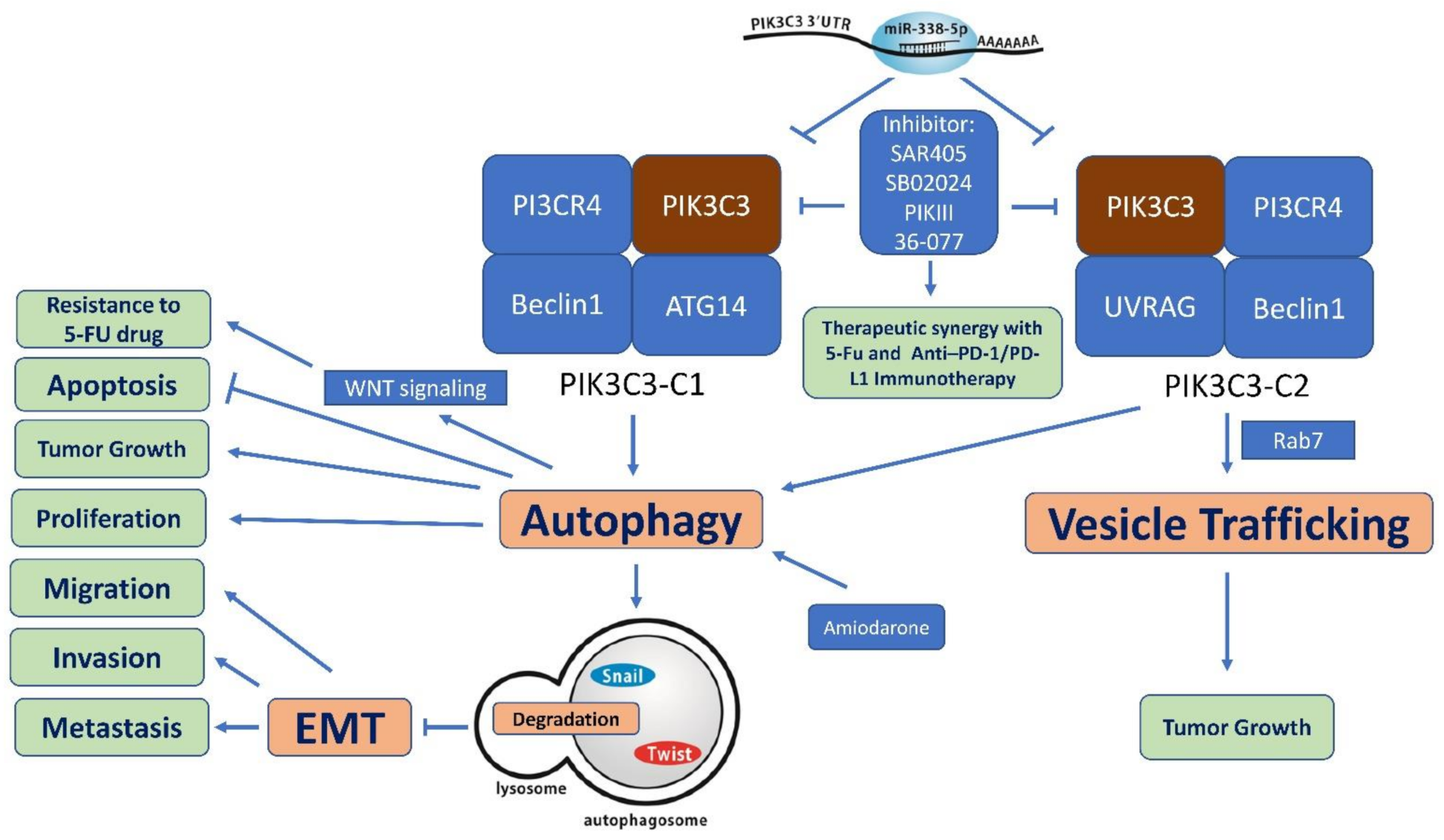
7. PIK3C3 as Therapeutic Target
7.1. SAR405
7.2. PIK-III
7.3. VPS34-IN1
7.4. Other Selective PIK3C3 Inhibitors
8. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wei, Y.; Liu, M.; Li, X.; Liu, J.; Li, H. Origin of the autophagosome membrane in mammals. Biomed. Res. Int. 2018, 2018, 1012789. [Google Scholar] [CrossRef] [PubMed]
- Yoshimori, T. Autophagy: A regulated bulk degradation process inside cells. Biochem. Biophys. Res. Commun. 2004, 313, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Ng, G.; Huang, J. The significance of autophagy in cancer. Mol. Carcinog. 2005, 43, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Hippert, M.M.; O’Toole, P.S.; Thorburn, A. Autophagy in cancer: Good, bad, or both? Cancer Res. 2006, 66, 9349–9351. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Kanzawa, T.; Sawaya, R.; Kondo, S. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer 2005, 5, 726–734. [Google Scholar] [CrossRef]
- Zhou, X.; Takatoh, J.; Wang, F. The mammalian class 3 PI3K (PIK3C3) is required for early embryogenesis and cell proliferation. PLoS ONE 2011, 6, e16358. [Google Scholar] [CrossRef]
- Itakura, E.; Kishi, C.; Inoue, K.; Mizushima, N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. J. Mol. Cell Biol. 2008, 19, 5360–5372. [Google Scholar] [CrossRef]
- Ohashi, Y. Class III phosphatidylinositol 3-kinase complex I subunit NRBF2/Atg38–from cell and structural biology to health and disease. Autophagy 2021, 17, 1–12. [Google Scholar] [CrossRef]
- Yu, X.; Long, Y.C.; Shen, H.M. Differential regulatory functions of three classes of phosphatidylinositol and phosphoinositide 3-kinases in autophagy. Autophagy 2015, 11, 1711–1728. [Google Scholar] [CrossRef]
- Qi, C.; Zou, L.; Wang, S.; Mao, X.; Hu, Y.; Shi, J.; Zhang, Z.; Wu, H. Vps34 inhibits hepatocellular carcinoma invasion by regulating endosome-lysosome trafficking via Rab7-RILP and Rab11. Cancer Res. Treat. 2021. Online ahead of print. [Google Scholar] [CrossRef]
- Schlafli, A.M.; Tokarchuk, I.; Parejo, S.; Jutzi, S.; Berezowska, S.; Engedal, N.; Tschan, M.P. ALK inhibition activates LC3B-independent, protective autophagy in EML4-ALK positive lung cancer cells. Sci. Rep. 2021, 11, 9011. [Google Scholar] [CrossRef]
- Chu, C.A.; Lee, C.T.; Lee, J.C.; Wang, Y.W.; Huang, C.T.; Lan, S.H.; Lin, P.C.; Lin, B.W.; Tian, Y.F.; Liu, H.S.; et al. MiR-338-5p promotes metastasis of colorectal cancer by inhibition of phosphatidylinositol 3-kinase, catalytic subunit type 3-mediated autophagy pathway. EBioMedicine 2019, 43, 270–281. [Google Scholar] [CrossRef]
- Dyczynski, M.; Yu, Y.; Otrocka, M.; Parpal, S.; Braga, T.; Henley, A.B.; Zazzi, H.; Lerner, M.; Wennerberg, K.; Viklund, J.; et al. Targeting autophagy by small molecule inhibitors of vacuolar protein sorting 34 (Vps34) improves the sensitivity of breast cancer cells to Sunitinib. Cancer Lett. 2018, 435, 32–43. [Google Scholar] [CrossRef]
- Swaminathan, G.; Zhu, W.; Plowey, E.D. BECN1/Beclin 1 sorts cell-surface APP/amyloid beta precursor protein for lysosomal degradation. Autophagy 2016, 12, 2404–2419. [Google Scholar] [CrossRef]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A.; Ojala, J.; Haapasalo, A.; Soininen, H.; Hiltunen, M. Impaired autophagy and APP processing in Alzheimer’s disease: The potential role of Beclin 1 interactome. Prog. Neurobiol. 2013, 106–107, 33–54. [Google Scholar] [CrossRef]
- Yang, G.; Van Kaer, L. PIK3C3/VPS34 links T-cell autophagy to autoimmunity. Cell Death. Dis. 2020, 11, 334. [Google Scholar] [CrossRef]
- Yang, G.; Song, W.; Postoak, J.L.; Chen, J.; Martinez, J.; Zhang, J.; Wu, L.; Van Kaer, L. Autophagy-related protein PIK3C3/VPS34 controls T cell metabolism and function. Autophagy 2021, 17, 1193–1204. [Google Scholar] [CrossRef]
- Zhang, X.W.; Zhou, J.C.; Peng, D.; Hua, F.; Li, K.; Yu, J.J.; Lv, X.X.; Cui, B.; Liu, S.S.; Yu, J.M.; et al. Disrupting the TRIB3-SQSTM1 interaction reduces liver fibrosis by restoring autophagy and suppressing exosome-mediated HSC activation. Autophagy 2020, 16, 782–796. [Google Scholar] [CrossRef]
- Reifler, A.; Li, X.; Archambeau, A.J.; McDade, J.R.; Sabha, N.; Michele, D.E.; Dowling, J.J. Conditional knockout of pik3c3 causes a murine muscular dystrophy. Am. J. Pathol. 2014, 184, 1819–1830. [Google Scholar] [CrossRef]
- O’Farrell, F.; Rusten, T.E.; Stenmark, H. Phosphoinositide 3-kinases as accelerators and brakes of autophagy. FEBS J. 2013, 280, 6322–6337. [Google Scholar] [CrossRef]
- Margaria, J.P.; Campa, C.C.; De Santis, M.C.; Hirsch, E.; Franco, I. The PI3K/Akt/mTOR pathway in polycystic kidney disease: A complex interaction with polycystins and primary cilium. Cell Signal. 2020, 66, 109468. [Google Scholar] [CrossRef]
- Margaria, J.P.; Ratto, E.; Gozzelino, L.; Li, H.; Hirsch, E. Class II PI3Ks at the intersection between signal transduction and membrane trafficking. Biomolecules 2019, 9, 104. [Google Scholar] [CrossRef]
- Vanhaesebroeck, B.; Guillermet-Guibert, J.; Graupera, M.; Bilanges, B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 2010, 11, 329–341. [Google Scholar] [CrossRef]
- Falasca, M.; Maffucci, T. Regulation and cellular functions of class II phosphoinositide 3-kinases. Biochem. J. 2012, 443, 587–601. [Google Scholar] [CrossRef]
- Gulluni, F.; Martini, M.; De Santis, M.C.; Campa, C.C.; Ghigo, A.; Margaria, J.P.; Ciraolo, E.; Franco, I.; Ala, U.; Annaratone, L.; et al. Mitotic spindle assembly and genomic stability in breast cancer require PI3K-C2alpha scaffolding function. Cancer Cell 2017, 32, 444–459. [Google Scholar] [CrossRef]
- Braccini, L.; Ciraolo, E.; Campa, C.C.; Perino, A.; Longo, D.L.; Tibolla, G.; Pregnolato, M.; Cao, Y.; Tassone, B.; Damilano, F.; et al. PI3K-C2gamma is a Rab5 effector selectively controlling endosomal Akt2 activation downstream of insulin signalling. Nat. Commun. 2015, 6, 7400. [Google Scholar] [CrossRef]
- Biswas, K.; Yoshioka, K.; Asanuma, K.; Okamoto, Y.; Takuwa, N.; Sasaki, T.; Takuwa, Y. Essential role of class II phosphatidylinositol-3-kinase-C2alpha in sphingosine 1-phosphate receptor-1-mediated signaling and migration in endothelial cells. J. Biol. Chem. 2013, 288, 2325–2339. [Google Scholar] [CrossRef]
- Franco, I.; Margaria, J.P.; De Santis, M.C.; Ranghino, A.; Monteyne, D.; Chiaravalli, M.; Pema, M.; Campa, C.C.; Ratto, E.; Gulluni, F.; et al. Phosphoinositide 3-kinase-C2alpha regulates polycystin-2 ciliary entry and protects against kidney cyst formation. J. Am. Soc. Nephrol. 2016, 27, 1135–1144. [Google Scholar] [CrossRef]
- Campa, C.C.; Margaria, J.P.; Derle, A.; Del Giudice, M.; De Santis, M.C.; Gozzelino, L.; Copperi, F.; Bosia, C.; Hirsch, E. Rab11 activity and PtdIns(3)P turnover removes recycling cargo from endosomes. Nat. Chem. Biol. 2018, 14, 801–810. [Google Scholar] [CrossRef]
- Tiosano, D.; Baris, H.N.; Chen, A.; Hitzert, M.M.; Schueler, M.; Gulluni, F.; Wiesener, A.; Bergua, A.; Mory, A.; Copeland, B.; et al. Mutations in PIK3C2A cause syndromic short stature, skeletal abnormalities, and cataracts associated with ciliary dysfunction. PLoS Genet. 2019, 15, e1008088. [Google Scholar] [CrossRef]
- Gulluni, F.; De Santis, M.C.; Margaria, J.P.; Martini, M.; Hirsch, E. Class II PI3K functions in cell biology and disease. Trends. Cell Biol. 2019, 29, 339–359. [Google Scholar] [CrossRef] [PubMed]
- Bougen-Zhukov, N.M.; Lee, Y.Y.; Lee, J.J.; Lee, P.; Loo, L.H. PI3K catalytic subunits alpha and beta modulate cell death and IL-6 secretion induced by talc particles in human lung carcinoma cells. Am. J. Respir. Cell Mol. Biol. 2020, 62, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Cisse, O.; Quraishi, M.; Gulluni, F.; Guffanti, F.; Mavrommati, I.; Suthanthirakumaran, M.; Oh, L.C.R.; Schlatter, J.N.; Sarvananthan, A.; Broggini, M.; et al. Downregulation of class II phosphoinositide 3-kinase PI3K-C2beta delays cell division and potentiates the effect of docetaxel on cancer cell growth. J. Exp. Clin. Cancer Res. 2019, 38, 472. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, X.; Ma, J.; Li, D.; Ju, H.; Liu, Y.; Chen, Y.; He, X.; Zhu, Y. Integrative analysis of the IQ motif-containing GTPase-activating protein family indicates that the IQGAP3-PIK3C2B axis promotes invasion in colon cancer. Onco. Targets Ther. 2020, 13, 8299–8311. [Google Scholar] [CrossRef]
- Li, A.; Chen, H.; Lin, M.; Zhang, C.; Tang, E.; Peng, J.; Wei, Q.; Li, H.; Yin, L. PIK3C2G copy number is associated with clinical outcomes of colorectal cancer patients treated with oxaliplatin. Int. J. Clin. Exp. Med. 2015, 8, 1137–1143. [Google Scholar]
- Pavlinov, I.; Salkovski, M.; Aldrich, L.N. Selective autophagy inhibition through disruption of the PIK3C3-containing complex I. Autophagy 2020, 16, 1547–1549. [Google Scholar] [CrossRef]
- Rostislavleva, K.; Soler, N.; Ohashi, Y.; Zhang, L.; Pardon, E.; Burke, J.E.; Masson, G.R.; Johnson, C.; Steyaert, J.; Ktistakis, N.T.; et al. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 2015, 350, aac7365. [Google Scholar] [CrossRef]
- Ma, M.; Liu, J.J.; Li, Y.; Huang, Y.; Ta, N.; Chen, Y.; Fu, H.; Ye, M.D.; Ding, Y.; Huang, W.; et al. Cryo-EM structure and biochemical analysis reveal the basis of the functional difference between human PI3KC3-C1 and -C2. Cell Res. 2017, 27, 989–1001. [Google Scholar] [CrossRef]
- Baskaran, S.; Carlson, L.A.; Stjepanovic, G.; Young, L.N.; Kim, D.J.; Grob, P.; Stanley, R.E.; Nogales, E.; Hurley, J.H. Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. Elife 2014, 3, e05115. [Google Scholar] [CrossRef]
- Ohashi, Y.; Tremel, S.; Masson, G.R.; McGinney, L.; Boulanger, J.; Rostislavleva, K.; Johnson, C.M.; Niewczas, I.; Clark, J.; Williams, R.L. Membrane characteristics tune activities of endosomal and autophagic human VPS34 complexes. Elife 2020, 9, e58281. [Google Scholar] [CrossRef]
- Chang, C.; Young, L.N.; Morris, K.L.; von Bulow, S.; Schoneberg, J.; Yamamoto-Imoto, H.; Oe, Y.; Yamamoto, K.; Nakamura, S.; Stjepanovic, G.; et al. Bidirectional Control of Autophagy by BECN1 BARA Domain Dynamics. Mol. Cell 2019, 73, 339–353. [Google Scholar] [CrossRef]
- Hu, Y.J.; Zhong, J.T.; Gong, L.; Zhang, S.C.; Zhou, S.H. Autophagy-related beclin 1 and head and neck cancers. Onco. Targets Ther. 2020, 13, 6213–6227. [Google Scholar] [CrossRef]
- Fan, W.; Nassiri, A.; Zhong, Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc. Natl. Acad. Sci. USA 2011, 108, 7769–7774. [Google Scholar] [CrossRef]
- Shan, H.; He, Y.; Hao, S.; Wang, B.; Xu, M.; Qi, H.; Liu, S.; Du, Y.; Yu, X. Vps15 is critical to mediate autophagy in AngII treated HUVECs probably by PDK1/PKC signaling pathway. Life Sci. 2019, 233, 116701. [Google Scholar] [CrossRef]
- Cheung, Y.W.S.; Nam, S.E.; Yip, C.K. Recent advances in single-particle electron microscopic analysis of autophagy degradation machinery. Int. J. Mol. Sci. 2020, 21, 8051. [Google Scholar] [CrossRef]
- Antonioli, M.; Di Rienzo, M.; Piacentini, M.; Fimia, G.M. Emerging mechanisms in initiating and terminating autophagy. Trends Biochem. Sci. 2017, 42, 28–41. [Google Scholar] [CrossRef]
- Xia, P.; Wang, S.; Huang, G.; Du, Y.; Zhu, P.; Li, M.; Fan, Z. RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy. Cell Res. 2014, 24, 943–958. [Google Scholar] [CrossRef]
- Antonioli, M.; Albiero, F.; Nazio, F.; Vescovo, T.; Perdomo, A.B.; Corazzari, M.; Marsella, C.; Piselli, P.; Gretzmeier, C.; Dengjel, J.; et al. AMBRA1 interplay with cullin E3 ubiquitin ligases regulates autophagy dynamics. Dev. Cell 2014, 31, 734–746. [Google Scholar] [CrossRef]
- Lu, Z.; Baquero, M.T.; Yang, H.; Yang, M.; Reger, A.S.; Kim, C.; Levine, D.A.; Clarke, C.H.; Liao, W.S.; Bast, R.C., Jr. DIRAS3 regulates the autophagosome initiation complex in dormant ovarian cancer cells. Autophagy 2014, 10, 1071–1092. [Google Scholar] [CrossRef]
- Vega-Rubin-de-Celis, S.; Kinch, L.; Pena-Llopis, S. Regulation of beclin 1-mediated autophagy by oncogenic tyrosine kinases. Int. J. Mol. Sci. 2020, 21, 9210. [Google Scholar] [CrossRef]
- Pattingre, S.; Tassa, A.; Qu, X.; Garuti, R.; Liang, X.H.; Mizushima, N.; Packer, M.; Schneider, M.D.; Levine, B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005, 122, 927–939. [Google Scholar] [CrossRef]
- Valente, G.; Morani, F.; Nicotra, G.; Fusco, N.; Peracchio, C.; Titone, R.; Alabiso, O.; Arisio, R.; Katsaros, D.; Benedetto, C.; et al. Expression and clinical significance of the autophagy proteins BECLIN 1 and LC3 in ovarian cancer. Biomed. Res. Int. 2014, 2014, 462658. [Google Scholar] [CrossRef]
- Liang, X.H.; Jackson, S.; Seaman, M.; Brown, K.; Kempkes, B.; Hibshoosh, H.; Levine, B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999, 402, 672–676. [Google Scholar] [CrossRef]
- Li, Z.; Chen, B.; Wu, Y.; Jin, F.; Xia, Y.; Liu, X. Genetic and epigenetic silencing of the beclin 1 gene in sporadic breast tumors. BMC Cancer 2010, 10, 98. [Google Scholar] [CrossRef]
- Aita, V.M.; Liang, X.H.; Murty, V.V.; Pincus, D.L.; Yu, W.; Cayanis, E.; Kalachikov, S.; Gilliam, T.C.; Levine, B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 1999, 59, 59–65. [Google Scholar] [CrossRef]
- Tang, H.; Sebti, S.; Titone, R.; Zhou, Y.; Isidoro, C.; Ross, T.S.; Hibshoosh, H.; Xiao, G.; Packer, M.; Xie, Y.; et al. Decreased BECN1 mRNA expression in human breast cancer is associated with estrogen receptor-negative subtypes and poor prognosis. EBioMedicine 2015, 2, 255–263. [Google Scholar] [CrossRef]
- Wijshake, T.; Zou, Z.; Chen, B.; Zhong, L.; Xiao, G.; Xie, Y.; Doench, J.G.; Bennett, L.; Levine, B. Tumor-suppressor function of Beclin 1 in breast cancer cells requires E-cadherin. Proc. Natl. Acad. Sci. USA 2021, 118, e2020478118. [Google Scholar] [CrossRef]
- Zhang, M.; Sun, Y.; Meng, J.; Zhang, L.; Liang, C.; Chang, C. Targeting AR-Beclin 1 complex-modulated growth factor signaling increases the antiandrogen enzalutamide sensitivity to better suppress the castration-resistant prostate cancer growth. Cancer Lett. 2019, 442, 483–490. [Google Scholar] [CrossRef]
- Zhou, W.H.; Tang, F.; Xu, J.; Wu, X.; Yang, S.B.; Feng, Z.Y.; Ding, Y.G.; Wan, X.B.; Guan, Z.; Li, H.G.; et al. Low expression of Beclin 1, associated with high Bcl-xL, predicts a malignant phenotype and poor prognosis of gastric cancer. Autophagy 2012, 8, 389–400. [Google Scholar] [CrossRef]
- Yu, W.X.; Lu, C.; Wang, B.; Ren, X.Y.; Xu, K. Effects of rapamycin on osteosarcoma cell proliferation and apoptosis by inducing autophagy. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 915–921. [Google Scholar]
- Wang, H.; Wang, Y.; Qian, L.; Wang, X.; Gu, H.; Dong, X.; Huang, S.; Jin, M.; Ge, H.; Xu, C.; et al. RNF216 contributes to proliferation and migration of colorectal cancer via suppressing BECN1-dependent autophagy. Oncotarget 2016, 7, 51174–51183. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Li, G.M.; Li, L.; Li, M.Q.; Chen, X.; Su, Q.; Deng, Z.J.; Liu, H.B.; Li, B.; Zhang, W.H.; Jia, Y.X.; et al. DAPK3 inhibits gastric cancer progression via activation of ULK1-dependent autophagy. Cell Death. Differ. 2021, 28, 952–967. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Al-Aidaroos, A.Q.; Yuen, H.F.; Zhang, S.D.; Shen, H.M.; Rozycka, E.; McCrudden, C.M.; Tergaonkar, V.; Gupta, A.; Lin, Y.B.; et al. A role of autophagy in PTP4A3-driven cancer progression. Autophagy 2014, 10, 1787–1800. [Google Scholar] [CrossRef]
- Yang, S.; Wang, M.; Yang, L.; Li, Y.; Ma, Y.; Peng, X.; Li, X.; Li, B.; Jin, H.; Li, H. MicroRNA-375 targets ATG14 to inhibit autophagy and sensitize hepatocellular carcinoma cells to sorafenib. Onco. Targets Ther. 2020, 13, 3557–3570. [Google Scholar] [CrossRef]
- Zhou, C.; Yi, C.; Yi, Y.; Qin, W.; Yan, Y.; Dong, X.; Zhang, X.; Huang, Y.; Zhang, R.; Wei, J.; et al. LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/beta-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol. Cancer 2020, 19, 118. [Google Scholar] [CrossRef]
- Hu, Z.; Cai, M.; Zhang, Y.; Tao, L.; Guo, R. miR-29c-3p inhibits autophagy and cisplatin resistance in ovarian cancer by regulating FOXP1/ATG14 pathway. Cell Cycle 2020, 19, 193–206. [Google Scholar] [CrossRef]
- Han, Y.; Zhou, S.; Wang, X.; Mao, E.; Huang, L. SNHG14 stimulates cell autophagy to facilitate cisplatin resistance of colorectal cancer by regulating miR-186/ATG14 axis. Biomed. Pharmacother. 2020, 121, 109580. [Google Scholar] [CrossRef]
- Chaikovsky, A.C.; Li, C.; Jeng, E.E.; Loebell, S.; Lee, M.C.; Murray, C.W.; Cheng, R.; Demeter, J.; Swaney, D.L.; Chen, S.H.; et al. The AMBRA1 E3 ligase adaptor regulates the stability of cyclin D. Nature 2021, 592, 794–798. [Google Scholar] [CrossRef]
- Maiani, E.; Milletti, G.; Nazio, F.; Holdgaard, S.G.; Bartkova, J.; Rizza, S.; Cianfanelli, V.; Lorente, M.; Simoneschi, D.; Di Marco, M.; et al. AMBRA1 regulates cyclin D to guard S-phase entry and genomic integrity. Nature 2021, 592, 799–803. [Google Scholar] [CrossRef]
- Di Leo, L.; Bodemeyer, V.; Bosisio, F.M.; Claps, G.; Carretta, M.; Rizza, S.; Faienza, F.; Frias, A.; Khan, S.; Bordi, M.; et al. Loss of Ambra1 promotes melanoma growth and invasion. Nat. Commun. 2021, 12, 2550. [Google Scholar] [CrossRef]
- Liang, C.; Lee, J.S.; Inn, K.S.; Gack, M.U.; Li, Q.; Roberts, E.A.; Vergne, I.; Deretic, V.; Feng, P.; Akazawa, C.; et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008, 10, 776–787. [Google Scholar] [CrossRef]
- Liang, C.; Feng, P.; Ku, B.; Dotan, I.; Canaani, D.; Oh, B.H.; Jung, J.U. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 2006, 8, 688–699. [Google Scholar] [CrossRef]
- Sencan, S.; Tanriover, M.; Ulasli, M.; Karakas, D.; Ozpolat, B. UV radiation resistance-associated gene (UVRAG) promotes cell proliferation, migration, invasion by regulating cyclin-dependent kinases (CDK) and integrin-beta/Src signaling in breast cancer cells. Mol. Cell Biochem. 2021, 476, 2075–2084. [Google Scholar] [CrossRef]
- Feng, X.; Jia, Y.; Zhang, Y.; Ma, F.; Zhu, Y.; Hong, X.; Zhou, Q.; He, R.; Zhang, H.; Jin, J.; et al. Ubiquitination of UVRAG by SMURF1 promotes autophagosome maturation and inhibits hepatocellular carcinoma growth. Autophagy 2019, 15, 1130–1149. [Google Scholar] [CrossRef]
- Quach, C.; Song, Y.; Guo, H.; Li, S.; Maazi, H.; Fung, M.; Sands, N.; O’Connell, D.; Restrepo-Vassalli, S.; Chai, B.; et al. A truncating mutation in the autophagy gene UVRAG drives inflammation and tumorigenesis in mice. Nat. Commun. 2019, 10, 5681. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Liu, K.; Gu, L.; Yu, L.; Han, L.; Meng, Z. Suppressing UVRAG induces radiosensitivity by triggering lysosomal membrane permeabilization in hypopharyngeal squamous cell carcinoma. Onco. Targets Ther. 2020, 13, 10275–10285. [Google Scholar] [CrossRef]
- Chang, V.H.; Tsai, Y.C.; Tsai, Y.L.; Peng, S.L.; Chen, S.L.; Chang, T.M.; Yu, W.C.; Ch’ang, H.J. Krupple-like factor 10 regulates radio-sensitivity of pancreatic cancer via UV radiation resistance-associated gene. Radiother. Oncol. 2017, 122, 476–484. [Google Scholar] [CrossRef]
- Jo, Y.K.; Park, N.Y.; Shin, J.H.; Jo, D.S.; Bae, J.E.; Choi, E.S.; Maeng, S.; Jeon, H.B.; Roh, S.A.; Chang, J.W.; et al. Up-regulation of UVRAG by HDAC1 inhibition attenuates 5FU-induced cell death in HCT116 colorectal cancer cells. Anticancer Res. 2018, 38, 271–277. [Google Scholar]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas--a tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Su, H.; Yang, F.; Wang, Q.; Shen, Q.; Huang, J.; Peng, C.; Zhang, Y.; Wan, W.; Wong, C.C.L.; Sun, Q.; et al. VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical Autophagy. Mol. Cell 2017, 67, 907–921. [Google Scholar] [CrossRef]
- Su, H.; Liu, W. PIK3C3/VPS34 control by acetylation. Autophagy 2018, 14, 1086–1087. [Google Scholar] [CrossRef]
- Tamargo-Gomez, I.; Marino, G. AMPK: Regulation of metabolic dynamics in the context of autophagy. Int. J. Mol. Sci. 2018, 19, 3812. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y. AMPK and autophagy. Adv. Exp. Med. Biol. 2019, 1206, 85–108. [Google Scholar]
- You, L.; Wang, Z.; Li, H.; Shou, J.; Jing, Z.; Xie, J.; Sui, X.; Pan, H.; Han, W. The role of STAT3 in autophagy. Autophagy 2015, 11, 729–739. [Google Scholar] [CrossRef]
- Yamada, E.; Bastie, C.C.; Koga, H.; Wang, Y.; Cuervo, A.M.; Pessin, J.E. Mouse skeletal muscle fiber-type-specific macroautophagy and muscle wasting are regulated by a Fyn/STAT3/Vps34 signaling pathway. Cell Rep. 2012, 1, 557–569. [Google Scholar] [CrossRef]
- Xie, W.; Jin, S.; Wu, Y.; Xian, H.; Tian, S.; Liu, D.A.; Guo, Z.; Cui, J. Auto-ubiquitination of NEDD4-1 recruits USP13 to facilitate autophagy through deubiquitinating VPS34. Cell Rep. 2020, 30, 2807–2819. [Google Scholar] [CrossRef]
- Yao, Y.; Li, H.; Da, X.; He, Z.; Tang, B.; Li, Y.; Hu, C.; Xu, C.; Chen, Q.; Wang, Q.K. SUMOylation of Vps34 by SUMO1 promotes phenotypic switching of vascular smooth muscle cells by activating autophagy in pulmonary arterial hypertension. Pulm. Pharmacol. Ther. 2019, 55, 38–49. [Google Scholar] [CrossRef]
- Yang, Y.; Fiskus, W.; Yong, B.; Atadja, P.; Takahashi, Y.; Pandita, T.K.; Wang, H.G.; Bhalla, K.N. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc. Natl. Acad. Sci. USA 2013, 110, 6841–6846. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gu, M.; Ju, Y.; Zhou, J. PIK3C3 acts as a tumor suppressor in esophageal squamous cell carcinoma and was regulated by miR-340-5p. Med. Sci. Monit. 2020, 26, e920642. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, H.; Li, Z.; Yang, P.; Li, Z. A positive feedback loop between GRP78 and VPS34 is critical for GRP78-mediated autophagy in cancer cells. Exp. Cell Res. 2017, 351, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Boutouja, F.; Brinkmeier, R.; Mastalski, T.; El Magraoui, F.; Platta, H.W. Regulation of the tumor-suppressor BECLIN 1 by distinct ubiquitination cascades. Int. J. Mol. Sci. 2017, 18, 2541. [Google Scholar] [CrossRef]
- Gillooly, D.J.; Morrow, I.C.; Lindsay, M.; Gould, R.; Bryant, N.J.; Gaullier, J.M.; Parton, R.G.; Stenmark, H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000, 19, 4577–4588. [Google Scholar] [CrossRef]
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef]
- Dall’Armi, C.; Devereaux, K.A.; Di Paolo, G. The role of lipids in the control of autophagy. Curr. Biol. 2013, 23, R33–R45. [Google Scholar] [CrossRef]
- Chen, D.; Fan, W.; Lu, Y.; Ding, X.; Chen, S.; Zhong, Q. A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol. Cell 2012, 45, 629–641. [Google Scholar] [CrossRef]
- Pankiv, S.; Alemu, E.A.; Brech, A.; Bruun, J.A.; Lamark, T.; Overvatn, A.; Bjorkoy, G.; Johansen, T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 2010, 188, 253–269. [Google Scholar] [CrossRef]
- Liu, C.C.; Lin, Y.C.; Chen, Y.H.; Chen, C.M.; Pang, L.Y.; Chen, H.A.; Wu, P.R.; Lin, M.Y.; Jiang, S.T.; Tsai, T.F.; et al. Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol. Cell 2016, 61, 84–97. [Google Scholar] [CrossRef]
- Jia, W.; He, M.X.; McLeod, I.X.; Guo, J.; Ji, D.; He, Y.W. Autophagy regulates T lymphocyte proliferation through selective degradation of the cell-cycle inhibitor CDKN1B/p27Kip1. Autophagy 2015, 11, 2335–2345. [Google Scholar] [CrossRef]
- Filippakis, H.; Belaid, A.; Siroky, B.; Wu, C.; Alesi, N.; Hougard, T.; Nijmeh, J.; Lam, H.C.; Henske, E.P. Vps34-mediated macropinocytosis in Tuberous Sclerosis Complex 2-deficient cells supports tumorigenesis. Sci. Rep. 2018, 8, 14161. [Google Scholar] [CrossRef]
- Kobylarz, M.J.; Goodwin, J.M.; Kang, Z.B.; Annand, J.W.; Hevi, S.; O’Mahony, E.; McAllister, G.; Reece-Hoyes, J.; Wang, Q.; Alford, J.; et al. An iron-dependent metabolic vulnerability underlies VPS34-dependence in RKO cancer cells. PLoS ONE 2020, 15, e0235551. [Google Scholar] [CrossRef]
- Huang, S.; Sinicrope, F.A. Celecoxib-induced apoptosis is enhanced by ABT-737 and by inhibition of autophagy in human colorectal cancer cells. Autophagy 2010, 6, 256–269. [Google Scholar] [CrossRef]
- Noman, M.Z.; Parpal, S.; Van Moer, K.; Xiao, M.; Yu, Y.; Viklund, J.; De Milito, A.; Hasmim, M.; Andersson, M.; Amaravadi, R.K.; et al. Inhibition of Vps34 reprograms cold into hot inflamed tumors and improves anti-PD-1/PD-L1 immunotherapy. Sci. Adv. 2020, 6, eaax7881. [Google Scholar] [CrossRef]
- Liu, F.; Wu, X.; Qian, Y.; Jiang, X.; Wang, Y.; Gao, J. PIK3C3 regulates the expansion of liver CSCs and PIK3C3 inhibition counteracts liver cancer stem cell activity induced by PI3K inhibitor. Cell Death Dis. 2020, 11, 427. [Google Scholar] [CrossRef]
- Young, C.D.; Arteaga, C.L.; Cook, R.S. Dual inhibition of Type I and Type III PI3 kinases increases tumor cell apoptosis in HER2+ breast cancers. Breast Cancer Res. 2015, 17, 148. [Google Scholar] [CrossRef]
- Jiang, X.; Bao, Y.; Liu, H.; Kou, X.; Zhang, Z.; Sun, F.; Qian, Z.; Lin, Z.; Li, X.; Liu, X.; et al. VPS34 stimulation of p62 phosphorylation for cancer progression. Oncogene 2017, 36, 6850–6862. [Google Scholar] [CrossRef]
- Liu, H.; Gong, X.; Yang, K. Overexpression of the clock gene Per2 suppresses oral squamous cell carcinoma progression by activating autophagy via the PI3K/AKT/mTOR pathway. J. Cancer 2020, 11, 3655–3666. [Google Scholar] [CrossRef]
- New, J.; Arnold, L.; Ananth, M.; Alvi, S.; Thornton, M.; Werner, L.; Tawfik, O.; Dai, H.; Shnayder, Y.; Kakarala, K.; et al. Secretory autophagy in cancer-associated fibroblasts promotes head and neck cancer progression and offers a novel therapeutic target. Cancer Res. 2017, 77, 6679–6691. [Google Scholar] [CrossRef]
- Dayde, D.; Guerard, M.; Perron, P.; Hatat, A.S.; Barrial, C.; Eymin, B.; Gazzeri, S. Nuclear trafficking of EGFR by Vps34 represses Arf expression to promote lung tumor cell survival. Oncogene 2016, 35, 3986–3994. [Google Scholar] [CrossRef]
- Verykiou, S.; Alexander, M.; Edwards, N.; Plummer, R.; Chaudhry, B.; Lovat, P.E.; Hill, D.S. Harnessing autophagy to overcome mitogen-activated protein kinase kinase inhibitor-induced resistance in metastatic melanoma. Br. J. Dermatol. 2019, 180, 346–356. [Google Scholar] [CrossRef]
- Zahedi, S.; Fitzwalter, B.E.; Morin, A.; Grob, S.; Desmarais, M.; Nellan, A.; Green, A.L.; Vibhakar, R.; Hankinson, T.C.; Foreman, N.K.; et al. Effect of early-stage autophagy inhibition in BRAF(V600E) autophagy-dependent brain tumor cells. Cell Death Dis. 2019, 10, 679. [Google Scholar] [CrossRef]
- Polak, R.; Bierings, M.B.; van der Leije, C.S.; Sanders, M.A.; Roovers, O.; Marchante, J.R.M.; Boer, J.M.; Cornelissen, J.J.; Pieters, R.; den Boer, M.L.; et al. Autophagy inhibition as a potential future targeted therapy for ETV6-RUNX1-driven B-cell precursor acute lymphoblastic leukemia. Haematologica 2019, 104, 738–748. [Google Scholar] [CrossRef]
- Chen, H.; Lu, C.; Lin, C.; Li, L.; Wang, Y.; Han, R.; Hu, C.; He, Y. VPS34 suppression reverses osimertinib resistance via simultaneously inhibiting glycolysis and autophagy. Carcinogenesis 2021, 42, 880–890. [Google Scholar] [CrossRef]
- Rong, X.; Liang, Y.; Han, Q.; Zhao, Y.; Jiang, G.; Zhang, X.; Lin, X.; Liu, Y.; Zhang, Y.; Han, X.; et al. Molecular mechanisms of tyrosine kinase inhibitor resistance induced by membranous/cytoplasmic/nuclear translocation of epidermal growth factor receptor. J. Thorac. Oncol. 2019, 14, 1766–1783. [Google Scholar] [CrossRef]
- Mohan, N.; Shen, Y.; Dokmanovic, M.; Endo, Y.; Hirsch, D.S.; Wu, W.J. VPS34 regulates TSC1/TSC2 heterodimer to mediate RheB and mTORC1/S6K1 activation and cellular transformation. Oncotarget 2016, 7, 52239–52254. [Google Scholar] [CrossRef]
- Kumar, B.; Ahmad, R.; Sharma, S.; Gowrikumar, S.; Primeaux, M.; Rana, S.; Natarajan, A.; Oupicky, D.; Hopkins, C.R.; Dhawan, P.; et al. PIK3C3 inhibition promotes sensitivity to colon cancer therapy by inhibiting cancer stem cells. Cancers 2021, 13, 2168. [Google Scholar] [CrossRef]
- Baquero, P.; Dawson, A.; Mukhopadhyay, A.; Kuntz, E.M.; Mitchell, R.; Olivares, O.; Ianniciello, A.; Scott, M.T.; Dunn, K.; Nicastri, M.C.; et al. Targeting quiescent leukemic stem cells using second generation autophagy inhibitors. Leukemia 2019, 33, 981–994. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef]
- Noman, M.Z.; Janji, B.; Kaminska, B.; Van Moer, K.; Pierson, S.; Przanowski, P.; Buart, S.; Berchem, G.; Romero, P.; Mami-Chouaib, F.; et al. Blocking hypoxia-induced autophagy in tumors restores cytotoxic T-cell activity and promotes regression. Cancer Res. 2011, 71, 5976–5986. [Google Scholar] [CrossRef] [PubMed]
- Mgrditchian, T.; Arakelian, T.; Paggetti, J.; Noman, M.Z.; Viry, E.; Moussay, E.; Van Moer, K.; Kreis, S.; Guerin, C.; Buart, S.; et al. Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner. Proc. Natl. Acad. Sci. USA 2017, 114, E9271–E9279. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Anaya, J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef]
- Hirsch, D.S.; Shen, Y.; Dokmanovic, M.; Wu, W.J. pp60c-Src phosphorylates and activates vacuolar protein sorting 34 to mediate cellular transformation. Cancer Res. 2010, 70, 5974–5983. [Google Scholar] [CrossRef][Green Version]
- Kristensen, L.; Kristensen, T.; Abildgaard, N.; Thomassen, M.; Frederiksen, M.; Mourits-Andersen, T.; Moller, M.B. High expression of PI3K core complex genes is associated with poor prognosis in chronic lymphocytic leukemia. Leuk. Res. 2015, 39, 555–560. [Google Scholar] [CrossRef]
- Pasquier, B. Autophagy inhibitors. Cell Mol. Life Sci. 2016, 73, 985–1001. [Google Scholar] [CrossRef]
- Schlutermann, D.; Skowron, M.A.; Berleth, N.; Bohler, P.; Deitersen, J.; Stuhldreier, F.; Wallot-Hieke, N.; Wu, W.; Peter, C.; Hoffmann, M.J.; et al. Targeting urothelial carcinoma cells by combining cisplatin with a specific inhibitor of the autophagy-inducing class III PtdIns3K complex. Urol. Oncol. 2018, 36, e1–e160. [Google Scholar] [CrossRef]
- Ronan, B.; Flamand, O.; Vescovi, L.; Dureuil, C.; Durand, L.; Fassy, F.; Bachelot, M.F.; Lamberton, A.; Mathieu, M.; Bertrand, T. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 2014, 10, 1013–1019. [Google Scholar] [CrossRef]
- Meunier, G.; Birsen, R.; Cazelles, C.; Belhadj, M.; Cantero-Aguilar, L.; Kosmider, O.; Fontenay, M.; Azar, N.; Mayeux, P.; Chapuis, N.; et al. Antileukemic activity of the VPS34-IN1 inhibitor in acute myeloid leukemia. Oncogenesis 2020, 9, 94. [Google Scholar] [CrossRef]
- Dowdle, W.E.; Nyfeler, B.; Nagel, J.; Elling, R.A.; Liu, S.; Triantafellow, E.; Menon, S.; Wang, Z.; Honda, A.; Pardee, G.; et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell. Biol. 2014, 16, 1069–1079. [Google Scholar] [CrossRef]
- Bago, R.; Malik, N.; Munson, M.J.; Prescott, A.R.; Davies, P.; Sommer, E.; Shpiro, N.; Ward, R.; Cross, D.; Ganley, I.G.; et al. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem. J. 2014, 463, 413–427. [Google Scholar] [CrossRef]
| Cancer Type | Cell Line | Role of PIK3C3 | Proliferation | Growth | Migration | Invasion | Metastasis | Apoptosis | Genetic Manipulation | PIK3C3 Inhibitor | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRC | RKO | Onco | Indu | Indu | - | - | - | - | KD | PIK-III | [103] |
| CRC | HCT116 | Onco | - | - | - | - | - | Inhi | KD | - | [104] |
| CRC | CT26 | Onco | - | Indu | - | - | - | - | KD | SB02024 SAR405 | [105] |
| CRC | SW480 | Sup | - | - | Inhi | Inhi | Inhi | - | Expression | - | [12] |
| HCC | Huh7, MHCC97H | Onco | - | Indu | - | - | - | - | KD | Vps34-IN1 | [106] |
| HCC | HepG2, SMMC-7721 | Sup | - | - | - | Inhi | - | - | Expression, KD | - | [10] |
| Breast carcinoma | BT474, SKBR3 | Onco | Indu | - | - | - | - | - | KD | SAR405 | [107] |
| Breast carcinoma | MCF-7, MDA-MB-231 | Onco | - | Indu | - | - | - | - | - | SB02024 | [13] |
| Breast carcinoma | MCF-7 | Onco | - | Indu | - | Indu | - | - | Expression, KD | - | [108] |
| Esophageal SCC | KYSE-150, TE-12 | Sup | Inhi | - | - | - | - | - | Expression | - | [92] |
| Oral SCC | TSCCA | Sup | Inhi | - | - | - | - | Indu | - | Autophinib | [109] |
| Head and neck SCC | UM-SCC-1 | Onco | - | Indu | - | - | - | - | - | SAR405 | [110] |
| Lung adenocarcinoma | H1975 | Onco | - | - | - | - | - | Inhi | KD | - | [111] |
| Melanoma | B16-F10 | Onco | - | Indu | - | - | - | - | KD | SB02024 SAR405 | [105] |
| Melanoma | A375 | Onco | - | - | - | Indu | - | - | - | PIK-III | [112] |
| Glioblastoma | AM38 | Onco | - | Indu | - | - | - | - | - | VPS34-IN1 | [113] |
| Acute lymphocytic leukemia | REH, REHS1 | Onco | Indu | - | - | - | - | - | KD | - | [114] |
| UALCAN | OncoLnc # | ||||||
|---|---|---|---|---|---|---|---|
| Cancer Type | Expression (vs. Normal) | Tumor Stage (vs. Stage 1) | N Stage (vs. N0) | Tumor Grade (vs. Grade 1) | Mucinous Type (vs. Conventional Type) | Poor Overall Survival | Poor Overall Survival |
| Colon adenocarcinoma | Down *** | ND | ND | - | Up * | ND | ND |
| Rectal adenocarcinoma | Down *** | ND | ND | - | ND | ND | ND |
| Adrenocortical carcinoma | - | ND | ND | - | - | ND | - |
| Acute myeloid leukemia | - | -- | - | - | - | ND | ND |
| Bladder urothelial carcinoma | ND | ND | ND | - | - | ND | Up * |
| Low-grade glioma | - | - | - | ND | - | Up ** | Up * |
| Breast invasive carcinoma | Down *** | ND | ND | - | ND | Up * | ND |
| Cervical carcinoma | ND | ND | ND | ND | Down * (mucinous vs. SCC) | ND | Up * |
| Cholangiocarcinoma | Up *** | ND | ND | ND | - | ND | - |
| Esophageal carcinoma | ND | ND | ND | ND | - | ND | ND |
| Glioblastoma | ND | - | - | -- | - | ND | ND |
| Head and neck SCC | Up *** | ND | ND | Up (grade 2 *, 3 * & 4 ***) | - | ND | ND |
| Renal clear cell carcinoma | Down *** | Down *** (stage 3, 4) | ND | Down (grade 3 * & 4 **) | - | Down *** | Down ** |
| Renal papillary cell carcinoma | Down *** | ND | ND | - | - | ND | ND |
| HCC | Up *** | Up ** (stage 3) | Up * | ND | - | Up ** | ND |
| Lung adenocarcinoma | ND | ND | ND | - | ND | ND | ND |
| Lung SCC | Up *** | ND | ND | - | - | ND | ND |
| Diffuse large B cell lymphoma | - | ND | - | - | - | ND | - |
| Mesothelioma | - | Down * (stage 2) | ND | - | - | Up * | - |
| Ovarian serous carcinoma | - | -- | - | ND | - | ND | ND |
| Pancreatic adenocarcinoma | ND | ND | ND | ND | - | ND | ND |
| Prostate adenocarcinoma | Down *** | - | ND | ND | - | ND | - |
| Skin melanoma | ND | ND | ND | - | - | ND | Down * |
| Stomach adenocarcinoma | Up *** | Up * (stage 4) | ND | ND | ND | Up * | ND |
| Thyroid carcinoma | Down *** | Down ** (stage 4) | ND | - | - | ND | - |
| Endometrial carcinoma | Down ** | Down * (stage 3) | - | - | - | ND | ND |
| Uterine carcinosarcoma | - | Up * (stage 3) | - | - | - | ND | |
| PIK3C3 Inhibitor | Cancer | Cell Line | Effect (When Used Alone) | Synergistic Drug | Reference |
|---|---|---|---|---|---|
| SAR405 | CRC | CT-26 | Inhibits tumor growth | Anti-PD-1/PD-L1 immunotherapy | [105] |
| Melanoma | B16-F10 | Inhibits tumor growth | Anti-PD-1/PD-L1 immunotherapy | [105] | |
| Breast carcinoma | BT474, SKBR3 | Inhibit proliferation | Lapatinib, BYL719 | [107] | |
| Head & neck SCC | UM-SCC-1 | Inhibits tumor growth | Cisplatin | [110] | |
| Bladder carcinoma | RT-112 | - | Cisplatin | [128] | |
| Lung large cell carcinoma | H1299 | - | Everolimus | [129] | |
| Renal cell carcinoma | ACHN, 860-O | - | Everolimus | [129] | |
| PIK-III | CRC | RKO | Inhibits proliferation and tumor growth | - | [103] |
| Acute myeloid leukemia | MOLM14, MV411, OCI-AML2, HL60, THP1, K562 | Induces cell death | - | [130] | |
| Melanoma | A375 | Inhibits invasion | Trametinib | [112] | |
| Breast carcinoma | MCF-7, MDA-MB-231 | - | Sunitinib, erlotinib | [13] | |
| Chronic myeloid leukemia | Leukemic cells from mice | - | Nilotinib | [119] | |
| VPS34-IN1 | HCC | Huh7, MHCC97H | Inhibits tumor growth | ZSTK474 | [106] |
| Glioblastoma | AM38 | Inhibits tumor growth | Vemurafenib | [113] | |
| Acute myeloid leukemia | MOLM14 | Induces cell death | L-asparaginase | [130] | |
| Lung adenocarcinoma | H3122 | - | Ceritinib | [11] | |
| SB02024 | CRC | CT-26 | Inhibits tumor growth | Anti-PD-1/PD-L1 immunotherapy | [105] |
| Melanoma | B16-F10 | Inhibits tumor growth | Anti-PD-1/PD-L1 immunotherapy | [105] | |
| Breast carcinoma | MCF-7, MDA-MB-231 | Inhibits tumor growth | Sunitinib, erlotinib | [13] | |
| Autophinib | Acute myeloid leukemia | MOLM14, MV411, OCI-AML2, HL60, THP1, K562 | Induces cell death | - | [130] |
| Oral SCC | TSCCA | Increases proliferation and decreases apoptosis | - | [109] | |
| 36-077 | CRC | HCT116 | - | 5-FU | [118] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, C.-A.; Wang, Y.-W.; Chen, Y.-L.; Chen, H.-W.; Chuang, J.-J.; Chang, H.-Y.; Ho, C.-L.; Chang, C.; Chow, N.-H.; Lee, C.-T. The Role of Phosphatidylinositol 3-Kinase Catalytic Subunit Type 3 in the Pathogenesis of Human Cancer. Int. J. Mol. Sci. 2021, 22, 10964. https://doi.org/10.3390/ijms222010964
Chu C-A, Wang Y-W, Chen Y-L, Chen H-W, Chuang J-J, Chang H-Y, Ho C-L, Chang C, Chow N-H, Lee C-T. The Role of Phosphatidylinositol 3-Kinase Catalytic Subunit Type 3 in the Pathogenesis of Human Cancer. International Journal of Molecular Sciences. 2021; 22(20):10964. https://doi.org/10.3390/ijms222010964
Chicago/Turabian StyleChu, Chien-An, Yi-Wen Wang, Yi-Lin Chen, Hui-Wen Chen, Jing-Jing Chuang, Hong-Yi Chang, Chung-Liang Ho, Chen Chang, Nan-Haw Chow, and Chung-Ta Lee. 2021. "The Role of Phosphatidylinositol 3-Kinase Catalytic Subunit Type 3 in the Pathogenesis of Human Cancer" International Journal of Molecular Sciences 22, no. 20: 10964. https://doi.org/10.3390/ijms222010964
APA StyleChu, C.-A., Wang, Y.-W., Chen, Y.-L., Chen, H.-W., Chuang, J.-J., Chang, H.-Y., Ho, C.-L., Chang, C., Chow, N.-H., & Lee, C.-T. (2021). The Role of Phosphatidylinositol 3-Kinase Catalytic Subunit Type 3 in the Pathogenesis of Human Cancer. International Journal of Molecular Sciences, 22(20), 10964. https://doi.org/10.3390/ijms222010964





