Investigation of NPB Analogs That Target Phosphorylation of BAD-Ser99 in Human Mammary Carcinoma Cells
Abstract
:1. Introduction
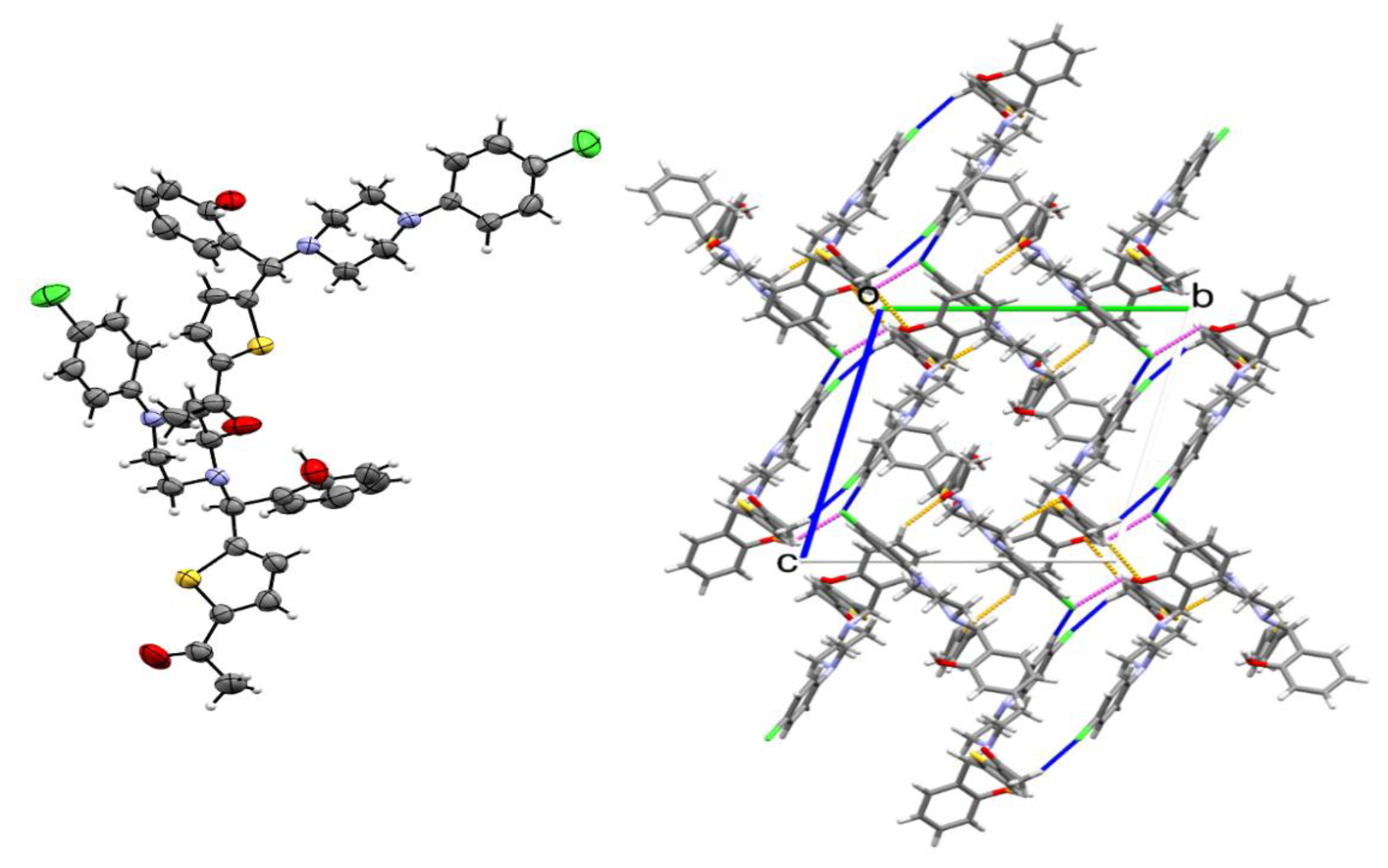
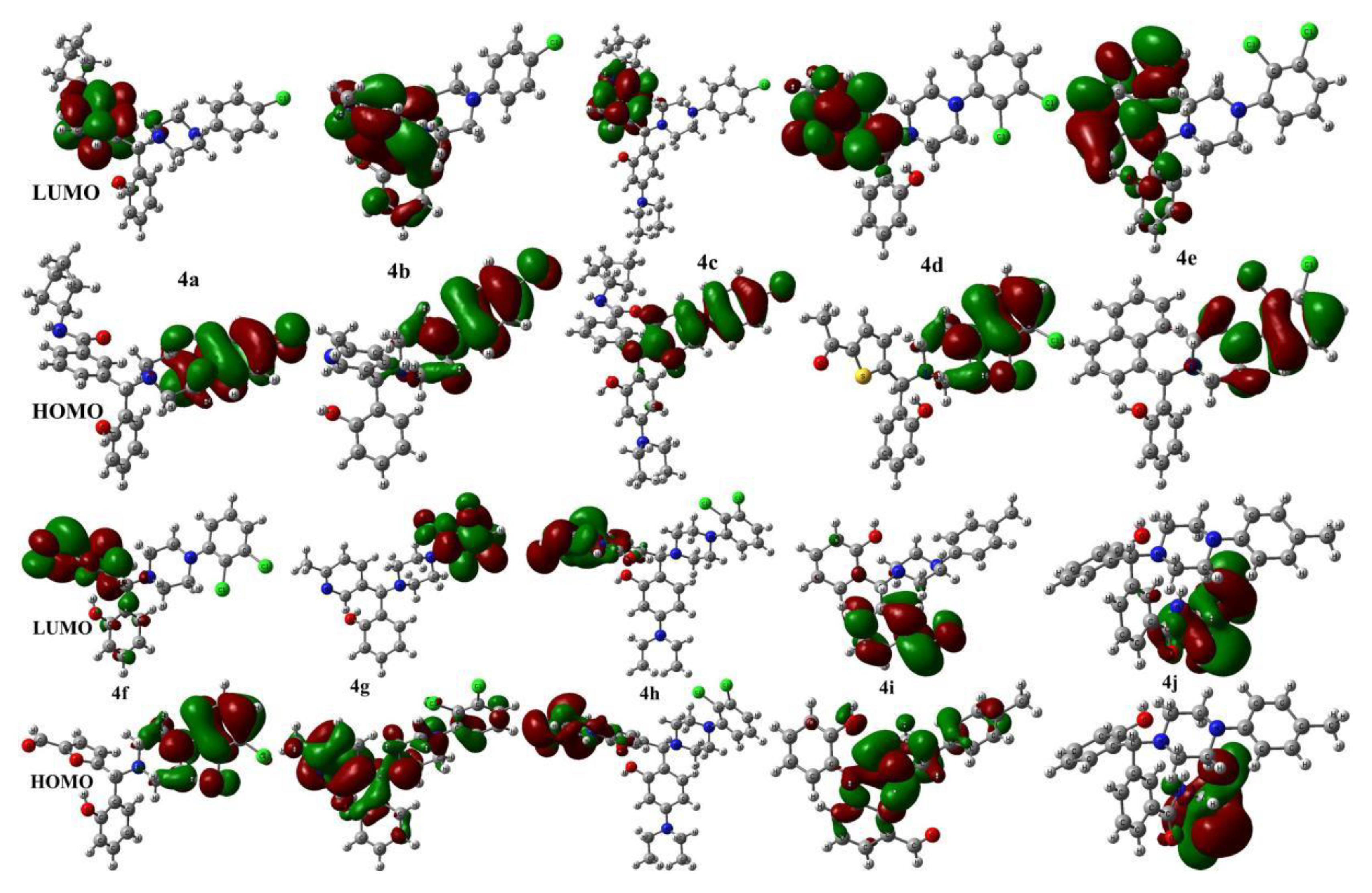
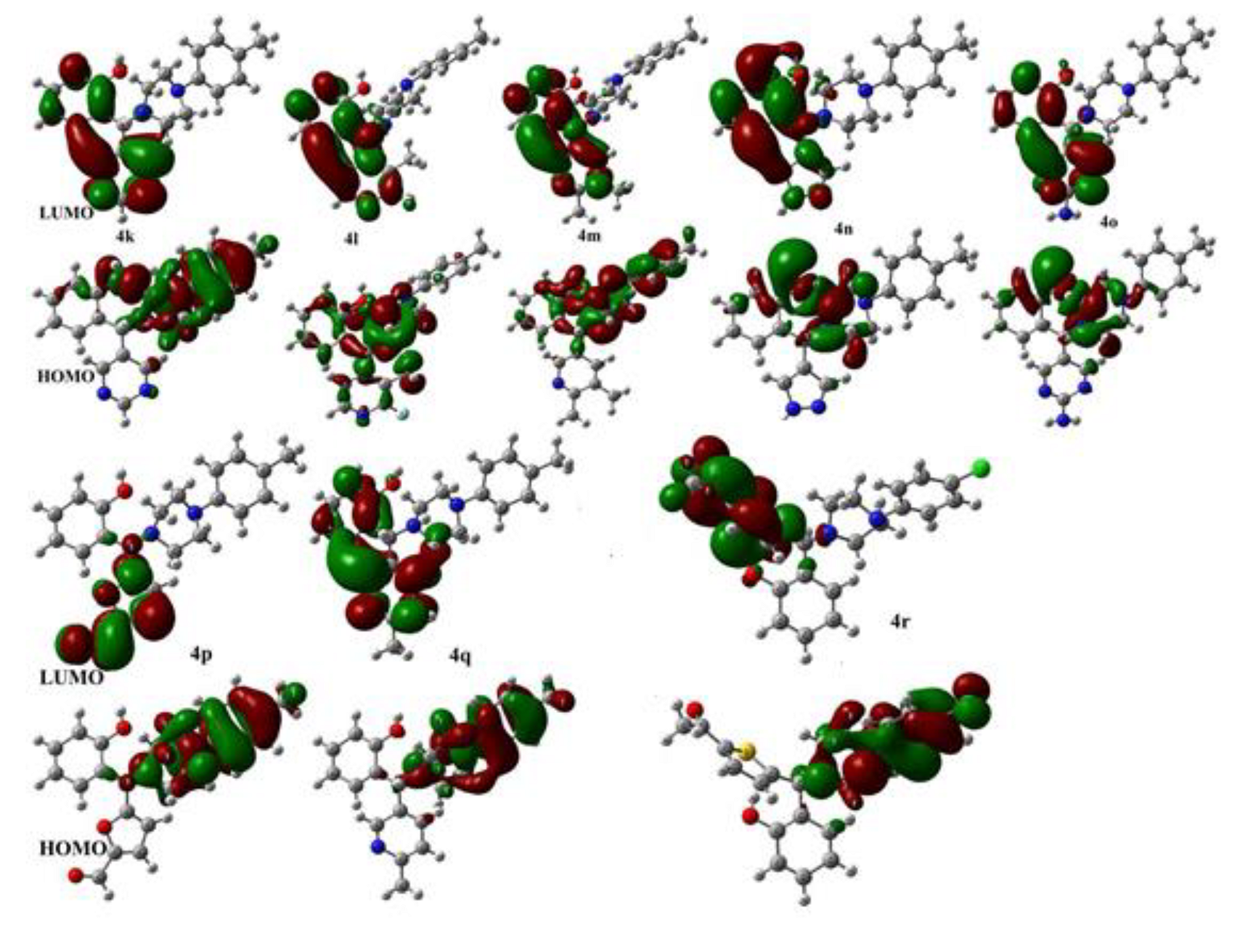
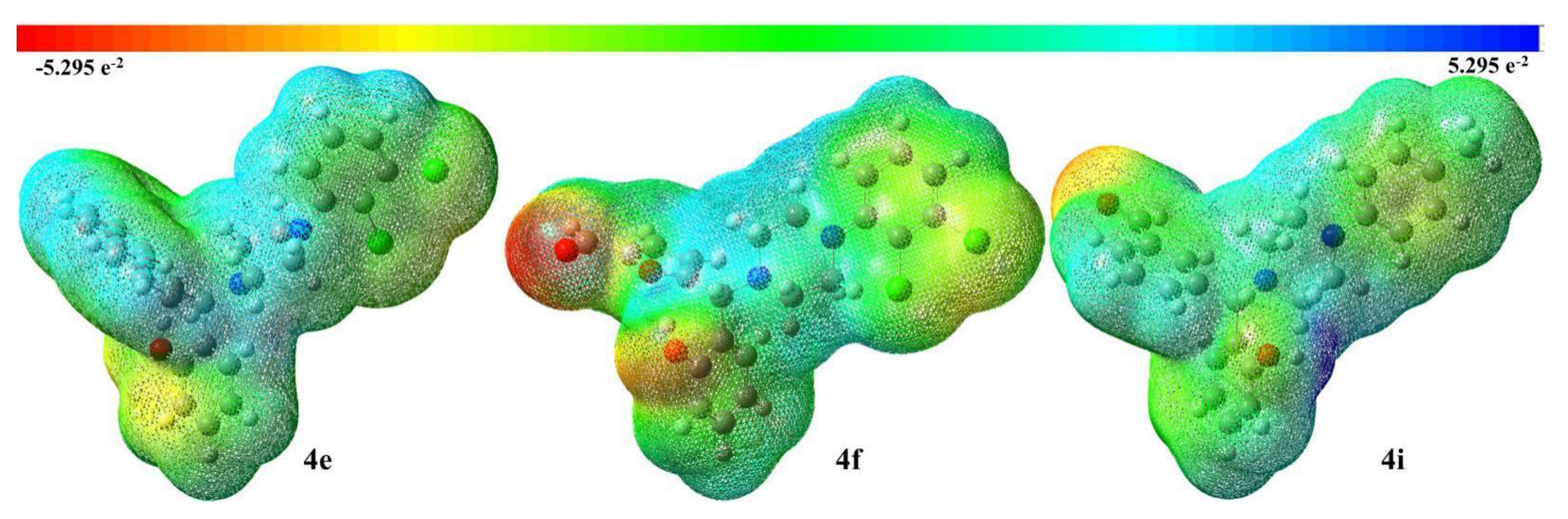
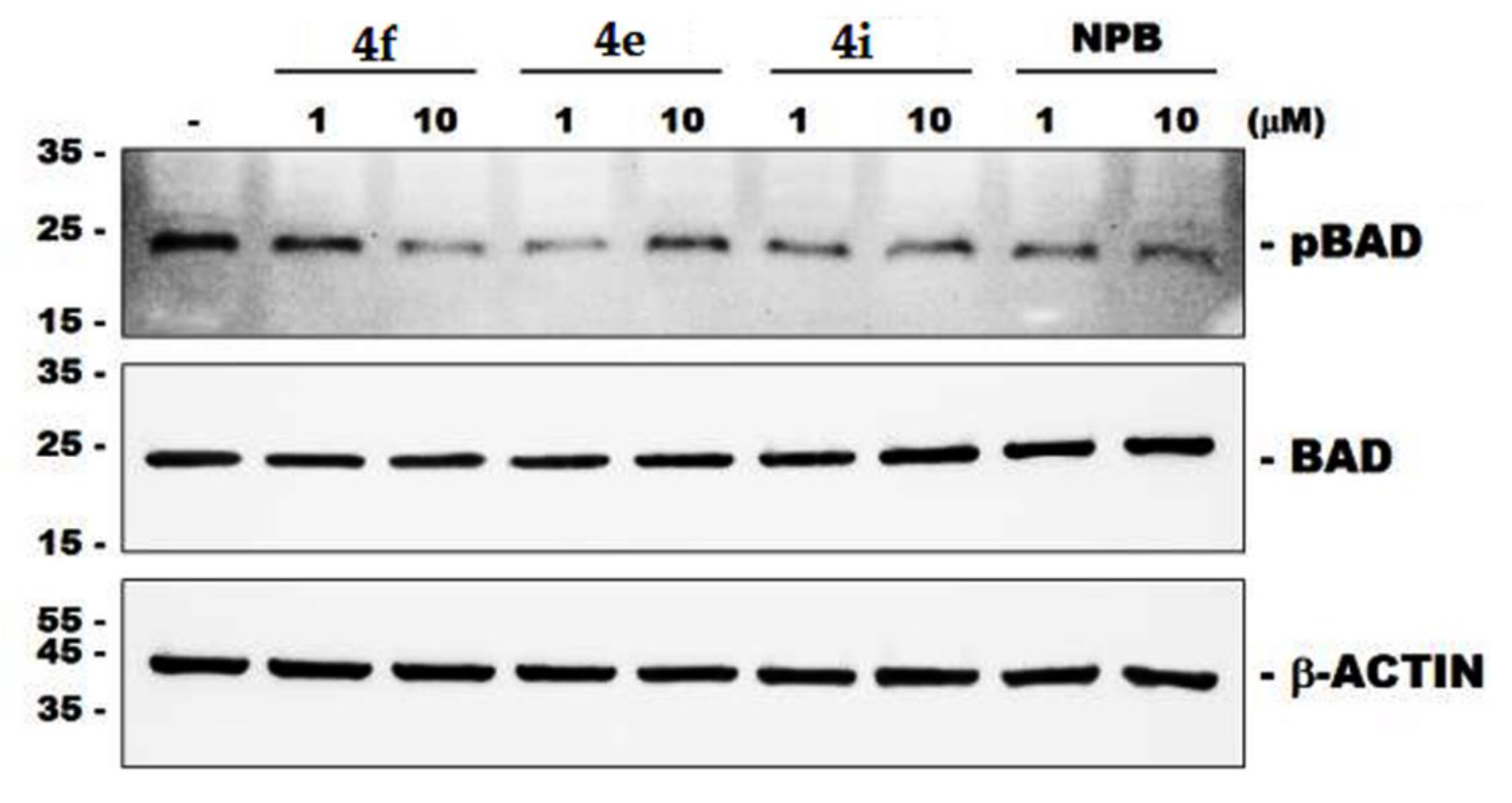
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fang, X.; Yu, S.; Eder, A.; Mao, M.; Bast, R.C., Jr.; Boyd, D.; Mills, G.B. Regulation of BAD phosphorylation at serine 112 by the Ras-mitogen-activated protein kinase pathway. Oncogene 1999, 18, 6635–6640. [Google Scholar] [CrossRef] [Green Version]
- Zha, J.; Harada, H.; Osipov, K.; Jockel, J.; Waksman, G.; Korsmeyer, S.J. BH3 domain of BAD is required for heterodimerization with BCL-XL and pro-apoptotic activity. J. Biol. Chem. 1997, 272, 24101–24104. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chiou, Y.S.; Chong, Q.Y.; Zhang, M.; Rangappa, K.S.; Ma, L.; Zhu, T.; Kumar, A.P.; Huang, R.Y.; Pandey, V.; et al. Pharmacological Inhibition of BAD Ser99 Phosphorylation Enhances the Efficacy of Cisplatin in Ovarian Cancer by Inhibition of Cancer Stem Cell-like Behavior. ACS Pharm. Transl. Sci. 2020, 3, 1083–1099. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.R.; Brunet, A.; Greenberg, M.E. Cellular survival: A play in three Akts. Genes Dev. 1999, 13, 2905–2927. [Google Scholar] [CrossRef] [PubMed]
- Bui, N.L.; Pandey, V.; Zhu, T.; Ma, L.; Basappa, B.; Lobie, P.E. Bad phosphorylation as a target of inhibition in oncology. Cancer Lett. 2018, 415, 177–186. [Google Scholar] [CrossRef]
- Moody, S.E.; Schinzel, A.C.; Singh, S.; Izzo, F.; Strickland, M.R.; Luo, L.; Thomas, S.R.; Boehm, J.S.; Kim, S.Y.; Wang, Z.C.; et al. PRKACA mediates resistance to HER2-targeted therapy in breast cancer cells and restores anti-apoptotic signaling. Oncogene 2015, 34, 2061–2071. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.; Demeter, M.R.; Ruan, H.; Comb, M.J. BAD Ser-155 phosphorylation regulates BAD/Bcl-XL interaction and cell survival. J. Biol. Chem. 2000, 275, 25865–25869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.M.; Liu, Y.; Payne, G.; Lutz, R.J.; Chittenden, T. Growth factors inactivate the cell death promoter BAD by phosphorylation of its BH3 domain on Ser155. J. Biol. Chem. 2000, 275, 25046–25051. [Google Scholar] [CrossRef] [Green Version]
- Pandey, V.; Wang, B.; Mohan, C.D.; Raquib, A.R.; Rangappa, S.; Srinivasa, V.; Fuchs, J.E.; Girish, K.S.; Zhu, T.; Bender, A.; et al. Discovery of a small-molecule inhibitor of specific serine residue BAD phosphorylation. Proc. Natl. Acad. Sci. USA 2018, 115, E10505–E10514. [Google Scholar] [CrossRef] [Green Version]
- Lobie, P.E.; Pandey, V.K.; Rangappa, K.S.; Basappa, M.; Chakrabhavi, D.; Rangappa, S.; Srinivasa, V. World Intellectual Property Organization. WO2018194520 A1, 25 October 2018. [Google Scholar]
- Anusha, S.; Mohan, C.D.; Ananda, H.; Baburajeev, C.P.; Rangappa, S.; Mathai, J.; Fuchs, J.E.; Li, F.; Shanmugam, M.K.; Bender, A.; et al. Adamantyl-tethered-biphenylic compounds induce apoptosis in cancer cells by targeting Bcl homologs. Bioorg. Med. Chem. Lett. 2016, 26, 1056–1060. [Google Scholar] [CrossRef]
- Priya, B.S.; Swamy, S.N.; Tejesvi, M.V.; Basappa, B.; Sarala, G.; Gaonkar, S.L.; Naveen, S.; Prasad, J.S.; Rangappa, K.S. Synthesis, characterization, antimicrobial and single crystal X-ray crystallographic studies of some new sulfonyl, 4-chloro phenoxy benzene and dibenzoazepine substituted benzamides. Eur. J. Med. Chem. 2006, 41, 1262–1270. [Google Scholar] [CrossRef]
- Bharathkumar, H.; Mohan, C.D.; Ananda, H.; Fuchs, J.E.; Li, F.; Rangappa, S.; Surender, M.; Bulusu, K.C.; Girish, K.S.; Sethi, G.; et al. Microwave-assisted synthesis, characterization and cytotoxic studies of novel estrogen receptor α ligands towards human breast cancer cells. Bioorg. Med. Chem. Lett. 2015, 25, 1804–1807. [Google Scholar] [CrossRef] [PubMed]
- Rangappa, K.; Basappa, B. New cholinesterase inhibitors: Synthesis and structure-activity relationship studies of 1,2-benzisoxazole series and novel imidazolyl-d 2-isoxazolines. J. Phys. Org. Chem. 2005, 18, 773–778. [Google Scholar] [CrossRef]
- Basappa, B.; Kavitha, C.V.; Rangappa, K.S. Simple and an efficient method for the synthesis of 1-[2-dimethylamino-1-(4-methoxy-phenyl)-ethyl]-cyclohexanol hydrochloride: (+/−) venlafaxine racemic mixtures. Bioorg. Med. Chem. Lett. 2004, 14, 3279–3281. [Google Scholar] [CrossRef]
- Rakesh, K.S.; Jagadish, S.; Vinayaka, A.C.; Hemshekhar, M.; Paul, M.; Thushara, R.M.; Sundaram, M.S.; Swaroop, T.R.; Mohan, C.D.; Basappa, B.; et al. A new ibuprofen derivative inhibits platelet aggregation and ROS mediated platelet apoptosis. PLoS ONE 2014, 9, e107182. [Google Scholar] [CrossRef] [Green Version]
- Naskar, D.; Roy, A.; Seibel, W.L.; Portlock, D.E. Novel Petasis boronic acid-Mannich reactions with tertiary aromatic amines. Tetrahedron Lett. 2003, 44, 5819–5821. [Google Scholar] [CrossRef]
- Petasis, N.A.; Akritopoulou, I. The boronic acid mannich reaction: A new method for the synthesis of geometrically pure allylamines. Tetrahedron Lett. 1992, 34, 583–586. [Google Scholar] [CrossRef]
- Rimpiläinen, T.; Nunes, A.; Calado, R.; Fernandes, A.S.; Andrade, J.; Ntungwe, E.; Spengler, G.; Szemerédi, N.; Rodrigues, J.; Gomes, J.P.; et al. Increased antibacterial properties of indoline-derived phenolic Mannich bases. Eur. J. Med. Chem. 2021, 220, 113459. [Google Scholar] [CrossRef] [PubMed]
- Hommelsheim, R.; Núñez Ponce, H.M.; Truong, K.N.; Rissanen, K.; Bolm, C. 2-Sulfoximidoyl Acetic Acids from Multicomponent Petasis Reactions and Their Use as Building Blocks in Syntheses of Sulfoximine Benzodiazepine Analogues. Org. Lett. 2021, 23, 3415–3420. [Google Scholar] [CrossRef]
- Potowski, M.; Esken, R.; Brunschweiger, A. Translation of the copper/bipyridine-promoted Petasis reaction to solid phase-coupled DNA for encoded library synthesis. Bioorg. Med. Chem. 2020, 28, 115441. [Google Scholar] [CrossRef]
- Basappa, B.; Satish Kumar, M.; Nanjunda Swamy, S.; Mahendra, M.; Shashidhara Prasad, J.; Viswanath, B.S.; Rangappa, K.S. Novel delta2-isoxazolines as group II phospholipase A2 inhibitors. Bioorg. Med. Chem. Lett. 2004, 14, 3679–3681. [Google Scholar] [CrossRef] [PubMed]
- A.P.E.X. Bruker; Bruker AXS Inc.: Madison, WI, USA, 2004.
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Speak, A.L. Platon, An integrated tool for the analysis of the results of a single crystal structure determination. Acta Crystallogr. Sect. A Found. Crystallogr. 1990, 46, C34. [Google Scholar]
- Gautam Desiraju, R.; Steiner, T. The Weak Hydrogen Bond in Structural Chemistry and Biology; Oxford University Press/International Union of Crystallography: Oxford, UK, 1999; Volume 9, pp. 1–507. [Google Scholar]
- Basappa, B.; Pookunoth, B.C.; Kempasiddegowda, M.S.; Subbegowda, R.K.; Lobie, P.; Pandey, V. Novel Biphenyl Amines Inhibit Oestrogen Receptor (ER)-α in ER-Positive Mammary Carcinoma Cells. Molecules 2021, 26, 783. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, B.; Chong, Q.Y.; Pandey, V.; Guo, Z.; Chen, R.M.; Wang, L.; Wang, Y.; Ma, L.; Kumar, A.P.; et al. A novel small-molecule inhibitor of trefoil factor 3 (TFF3) potentiates MEK1/2 inhibition in lung adenocarcinoma. Oncogenesis 2019, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Guéniche, N.; Huguet, A.; Bruyere, A.; Habauzit, D.; Le Hégarat, L.; Fardel, O. Comparative in silico prediction of P-glycoprotein-mediated transport for 2010–2020 US FDA-approved drugs using six Web-tools. Biopharm. Drug Dispos. 2021, 42, 393–398. [Google Scholar] [CrossRef]
- Poh, H.M.; Chiou, Y.S.; Chong, Q.Y.; Chen, R.M.; Rangappa, K.S.; Ma, L.; Zhu, T.; Kumar, A.P.; Pandey, V.; Basappa, B.; et al. Inhibition of TFF3 Enhances Sensitivity-and Overcomes Acquired Resistance-to Doxorubicin in Estrogen Receptor-Positive Mammary Carcinoma. Cancers 2019, 11, 1528. [Google Scholar] [CrossRef] [Green Version]
- Gilandoust, M.; Harsha, K.B.; Mohan, C.D.; Raquib, A.R.; Rangappa, S.; Pandey, V.; Lobie, P.E.; Basappa, B.; Rangappa, K.S. Synthesis, characterization and cytotoxicity studies of 1,2,3-triazoles and 1,2,4-triazolo [1,5-a] pyrimidines in human breast cancer cells. Bioorg. Med. Chem. Lett. 2018, 28, 2314–2319. [Google Scholar] [CrossRef]
- Chong, Q.Y.; You, M.L.; Pandey, V.; Banerjee, A.; Chen, Y.J.; Poh, H.M.; Zhang, M.; Ma, L.; Zhu, T.; Basappa, S.; et al. Release of HER2 repression of trefoil factor 3 (TFF3) expression mediates trastuzumab resistance in HER2+/ER+ mammary carcinoma. Oncotarget 2017, 8, 74188–74208. [Google Scholar] [CrossRef] [Green Version]
- Kumar, C.A.; Jayarama, S.; Basappa, B.; Salimath, B.P.; Rangappa, K.S. Pro-apoptotic activity of imidazole derivatives mediated by up-regulation of Bax and activation of CAD in Ehrlich Ascites Tumor cells. Invest. New Drugs 2007, 25, 343–350. [Google Scholar] [CrossRef]
- Anusha, S.; Anandakumar, B.S.; Mohan, C.D.; Nagabhushana, G.P.; Priya, B.S.; Rangappa, K.S.; Basappa, B.; Chandrappa, G.T. Preparation and use of combustion-derived Bi2O3 for the synthesis of heterocycles with anti-cancer properties by Suzuki-coupling reactions. RSC Adv. 2014, 4, 52181–52188. [Google Scholar]
- Ravi Kumar, K.R.; Mallesha, H.; Basappa, R.K.S. Synthesis of novel isoxazolidine derivatives and studies for their antifungal properties. Eur. J. Med. Chem. 2003, 38, 613–619. [Google Scholar] [CrossRef]
- Bharathkumar, H.; Paricharak, S.; Dinesh, K.R.; Siveen, K.S.; Fuchs, J.E.; Rangappa, S.; Mohan, C.D.; Mohandas, N.; Kumar, A.P.; Sethi, G.; et al. Synthesis, biological evaluation and in silico and in vitro mode-of-action analysis of novel dihydropyrimidones targeting PPAR-γ. RSC Adv. 2014, 4, 45143–45146. [Google Scholar] [CrossRef] [Green Version]
- Basappa, B.; Rangappa, K.S.; Sugahara, K. Roles of glycosaminoglycans and glycanmimetics in tumor progression and metastasis. Glycoconj. J. 2014, 31, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, V.; Chevalier, F.; Imberty, A.; Leeflang, B.R.; Basappa, B.; Sugahara, K.; Kamerling, J.P. Conformational studies on five octasaccharides isolated from chondroitin sulfate using NMR spectroscopy and molecular modeling. Biochemistry 2007, 46, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision, C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]

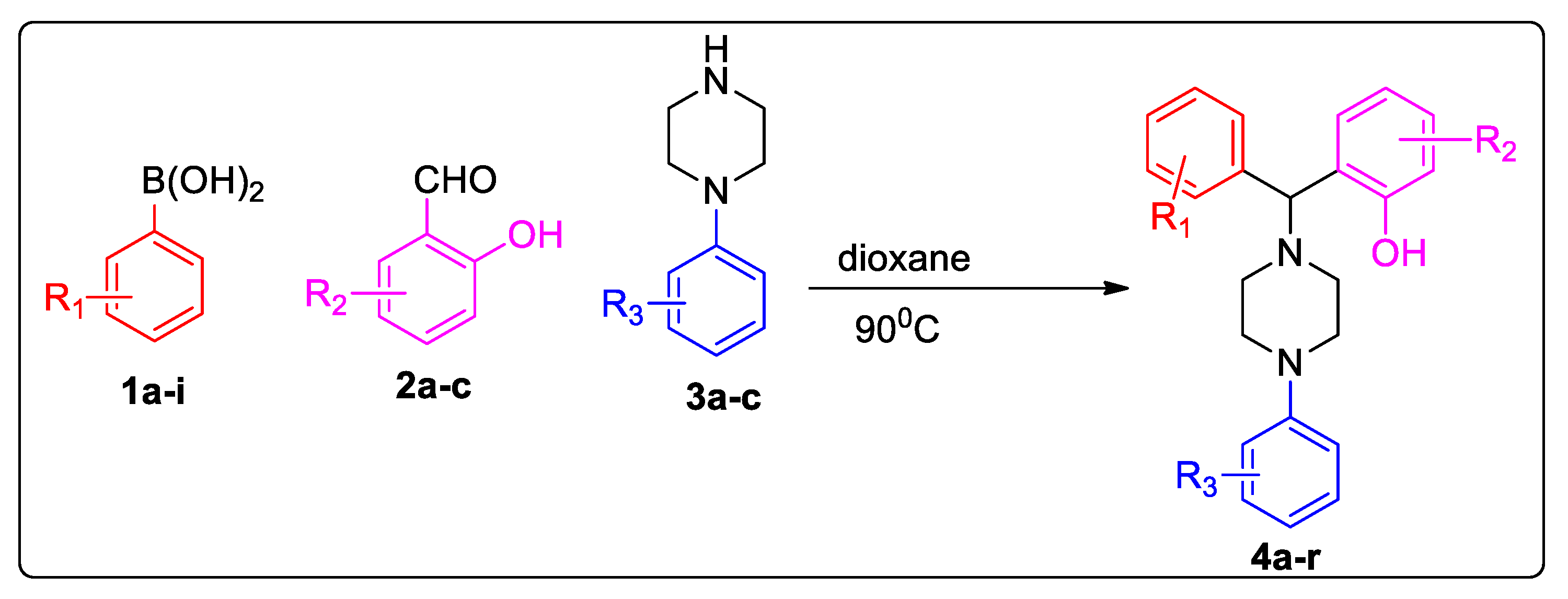
| Entry | Structure | MCF-7 IC50 (µM) | Yield (%) | M.P (°C) |
|---|---|---|---|---|
| 4a | 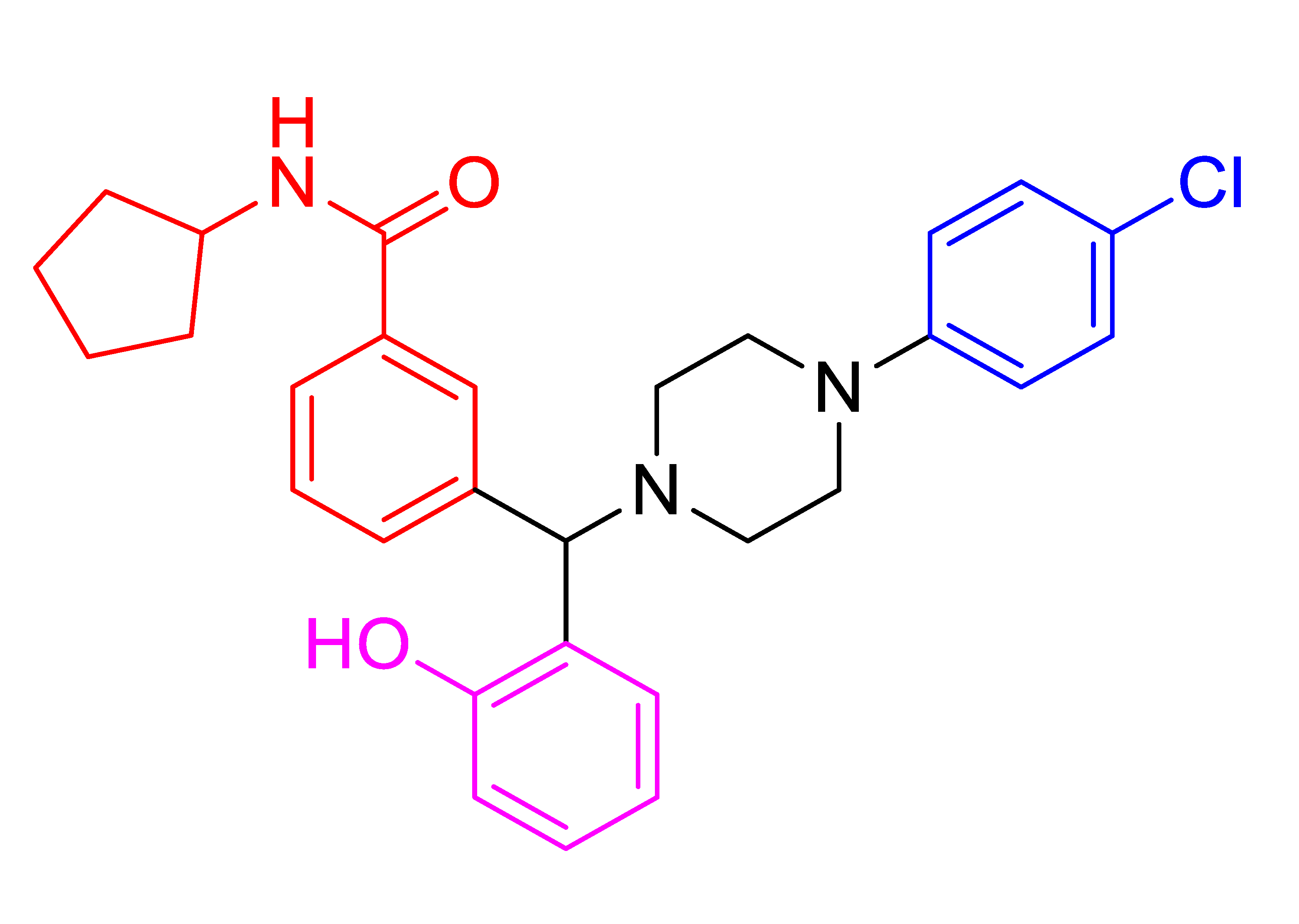 | 605.6 | 91 | 128–130 |
| 4b | 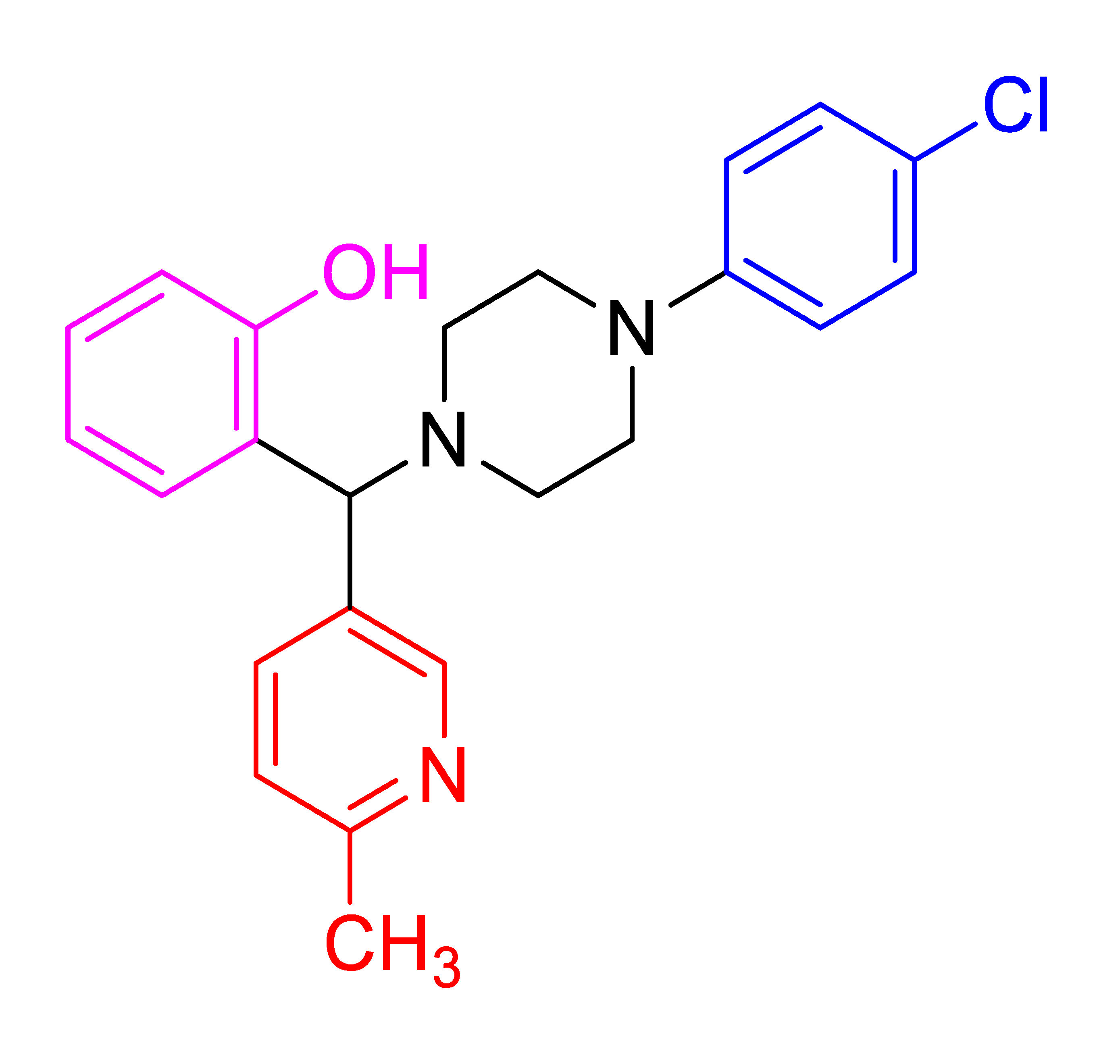 | 20.91 | 88 | 210–212 |
| 4c | 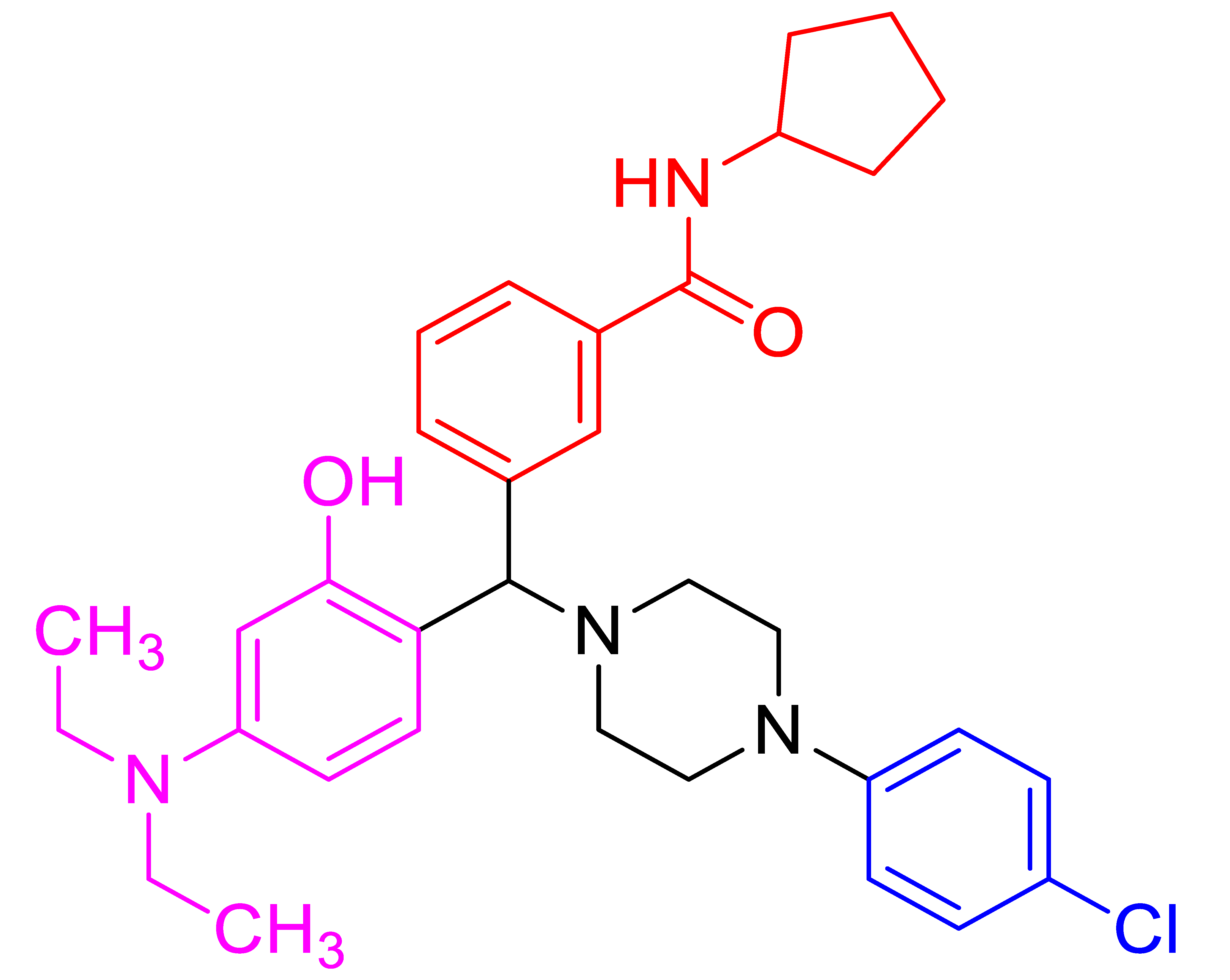 | 23.83 | 81 | 178–180 |
| 4d | 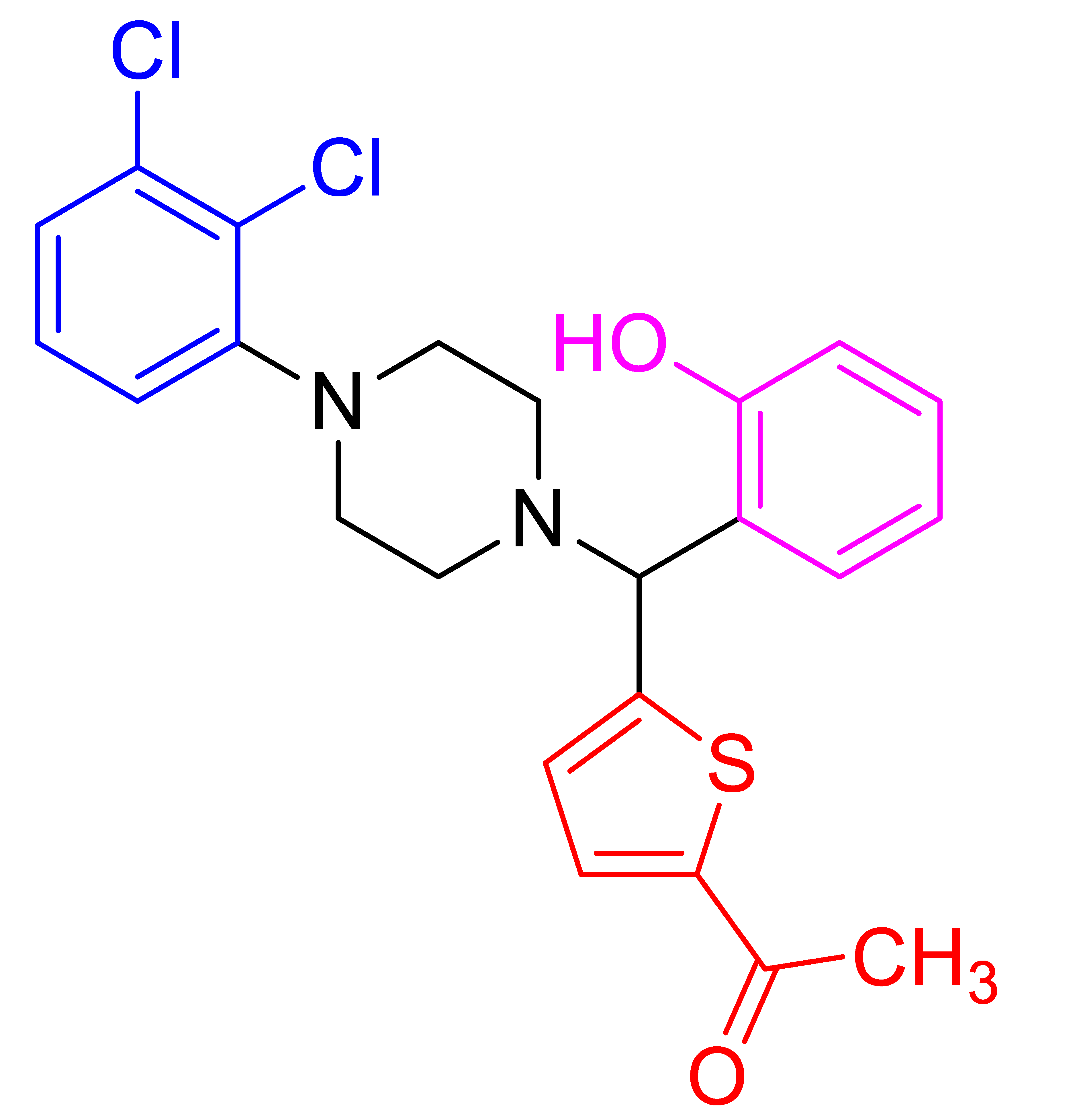 | 270.6 | 89 | 130–132 |
| 4e | 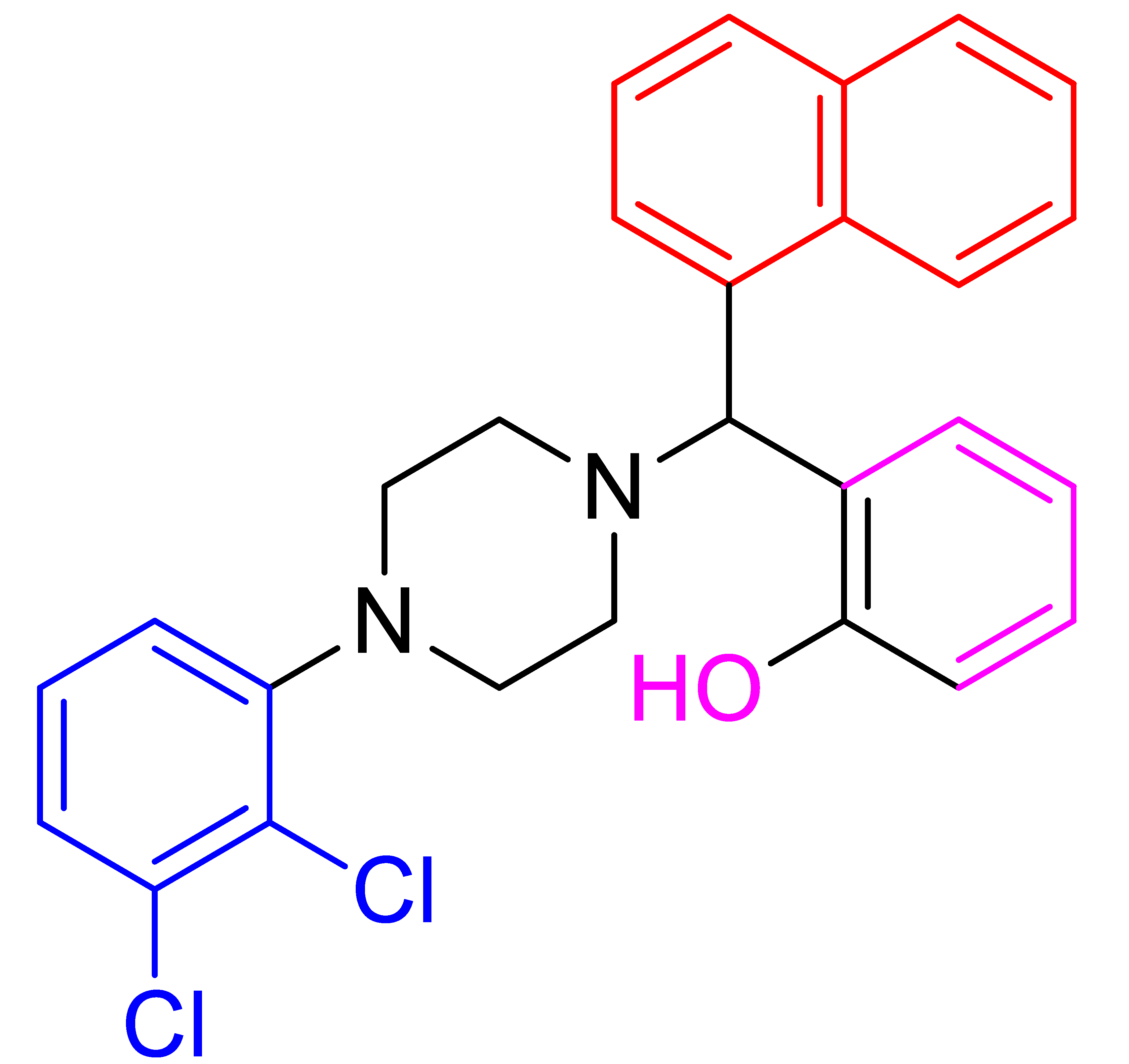 | 5.90 | 78 | 78–80 |
| 4f | 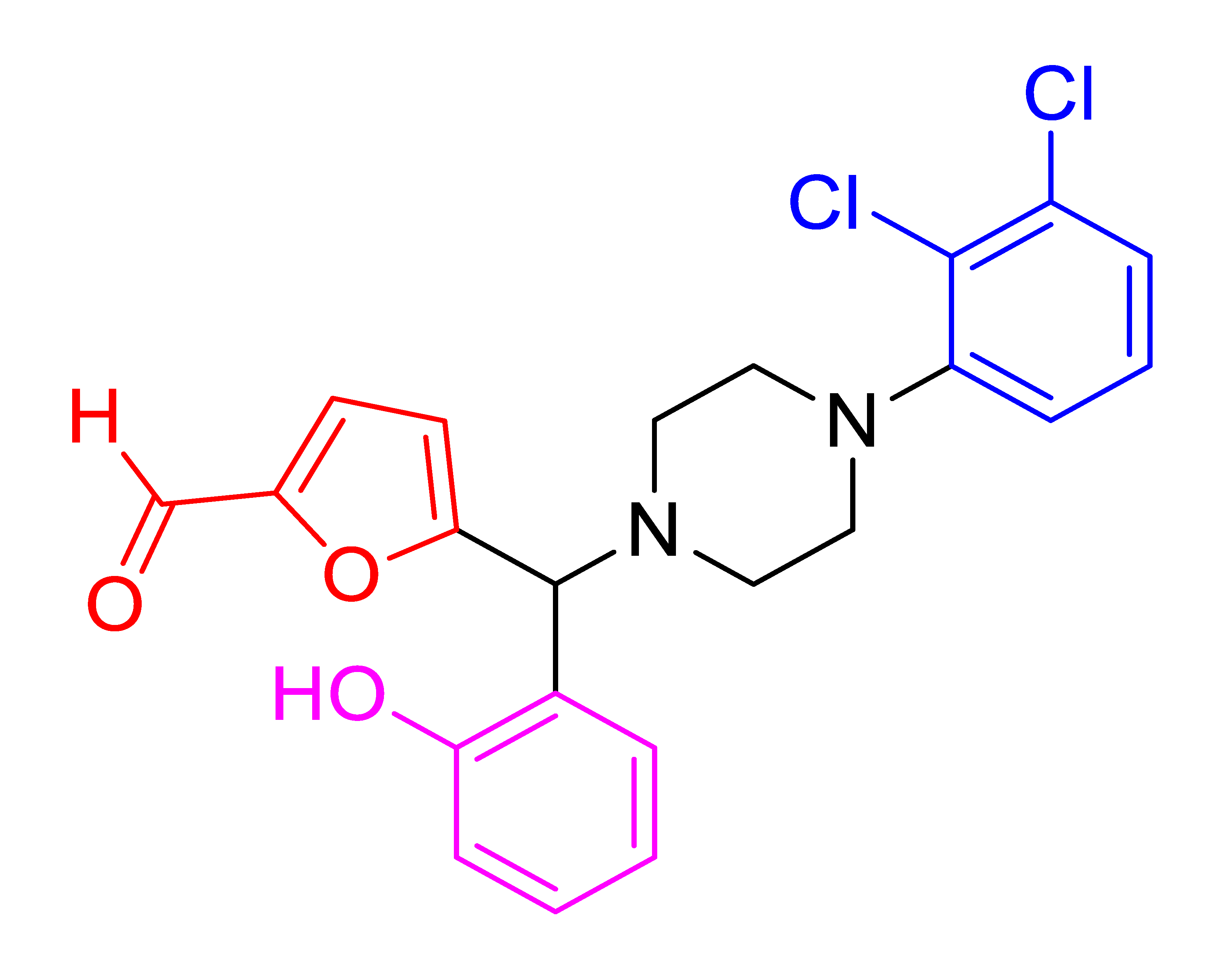 | 3.11 | 83 | 128–130 |
| 4g | 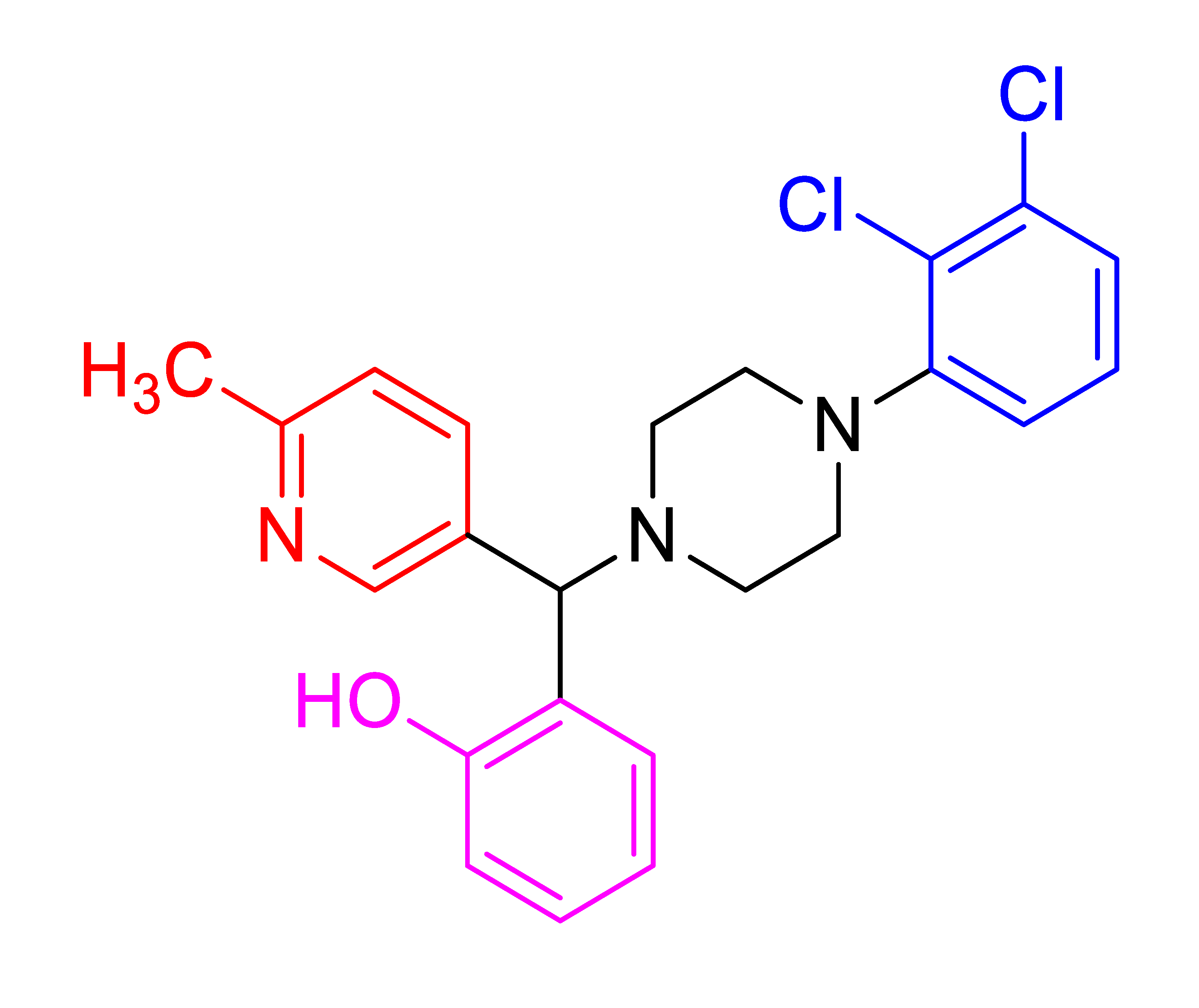 | - | 83 | 208–210 |
| 4h | 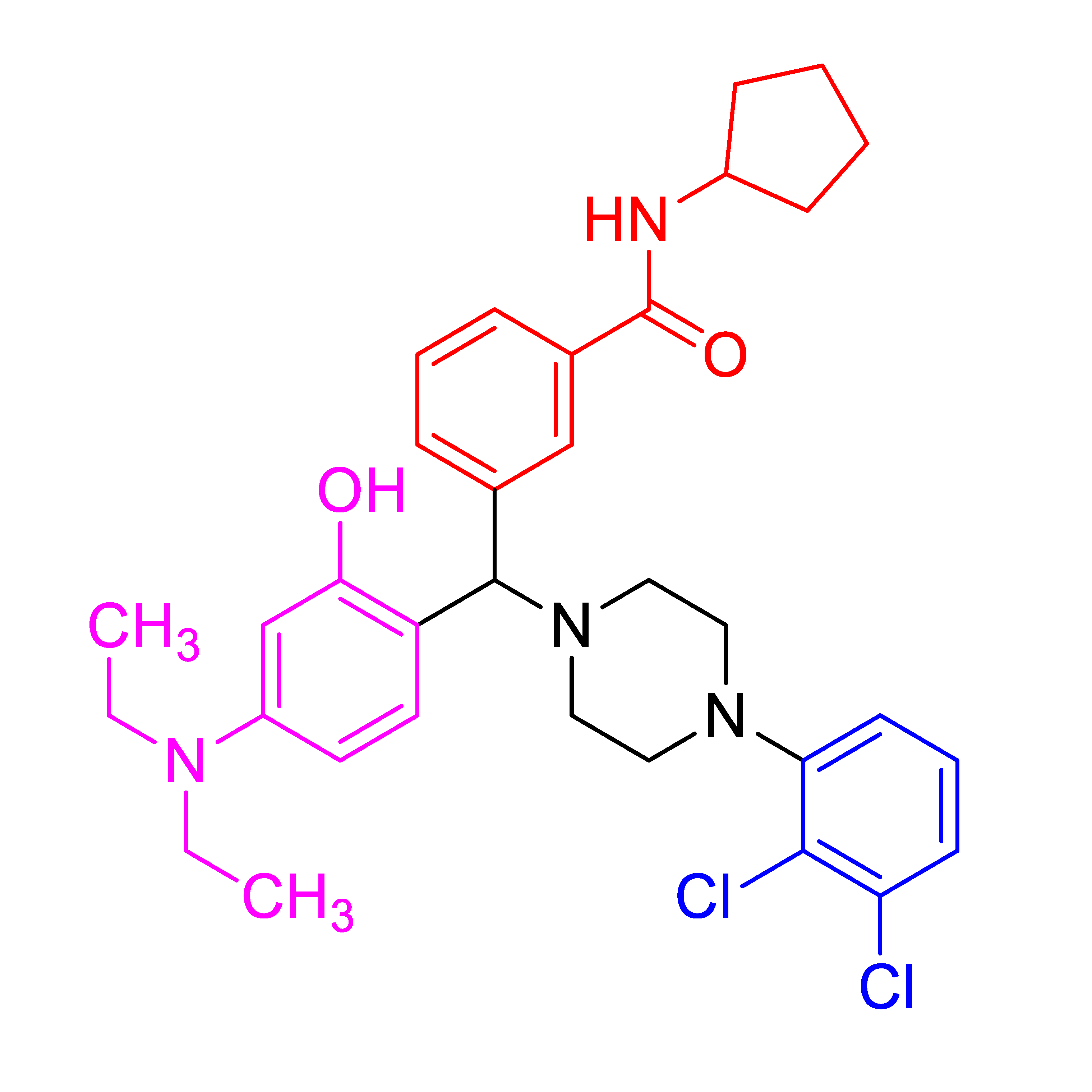 | 20.93 | 81 | 160–162 |
| 4i | 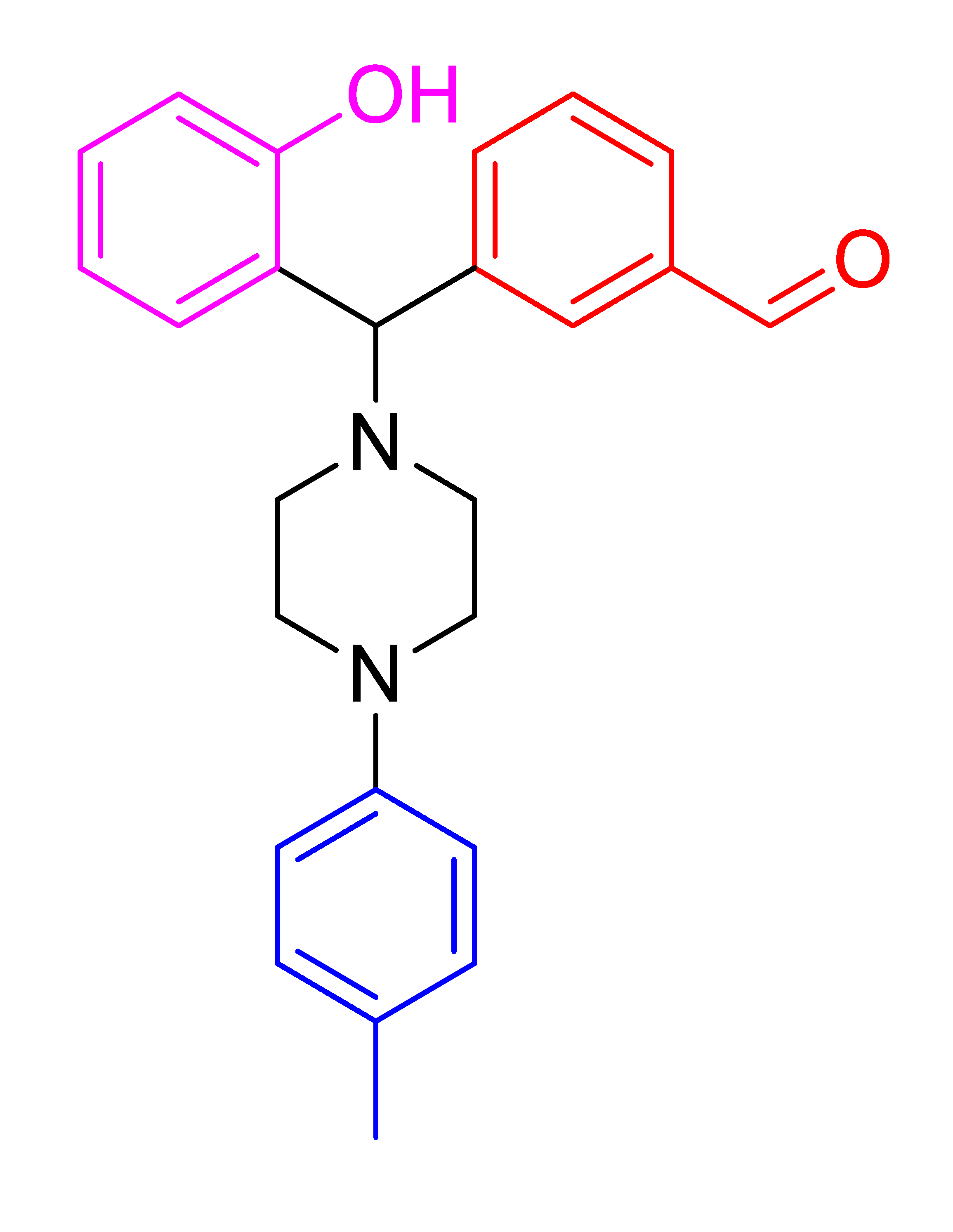 | 7.68 | 88 | 120–123 |
| 4j | 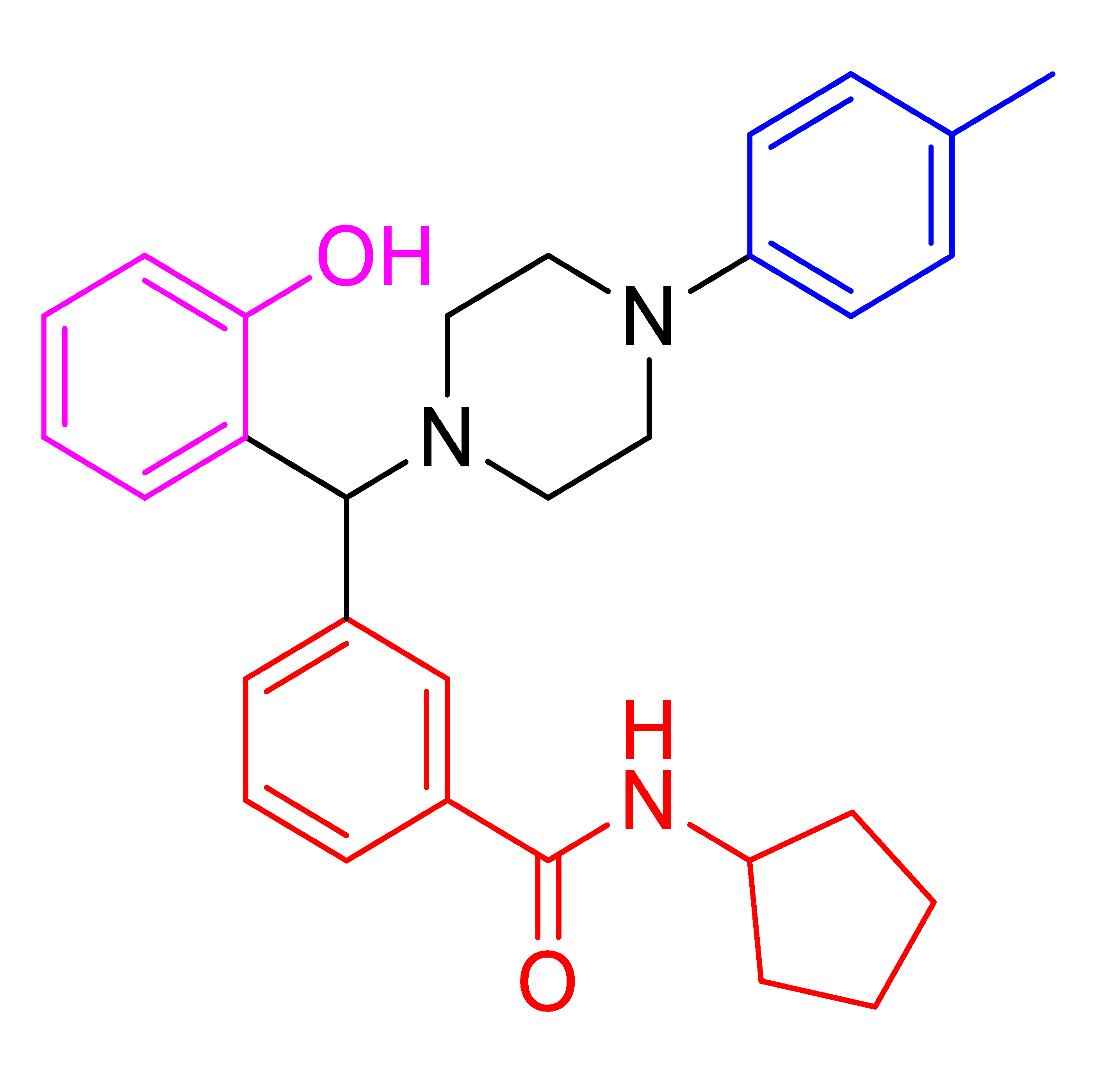 | 326.40 | 87 | 114–117 |
| 4k | 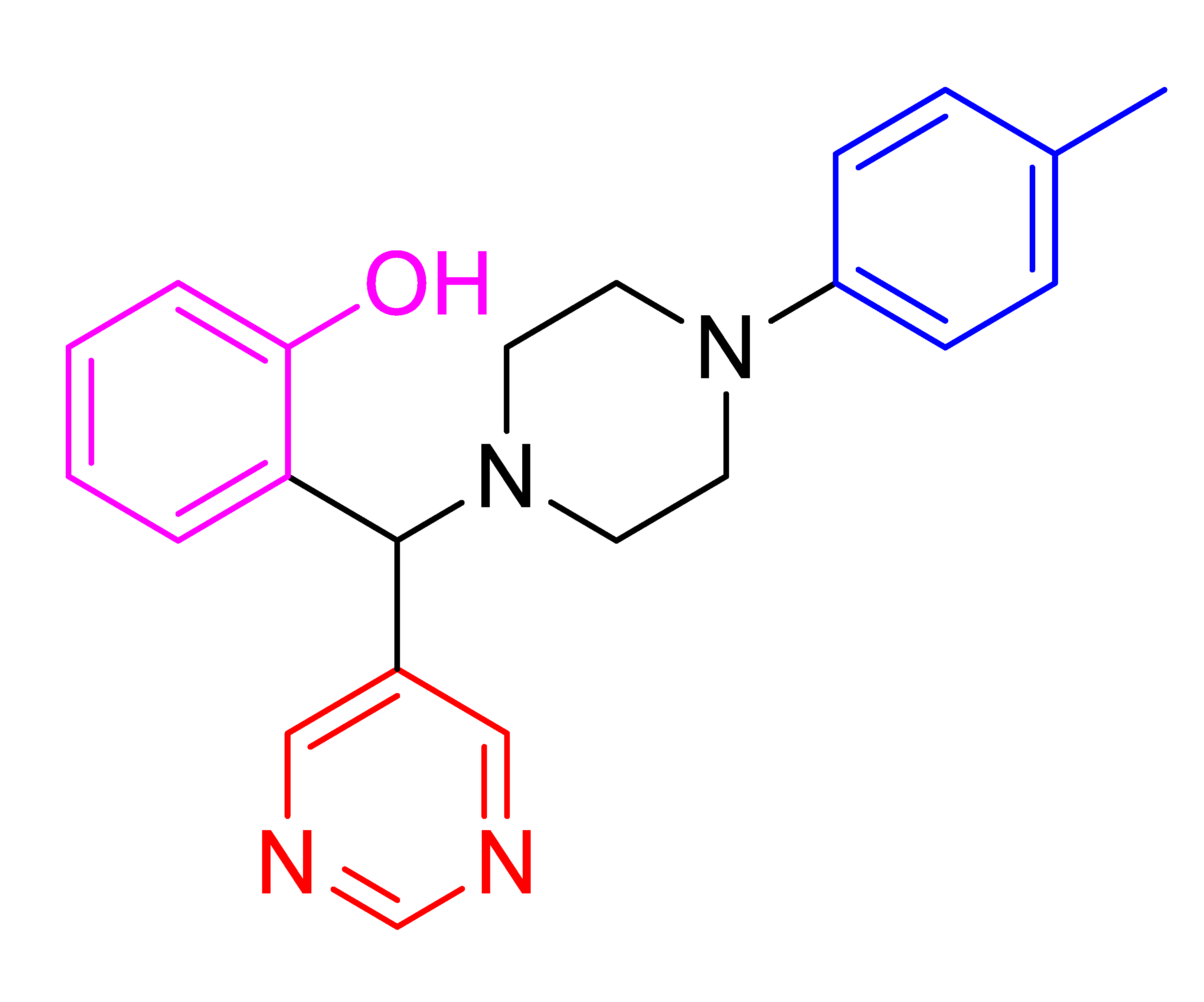 | 221.50 | 84 | 157–161 |
| 4l | 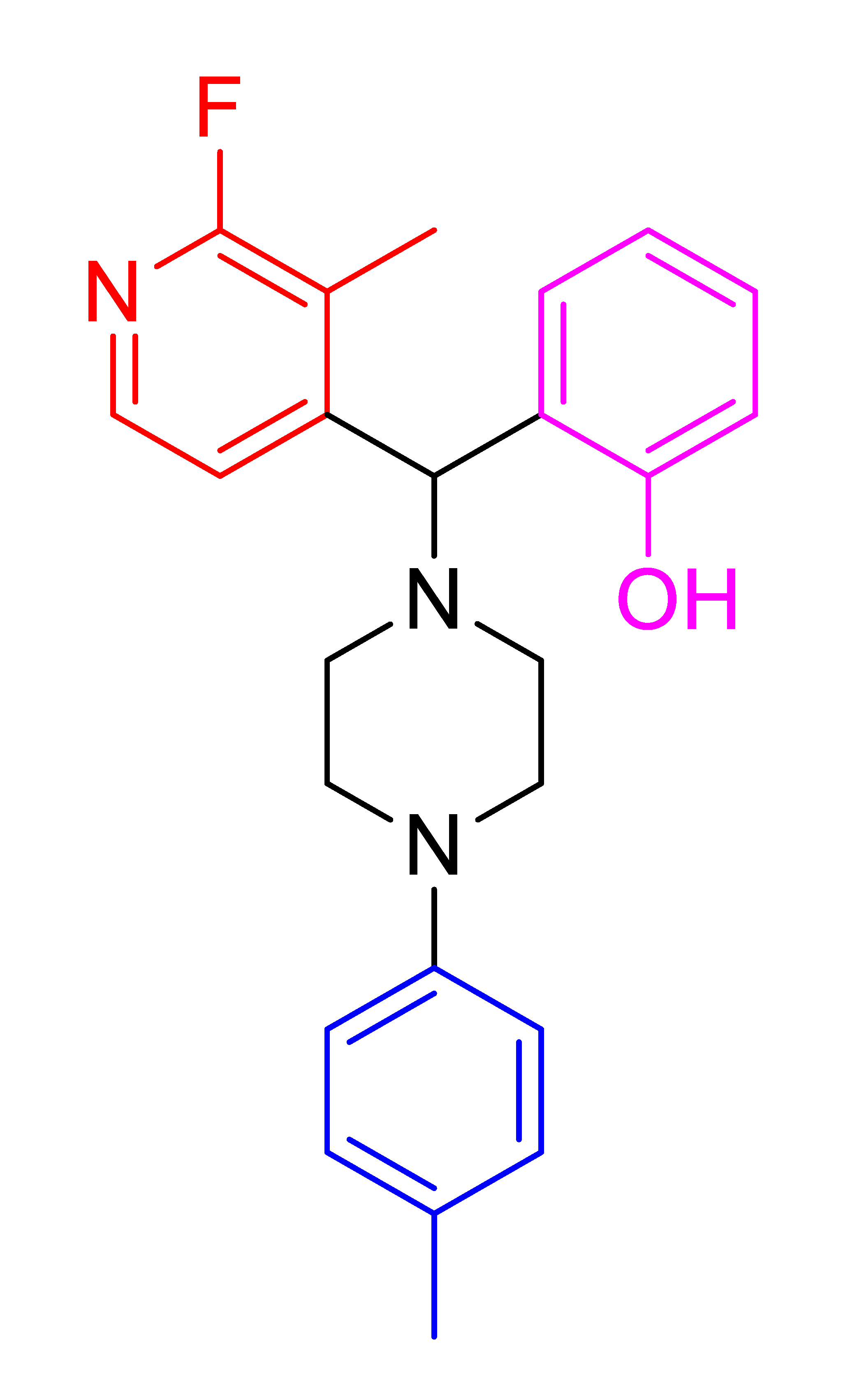 | 63.66 | 80 | 102–104 |
| 4m | 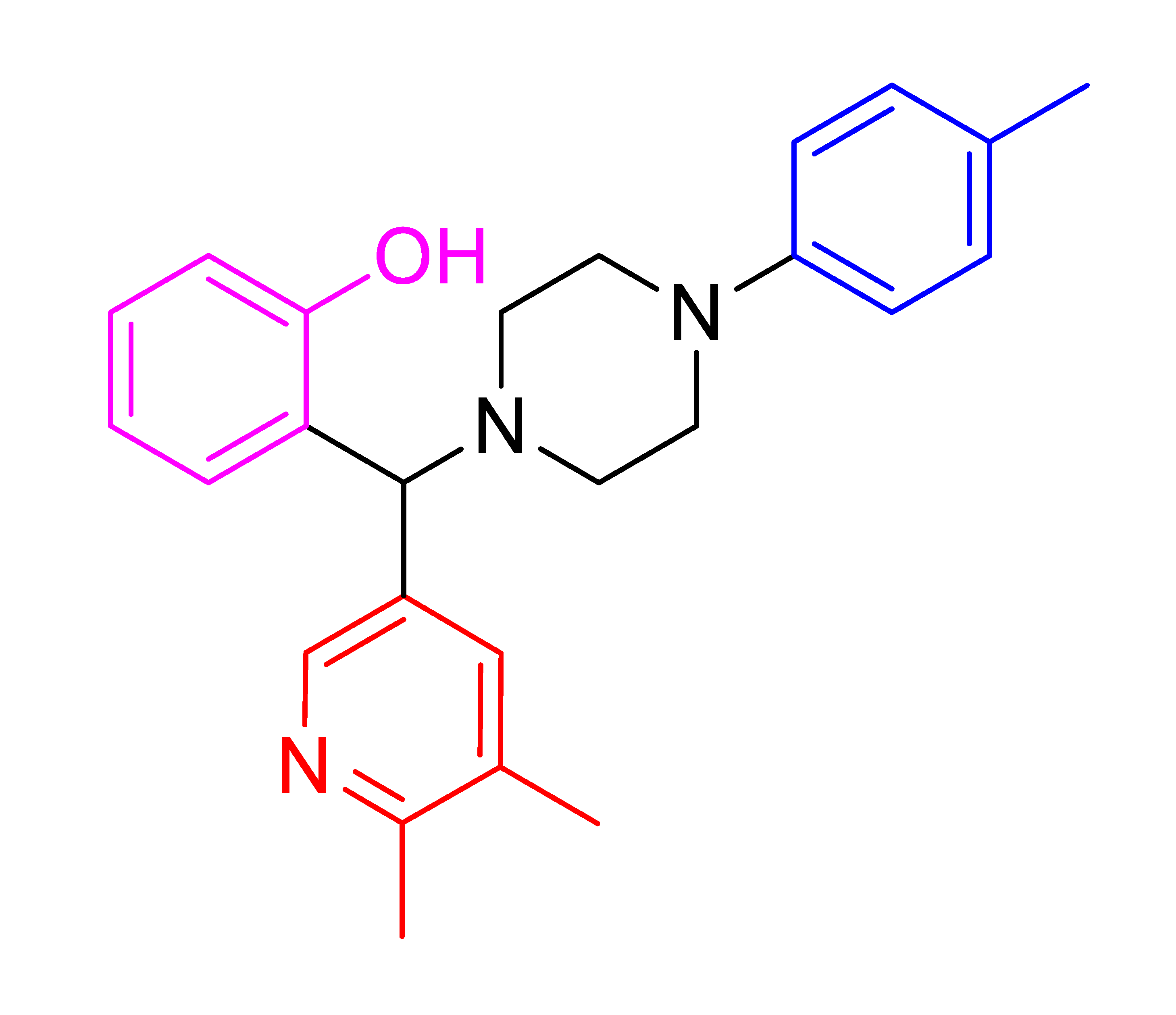 | 45.73 | 86 | 147–159 |
| 4n | 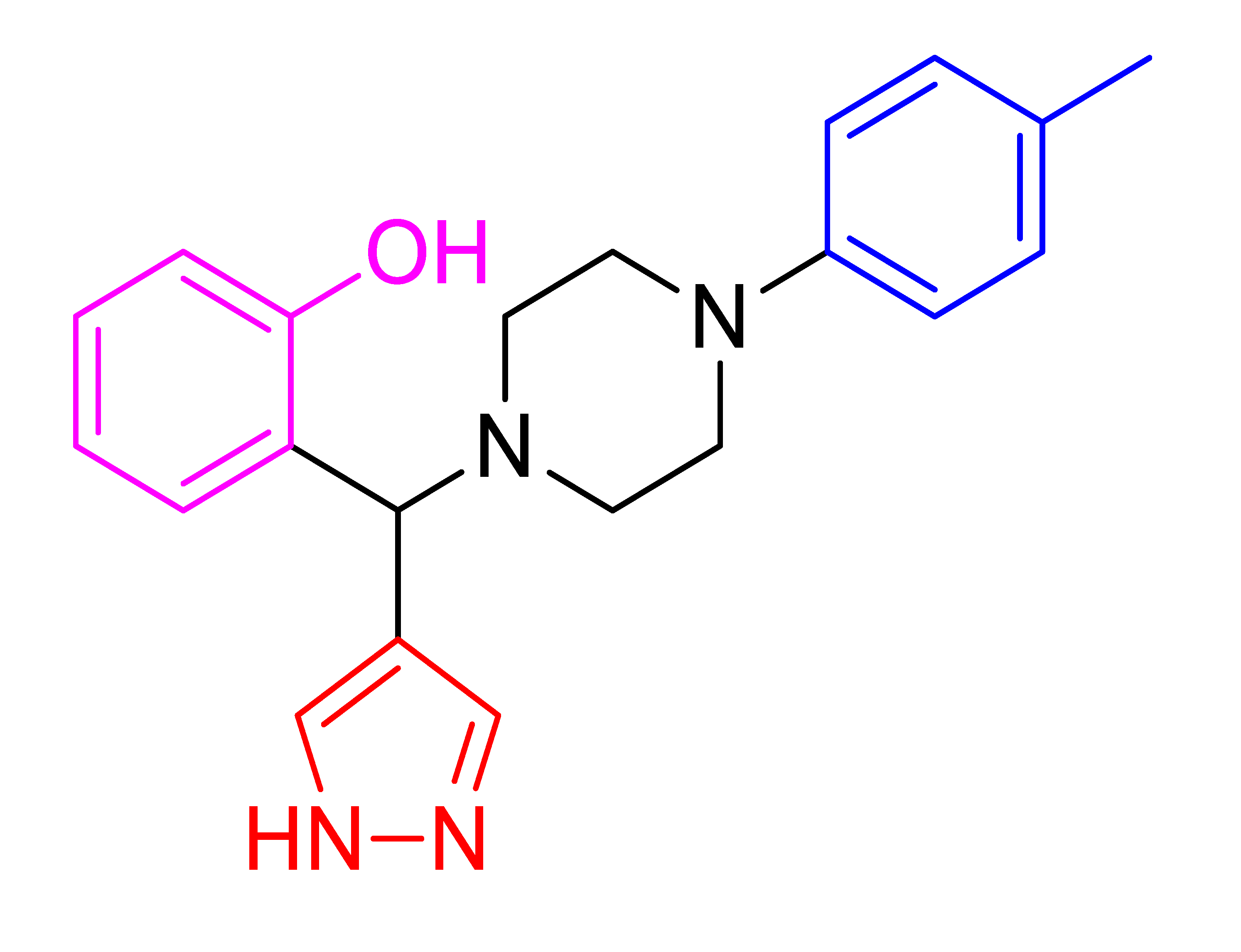 | 817.30 | 83 | 151–153 |
| 4o | 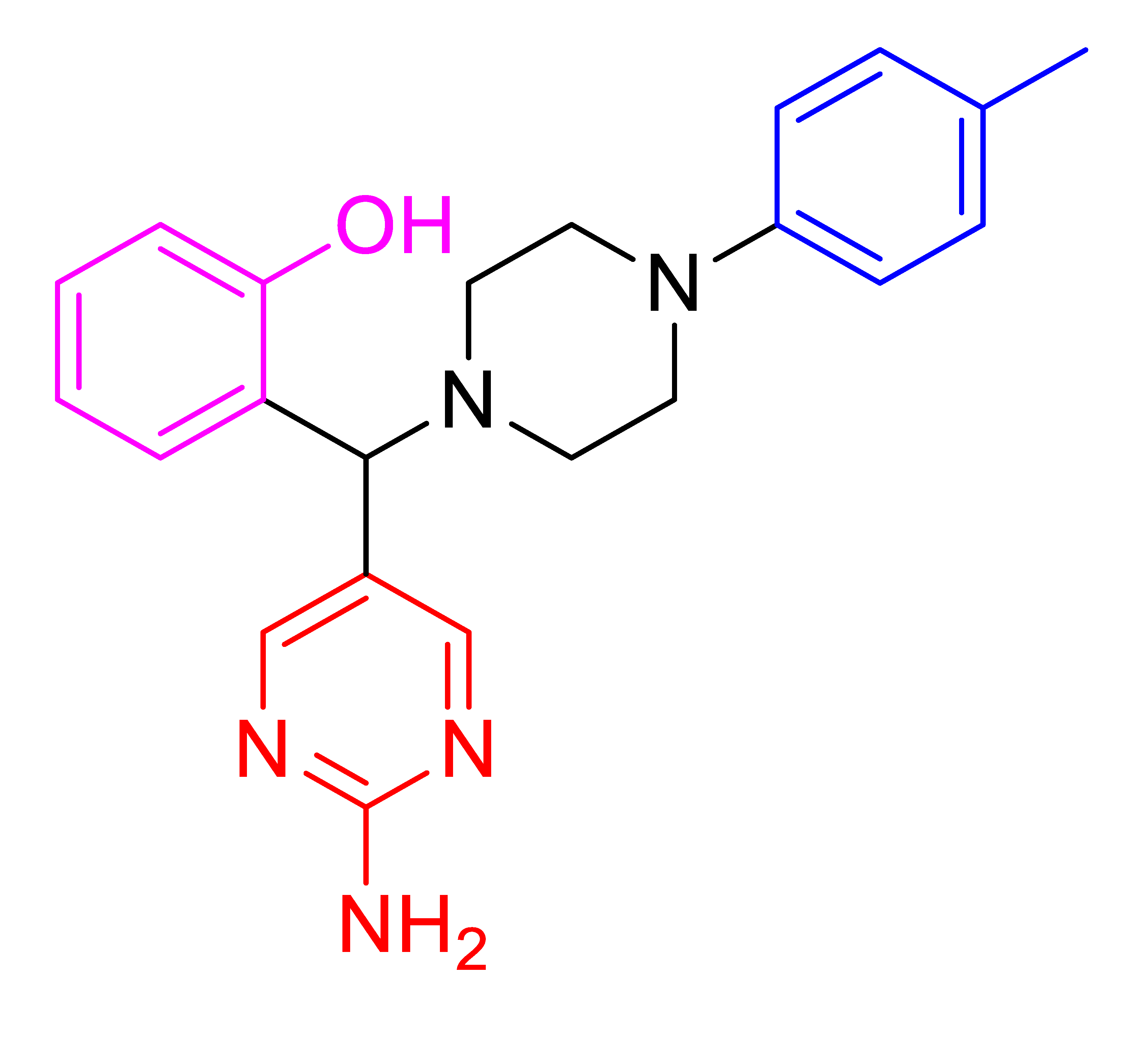 | 1113.00 | 91 | 131–134 |
| 4p |  | 156.00 | 87 | 94–97 |
| 4q | 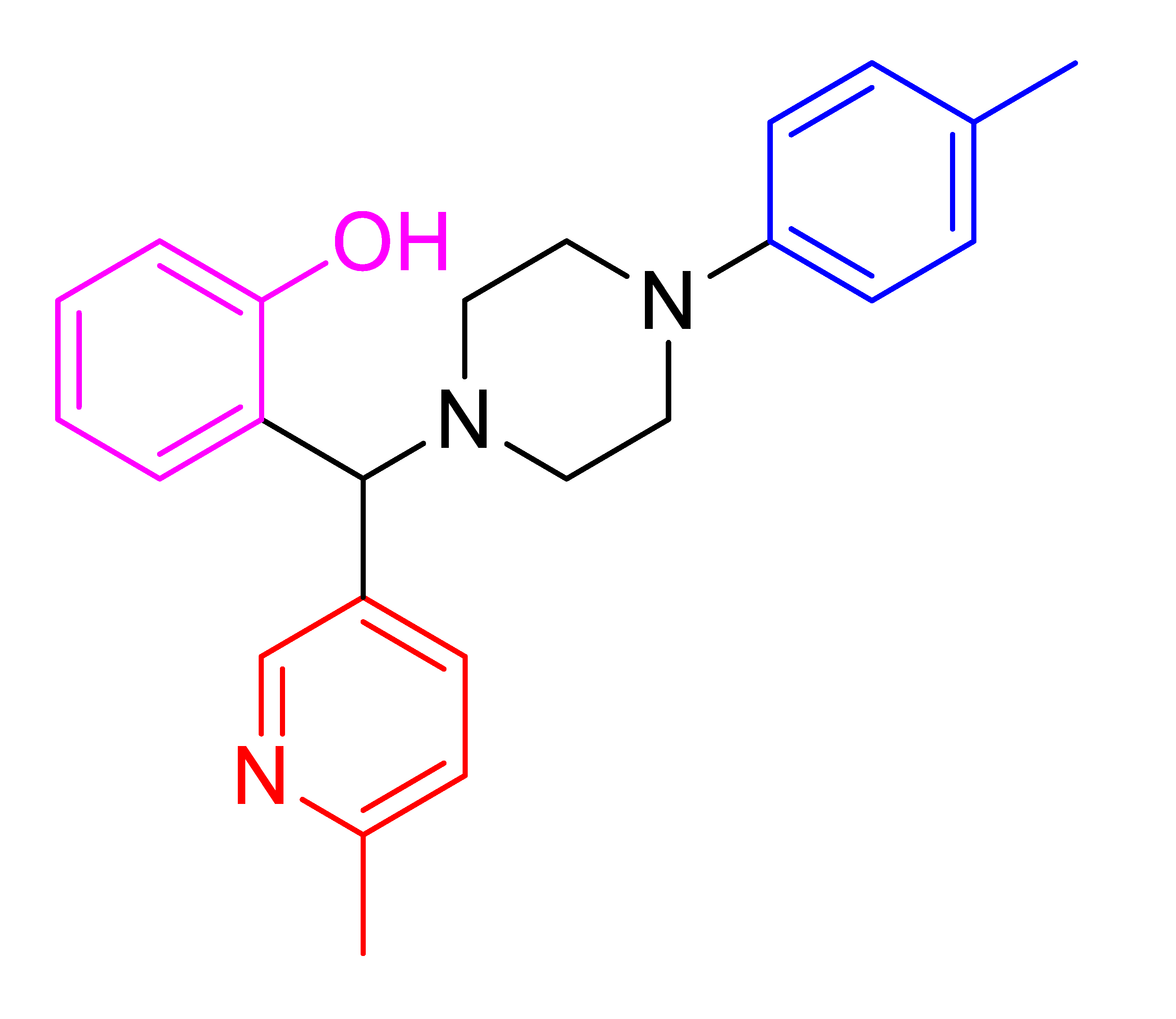 | 60.00 | 93 | 152–154 |
| 4r |  | - | 89 | 140–142 |
| Doxorubicin | 0.27 | |||
| NPB | 6.53 | |||
| CCDC No | 2027110 |
|---|---|
| Empirical formula | C23 H23 Cl N2 O2 S |
| Formula weight | 426.94 |
| Temperature | 293 K |
| Wavelength | 0.71073Å |
| Reflns. for cell determination | 1802 |
| θrange for above | 3.643° to 58.989° |
| Crystal system | P-1 |
| Space group | Triclinic |
| Cell dimensions | a = 11.4518(8)Å, b = 13.5450(10)Å, c = 14.7351(7)Å α = 101.790(4)°, β = 102.701(4)°, γ = 96.286(4)° |
| Volume | 2153.8(2) Å3 |
| Z | 4 |
| F000 | 896 |
| θ range for data collection | 2.215° to 25.827° |
| Index ranges | −14< = h< = 14; −16< = k< = 16; −18< = l< = 18 |
| Reflections collected | 43,360 |
| Independent reflections | 8277 |
| Refinement method | Full-matrix least-squares on F^2 |
| Data/restraints/parameters | 8277/36/538 |
| Goodness-of-fit on F2 | 1.015 |
| Final [I > 2σ(I)] | R1 = 0.0545, wR2 = 0.1249 |
| R indices (all data) | R1 = 0.1243, wR2 = 0.1639 |
| Largest diff. peak and hole | 0.333 and −0.536 eA°−3 |
| Entry | EHOMO | ELUMO | ∆ | η | χ | ω |
|---|---|---|---|---|---|---|
| 4a | −5.87 | −1.55 | 4.32 | 2.16 | 3.71 | 3.19 |
| 4b | −6.16 | −2.68 | 3.48 | 1.74 | 4.42 | 5.60 |
| 4c | −5.71 | −1.52 | 4.19 | 2.10 | 3.61 | 3.11 |
| 4r | −5.91 | −2.25 | 3.66 | 1.83 | 4.08 | 4.55 |
| 4d | −5.36 | −1.96 | 3.40 | 1.70 | 3.66 | 3.93 |
| 4e | −6.48 | −4.49 | 1.98 | 0.99 | 5.48 | 15.16 |
| 4f | −6.50 | −4.89 | 1.61 | 0.80 | 5.69 | 20.14 |
| 4g | −5.24 | −1.18 | 4.05 | 2.03 | 3.21 | 2.54 |
| 4h | −5.75 | −2.90 | 2.85 | 1.43 | 4.32 | 6.55 |
| 4i | −5.92 | −2.65 | 3.27 | 1.64 | 4.28 | 5.61 |
| 4j | −4.97 | −1.64 | 3.33 | 1.66 | 3.31 | 3.29 |
| 4k | −5.26 | −1.65 | 3.60 | 1.80 | 3.46 | 3.31 |
| 4l | −5.64 | −2.06 | 3.58 | 1.79 | 3.85 | 4.14 |
| 4m | −5.67 | −2.10 | 3.56 | 1.78 | 3.88 | 4.23 |
| 4n | −4.68 | −1.58 | 3.10 | 1.55 | 3.13 | 3.16 |
| 4o | −3.97 | −1.42 | 2.55 | 1.27 | 2.69 | 2.84 |
| 4p | −5.51 | −2.02 | 3.49 | 1.74 | 3.77 | 4.07 |
| 4q | −5.69 | −2.11 | 3.57 | 1.79 | 3.90 | 4.26 |
| Query | Liver Toxicity | Metabolism | ||||||
|---|---|---|---|---|---|---|---|---|
| CYP Inhibitors for | ||||||||
| DILI | CT | HLM | 1A2 | 3A4 | 2D6 | 2C9 | 2C19 | |
| 4e | Na | Yb | N | N | N | N | N | N |
| 4f | N | Y | Y | N | N | N | N | N |
| 4i | N | N | Y | N | N | N | N | N |
| NPB | Y | N | Y | N | N | N | N | N |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Girimanchanaika, S.S.; Dukanya, D.; Swamynayaka, A.; Govindachar, D.M.; Madegowda, M.; Periyasamy, G.; Rangappa, K.S.; Pandey, V.; Lobie, P.E.; Basappa, B. Investigation of NPB Analogs That Target Phosphorylation of BAD-Ser99 in Human Mammary Carcinoma Cells. Int. J. Mol. Sci. 2021, 22, 11002. https://doi.org/10.3390/ijms222011002
Girimanchanaika SS, Dukanya D, Swamynayaka A, Govindachar DM, Madegowda M, Periyasamy G, Rangappa KS, Pandey V, Lobie PE, Basappa B. Investigation of NPB Analogs That Target Phosphorylation of BAD-Ser99 in Human Mammary Carcinoma Cells. International Journal of Molecular Sciences. 2021; 22(20):11002. https://doi.org/10.3390/ijms222011002
Chicago/Turabian StyleGirimanchanaika, Swamy Savvemala, Dukanya Dukanya, Ananda Swamynayaka, Divya Maldepalli Govindachar, Mahendra Madegowda, Ganga Periyasamy, Kanchugarakoppal Subbegowda Rangappa, Vijay Pandey, Peter E. Lobie, and Basappa Basappa. 2021. "Investigation of NPB Analogs That Target Phosphorylation of BAD-Ser99 in Human Mammary Carcinoma Cells" International Journal of Molecular Sciences 22, no. 20: 11002. https://doi.org/10.3390/ijms222011002
APA StyleGirimanchanaika, S. S., Dukanya, D., Swamynayaka, A., Govindachar, D. M., Madegowda, M., Periyasamy, G., Rangappa, K. S., Pandey, V., Lobie, P. E., & Basappa, B. (2021). Investigation of NPB Analogs That Target Phosphorylation of BAD-Ser99 in Human Mammary Carcinoma Cells. International Journal of Molecular Sciences, 22(20), 11002. https://doi.org/10.3390/ijms222011002








