CELF Family Proteins in Cancer: Highlights on the RNA-Binding Protein/Noncoding RNA Regulatory Axis
Abstract
:1. Introduction
2. General Characteristics of the CELF Protein Family
3. CELF Targets in Cancer
3.1. CELF1
3.2. CELF2
3.3. Other CELF Members
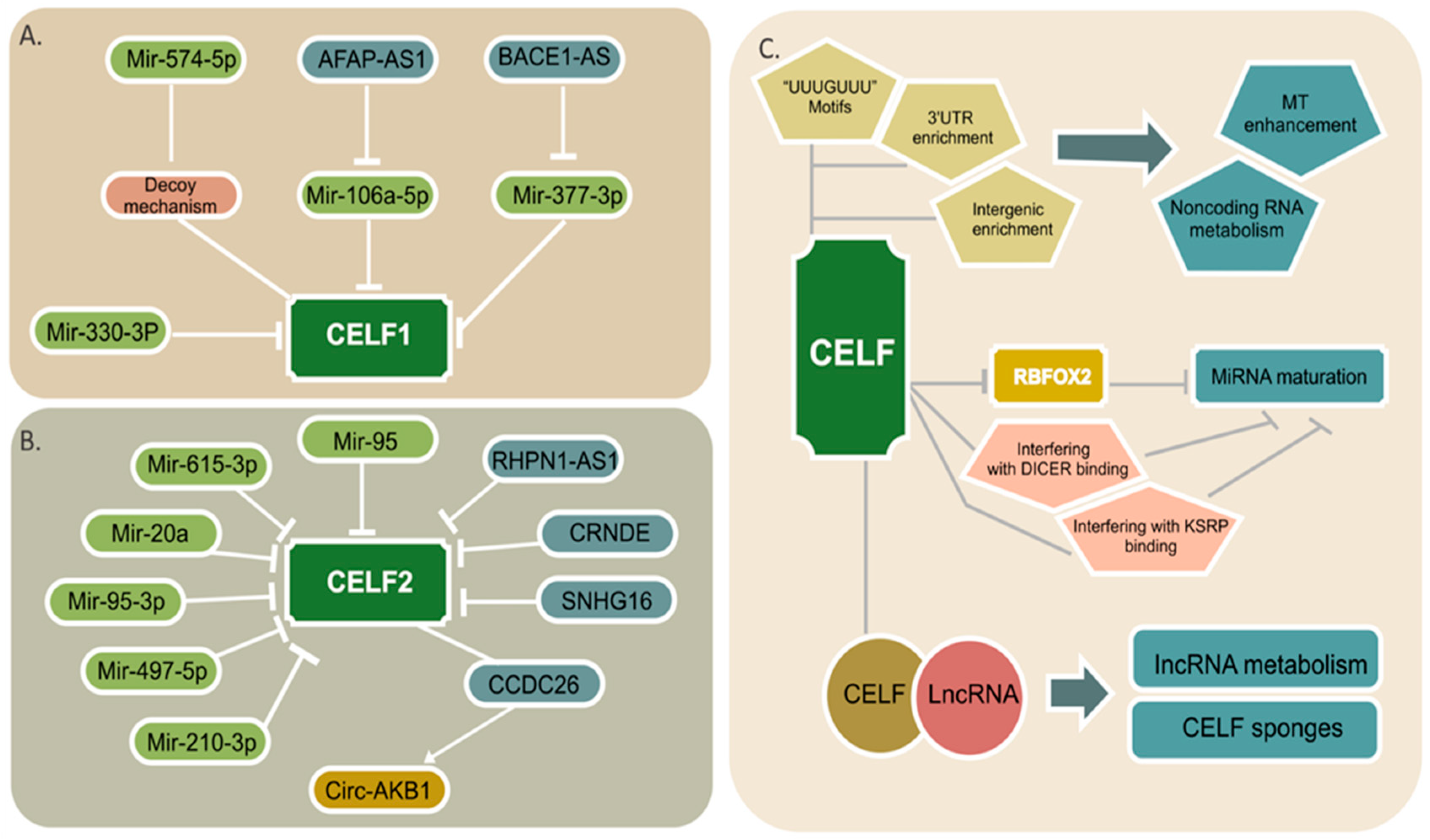
4. CELF Regulation by Noncoding RNAs
5. Noncoding RNA Regulation by CELF
6. Pharmacological Insights of CELF/Noncoding RNA Axis
7. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sternburg, E.L.; Karginov, F.V. Global Approaches in Studying RNA-Binding Protein Interaction Networks. Trends Biochem. Sci. 2020, 45, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, S.; Chang, H.R.; Kim, D.; Park, J.; Son, N.; Park, J.; Yoon, M.; Chae, G.; Kim, Y.-K.; et al. The regulatory impact of RNA-binding proteins on microRNA targeting. Nat. Commun. 2021, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sebestyén, E.; Singh, B.; Miñana, B.; Pagès, A.; Mateo, F.; Pujana, M.A.; Valcárcel, J.; Eyras, E. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 2016, 26, 732–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kechavarzi, B.; Janga, S.C. Dissecting the expression landscape of RNA-binding proteins in human cancers. Genome Biol. 2014, 15, R14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasgupta, T.; Ladd, A.N. The importance of CELF control: Molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip. Rev. RNA 2011, 3, 104–121. [Google Scholar] [CrossRef] [Green Version]
- Louis, I.V.-S.; Dickson, A.M.; Bohjanen, P.; Wilusz, C.J. CELFish ways to modulate mRNA decay. Biochim. Biophys. Acta Bioenerg. 2013, 1829, 695–707. [Google Scholar] [CrossRef]
- Ajith, S.; Gazzara, M.R.; Cole, B.S.; Shankarling, G.; Martinez, N.; Mallory, M.J.; Lynch, K.W. Position-dependent activity of CELF2 in the regulation of splicing and implications for signal-responsive regulation in T cells. RNA Biol. 2016, 13, 569–581. [Google Scholar] [CrossRef]
- Xia, H.; Chen, D.; Wu, Q.; Wu, G.; Zhou, Y.; Zhang, Y.; Zhang, L. CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in HeLa cells. Biochim. Biophys. Acta Bioenerg. 2017, 1860, 911–921. [Google Scholar] [CrossRef]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2020, 21, 22–36. [Google Scholar] [CrossRef]
- Cursons, J.; Pillman, K.A.; Scheer, K.G.; Gregory, P.A.; Foroutan, M.; Hediyeh-Zadeh, S.; Toubia, J.; Crampin, E.J.; Goodall, G.J.; Bracken, C.P.; et al. Combinatorial Targeting by MicroRNAs Co-ordinates Post-transcriptional Control of EMT. Cell Syst. 2018, 7, 77–91.e7. [Google Scholar] [CrossRef]
- Maloney, S.E.; Khangura, E.; Dougherty, J.D. The RNA-binding protein Celf6 is highly expressed in diencephalic nuclei and neuromodulatory cell populations of the mouse brain. Brain Struct. Funct. 2015, 221, 1809–1831. [Google Scholar] [CrossRef] [Green Version]
- Rieger, M.A.; King, D.M.; Crosby, H.; Liu, Y.; Cohen, B.A.; Dougherty, J.D. CLIP and Massively Parallel Functional Analysis of CELF6 Reveal a Role in Destabilizing Synaptic Gene mRNAs through Interaction with 3′ UTR Elements. Cell Rep. 2020, 33, 108531. [Google Scholar] [CrossRef]
- Ladd, A.; Cooper, T.A. Multiple domains control the subcellular localization and activity of ETR-3, a regulator of nuclear and cytoplasmic RNA processing events. J. Cell Sci. 2004, 117, 3519–3529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itai, T.; Hamanaka, K.; Sasaki, K.; Wagner, M.; Kotzaeridou, U.; Brösse, I.; Ries, M.; Kobayashi, Y.; Tohyama, J.; Kato, M.; et al. De novo variants in CELF2 that disrupt the nuclear localization signal cause developmental and epileptic encephalopathy. Hum. Mutat. 2020, 42, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Salisbury, E.; Sakai, K.; Schoser, B.; Huichalaf, C.; Schneider-Gold, C.; Nguyen, H.; Wang, G.-L.; Albrecht, J.H.; Timchenko, L.T. Ectopic expression of cyclin D3 corrects differentiation of DM1 myoblasts through activation of RNA CUG-binding protein, CUGBP. Exp. Cell Res. 2008, 314, 2266–2278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beisang, D.; Rattenbacher, B.; Louis, I.A.V.-S.; Bohjanen, P.R. Regulation of CUG-binding Protein 1 (CUGBP1) Binding to Target Transcripts upon T Cell Activation. J. Biol. Chem. 2012, 287, 950–960. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef] [Green Version]
- Gnad, F.; Gunawardena, J.; Mann, M. PHOSIDA 2011: The posttranslational modification database. Nucleic Acids Res. 2010, 39, D253–D260. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.; Maegawa, S.; Nishibu, T.; Sugiyama, T.; Yasuda, K.; Inoue, K. Vegetal localization of the maternal mRNA encoding an EDEN-BP/Bruno-like protein in zebrafish. Mech. Dev. 2000, 93, 205–209. [Google Scholar] [CrossRef]
- Choi, D.-K.; Yoo, K.-W.; Hong, S.-K.; Rhee, M.; Sakaki, Y.; Kim, C.-H. Isolation and expression of Napor/CUG-BP2 in embryo development. Biochem. Biophys. Res. Commun. 2003, 305, 448–454. [Google Scholar] [CrossRef]
- Ramalingam, S.; Natarajan, G.; Schafer, C.; Subramaniam, D.; May, R.; Ramachandran, I.; Queimado, L.; Houchen, C.W.; Anant, S. Novel intestinal splice variants of RNA-binding protein CUGBP2: Isoform-specific effects on mitotic catastrophe. Am. J. Physiol. Liver Physiol. 2008, 294, G971–G981. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.; Takeuchi, M.; Sugiyama, A.; Alam, A.K.; Vu, L.T.; Sekiyama, Y.; Dam, H.C.; Ohki, S.-Y.; Tsukahara, T. Alternative splicing produces structural and functional changes in CUGBP. BMC Biochem. 2012, 13, 6. [Google Scholar] [CrossRef] [Green Version]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2014, 43, D512–D520. [Google Scholar] [CrossRef] [Green Version]
- Fujimura, K.; Kano, F.; Murata, M. Dual localization of the RNA binding protein CUGBP-1 to stress granule and perinucleolar compartment. Exp. Cell Res. 2008, 314, 543–553. [Google Scholar] [CrossRef]
- Wagnon, J.L.; Briese, M.; Sun, W.; Mahaffey, C.L.; Curk, T.; Rot, G.; Ule, J.; Frankel, W.N. CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function. PLoS Genet. 2012, 8, e1003067. [Google Scholar] [CrossRef] [Green Version]
- Cox, D.C.; Guan, X.; Xia, Z.; Cooper, T.A. Increased nuclear but not cytoplasmic activities of CELF1 protein leads to muscle wasting. Hum. Mol. Genet. 2020, 29, 1729–1744. [Google Scholar] [CrossRef] [PubMed]
- Mallory, M.J.; Jackson, J.; Weber, B.; Chi, A.; Heyd, F.; Lynch, K.W. Signal- and Development-Dependent Alternative Splicing of LEF1 in T Cells Is Controlled by CELF. Mol. Cell. Biol. 2011, 31, 2184–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikonova, E.; Kao, S.-Y.; Ravichandran, K.; Wittner, A.; Spletter, M.L. Conserved functions of RNA-binding proteins in muscle. Int. J. Biochem. Cell Biol. 2019, 110, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Blech-Hermoni, Y.; Dasgupta, T.; Coram, R.J.; Ladd, A.N. Identification of Targets of CUG-BP, Elav-Like Family Member 1 (CELF1) Regulation in Embryonic Heart Muscle. PLoS ONE 2016, 11, e0149061. [Google Scholar] [CrossRef] [PubMed]
- Emmerich, A.C.; Wellstein, J.; Ossipova, E.; Baumann, I.; Lengqvist, J.; Kultima, K.; Jakobsson, P.-J.; Steinhilber, D.; Saul, M.J. Proteomics-Based Characterization of miR-574-5p Decoy to CUGBP1 Suggests Specificity for mPGES-1 Regulation in Human Lung Cancer Cells. Front. Pharmacol. 2020, 11, 196. [Google Scholar] [CrossRef] [Green Version]
- Gazzara, M.R.; Mallory, M.J.; Roytenberg, R.; Lindberg, J.P.; Jha, A.; Lynch, K.W.; Barash, Y. Ancient antagonism between CELF and RBFOX families tunes mRNA splicing outcomes. Genome Res. 2017, 27, 1360–1370. [Google Scholar] [CrossRef] [Green Version]
- Chatrikhi, R.; Mallory, M.J.; Gazzara, M.R.; Agosto, L.M.; Zhu, W.S.; Litterman, A.J.; Ansel, K.M.; Lynch, K.W. RNA Binding Protein CELF2 Regulates Signal-Induced Alternative Polyadenylation by Competing with Enhancers of the Polyadenylation Machinery. Cell Rep. 2019, 28, 2795–2806.e3. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Eggerman, T.L.; Patterson, A.P. ApoB mRNA editing is mediated by a coordinated modulation of multiple apoB mRNA editing enzyme components. Am. J. Physiol. Liver Physiol. 2007, 292, G53–G65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallory, M.J.; McClory, S.P.; Chatrikhi, R.; Gazzara, M.R.; Ontiveros, R.J.; Lynch, K.W. Reciprocal regulation of hnRNP C and CELF2 through translation and transcription tunes splicing activity in T cells. Nucleic Acids Res. 2020, 48, 5710–5719. [Google Scholar] [CrossRef] [PubMed]
- Moraes, K.C.; Wilusz, C.J.; Wilusz, J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA 2006, 12, 1084–1091. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.S.J.; Wu, M.K.; Zhu, T.H.; Zhao, H.; Cheung, S.T.; Chamberlain, T.; Mui, A.L.-F. Interleukin-10 control of pre-miR155 maturation involves CELF. PLoS ONE 2020, 15, e0231639. [Google Scholar] [CrossRef]
- Dejene, E.A.; Li, Y.; Showkatian, Z.; Ling, H.; Seto, E. Regulation of poly(a)-specific ribonuclease activity by reversible lysine acetylation. J. Biol. Chem. 2020, 295, 10255–10270. [Google Scholar] [CrossRef]
- Chaudhury, A.; Cheema, S.; Fachini, J.M.; Kongchan, N.; Lu, G.; Simon, L.M.; Wang, T.; Mao, S.; Rosen, D.G.; Ittmann, M.M.; et al. CELF1 is a central node in post-transcriptional regulatory programmes underlying EMT. Nat. Commun. 2016, 7, 13362. [Google Scholar] [CrossRef]
- Popovitchenko, T.; Park, Y.; Page, N.F.; Luo, X.; Krsnik, Z.; Liu, Y.; Salamon, I.; Stephenson, J.D.; Kraushar, M.L.; Volk, N.L.; et al. Translational derepression of Elavl4 isoforms at their alternative 5′ UTRs determines neuronal development. Nat. Commun. 2020, 11, 1674. [Google Scholar] [CrossRef]
- Gareau, C.; Fournier, M.-J.; Filion, C.; Coudert, L.; Martel, D.; Labelle, Y.; Mazroui, R. p21WAF1/CIP1 Upregulation through the Stress Granule-Associated Protein CUGBP1 Confers Resistance to Bortezomib-Mediated Apoptosis. PLoS ONE 2011, 6, e20254. [Google Scholar] [CrossRef]
- Subramaniam, D.; Natarajan, G.; Ramalingam, S.; Ramachandran, I.; May, R.; Queimado, L.; Houchen, C.W.; Anant, S. Translation inhibition during cell cycle arrest and apoptosis: Mcl-1 is a novel target for RNA binding protein CUGBP. Am. J. Physiol. Liver Physiol. 2008, 294, G1025–G1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, J.T.; Stover, P.J. Mechanism of the Internal Ribosome Entry Site-mediated Translation of Serine Hydroxymethyltransferase 1. J. Biol. Chem. 2009, 284, 31085–31096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treiber, T.; Treiber, N.; Plessmann, U.; Harlander, S.; Daiß, J.-L.; Eichner, N.; Lehmann, G.; Schall, K.; Urlaub, H.; Meister, G. A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol. Cell 2017, 66, 270–284.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amen, T.; Guihur, A.; Zelent, C.; Ursache, R.; Wilting, J.; Kaganovich, D. Resveratrol and related stilbene-derivatives induce Stress Granules with distinct clearance kinetics. Mol. Biol. Cell 2021. mbc.E21–02. [Google Scholar] [CrossRef]
- Moraes, K.C.M.; Monteiro, C.J.; Pacheco-Soares, C. A novel function for CUGBP2 in controlling the pro-inflammatory stimulus in H9c2 cells: Subcellular trafficking of messenger molecules. Cell Biol. Int. 2013, 37, 1129–1138. [Google Scholar] [CrossRef]
- Qian, Y.; Lv, S.; Dong, X. Detection and Significance of PNC in Breast Cancer Tissues. Clin. Lab. 2021, 67. [Google Scholar] [CrossRef]
- Vilimas, T.; Wang, A.Q.; Patnaik, S.; Hughes, E.A.; Singleton, M.D.; Knotts, Z.; Li, D.; Frankowski, K.; Schlomer, J.J.; Guerin, T.M.; et al. Pharmacokinetic evaluation of the PNC disassembler metarrestin in wild-type and Pdx1-Cre;LSL-KrasG12D/+;Tp53R172H/+ (KPC) mice, a genetically engineered model of pancreatic cancer. Cancer Chemother. Pharmacol. 2018, 82, 1067–1080. [Google Scholar] [CrossRef] [Green Version]
- Pollock, C.; Daily, K.; Nguyen, V.T.; Wang, C.; Lewandowska, M.A.; Bensaude, O.; Huang, S. Characterization of MRP RNA–protein interactions within the perinucleolar compartment. Mol. Biol. Cell 2011, 22, 858–866. [Google Scholar] [CrossRef]
- Pereira, B.; Billaud, M.; Almeida, R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 2017, 3, 506–528. [Google Scholar] [CrossRef]
- Schuschel, K.; Helwig, M.; Hüttelmaier, S.; Heckl, D.; Klusmann, J.-H.; Hoell, J.I. RNA-Binding Proteins in Acute Leukemias. Int. J. Mol. Sci. 2020, 21, 3409. [Google Scholar] [CrossRef]
- Philips, A.V.; Timchenko, L.T.; Cooper, T.A. Disruption of Splicing Regulated by a CUG-Binding Protein in Myotonic Dystrophy. Science 1998, 280, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Timchenko, L.T.; Miller, J.W.; Timchenko, N.A.; Devore, D.R.; Datar, K.V.; Lin, L.; Roberts, R.; Caskey, C.T.; Swanson, M.S. Identification of a (CUG)n Triplet Repeat RNA-Binding Protein and Its Expression in Myotonic Dystrophy. Nucleic Acids Res. 1996, 24, 4407–4414. [Google Scholar] [CrossRef] [Green Version]
- Chau, A.; Kalsotra, A. Developmental insights into the pathology of and therapeutic strategies for DM1: Back to the basics. Dev. Dyn. 2015, 244, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Shulman, J.M.; Imboywa, S.; Giagtzoglou, N.; Powers, M.P.; Hu, Y.; Devenport, D.; Chipendo, P.; Chibnik, L.B.; Diamond, A.; Perrimon, N.; et al. Functional screening in Drosophila identifies Alzheimer’s disease susceptibility genes and implicates Tau-mediated mechanisms. Hum. Mol. Genet. 2013, 23, 870–877. [Google Scholar] [CrossRef] [Green Version]
- Lambert, J.C.; Ibrahim-Verbaas, C.A.; Harold, D.; Naj, A.C.; Sims, R.; Bellenguez, C.; Jun, G.; DeStefano, A.L.; Bis, J.C.; Beecham, G.W.; et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef] [Green Version]
- Masuda, A.; Andersen, H.S.; Doktor, T.; Okamoto, T.; Ito, M.; Andresen, B.S.; Ohno, K. CUGBP1 and MBNL1 preferentially bind to 3′ UTRs and facilitate mRNA decay. Sci. Rep. 2012, 2, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Tonquèze, O.; Gschloessl, B.; Legagneux, V.; Paillard, L.; Audic, Y. Identification of CELF1 RNA targets by CLIP-seq in human HeLa cells. Genom. Data 2016, 8, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Cifdaloz, M.; Osterloh, L.; Graña, O.; Riveiro-Falkenbach, E.; Ximénez-Embún, P.; Muñoz, J.; Tejedo, C.; Calvo, T.G.; Karras, P.; Olmeda, D.; et al. Systems analysis identifies melanoma-enriched pro-oncogenic networks controlled by the RNA binding protein CELF. Nat. Commun. 2017, 8, 2249. [Google Scholar] [CrossRef] [Green Version]
- House, R.P.; Talwar, S.; Hazard, E.S.; Hill, E.G.; Palanisamy, V. RNA-binding protein CELF1 promotes tumor growth and alters gene expression in oral squamous cell carcinoma. Oncotarget 2015, 6, 43620–43634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talwar, S.; Balasubramanian, S.; Sundaramurthy, S.; House, R.; Wilusz, C.J.; Kuppuswamy, D.; D’Silva, N.; Gillespie, M.B.; Hill, E.G.; Palanisamy, V. Overexpression of RNA-binding protein CELF1 prevents apoptosis and destabilizes pro-apoptotic mRNAs in oral cancer cells. RNA Biol. 2013, 10, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Buyuk, B.; Jin, S.; Ye, K. Epithelial-to-Mesenchymal Transition Signaling Pathways Responsible for Breast Cancer Metastasis. Cell. Mol. Bioeng. 2021, 1–13. [Google Scholar] [CrossRef]
- Jiao, W.; Zhao, J.; Wang, M.; Wang, Y.; Luo, Y.; Zhao, Y.; Tang, D.; Shen, Y. CUG-binding protein 1 (CUGBP1) expression and prognosis of non-small cell lung cancer. Clin. Transl. Oncol. 2013, 15, 789–795. [Google Scholar] [CrossRef]
- Wu, L.-N.; Xue, Y.-J.; Zhang, L.-J.; Ma, X.-M.; Chen, J.-F. Si-RNA mediated knockdown of CELF1 gene suppressed the proliferation of human lung cancer cells. Cancer Cell Int. 2013, 13, 115. [Google Scholar] [CrossRef] [Green Version]
- Gao, C.; Yu, Z.; Liu, S.; Xin, H.; Li, X. Overexpression of CUGBP1 is associated with the progression of non-small cell lung cancer. Tumor Biol. 2015, 36, 4583–4589. [Google Scholar] [CrossRef] [PubMed]
- Huichalaf, C.; Sakai, K.; Jin, B.; Jones, K.; Wang, G.; Schoser, B.; Schneider-Gold, C.; Sarkar, P.; Pereira-Smith, O.M.; Timchenko, N.; et al. Expansion of CUG RNA repeats causes stress and inhibition of translation in myotonic dystrophy 1 (DM1) cells. FASEB J. 2010, 24, 3706–3719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, M.; Bode, A.M.; Dong, Z.; Lee, M.-H. AKT as a Therapeutic Target for Cancer. Cancer Res. 2019, 79, 1019–1031. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Leone, G.W.; Wang, H. Cyclin D-CDK4/6 functions in cancer. Adv. Cancer Res. 2020, 148, 147–169. [Google Scholar] [CrossRef] [PubMed]
- Bohjanen, P.R.; Moua, M.L.; Guo, L.; Taye, A.; Louis, I.V.-S. Altered CELF1 binding to target transcripts in malignant T cells. RNA 2015, 21, 1757–1769. [Google Scholar] [CrossRef] [Green Version]
- Valanejad, L.; Cast, A.; Wright, M.; Bissig, K.-D.; Karns, R.; Weirauch, M.T.; Timchenko, N. PARP1 activation increases expression of modified tumor suppressors and pathways underlying development of aggressive hepatoblastoma. Commun. Biol. 2018, 1, 67. [Google Scholar] [CrossRef]
- Lewis, K.; Valanejad, L.; Cast, A.; Wright, M.; Wei, C.; Iakova, P.; Stock, L.; Karns, R.; Timchenko, L.; Timchenko, N. RNA Binding Protein CUGBP1 Inhibits Liver Cancer in a Phosphorylation-Dependent Manner. Mol. Cell. Biol. 2017, 37. [Google Scholar] [CrossRef] [Green Version]
- Xia, L.; Sun, C.; Li, Q.; Feng, F.; Qiao, E.; Jiang, L.; Wu, B.; Ge, M. CELF1 is Up-Regulated in Glioma and Promotes Glioma Cell Proliferation by Suppression of CDKN1B. Int. J. Biol. Sci. 2015, 11, 1314–1324. [Google Scholar] [CrossRef]
- Huang, G.; Song, C.; Wang, N.; Qin, T.; Sui, S.; Obr, A.; Zeng, L.; Wood, T.L.; Leroith, D.; Li, M.; et al. RNA-binding protein CUGBP1 controls the differential INSR splicing in molecular subtypes of breast cancer cells and affects cell aggressiveness. Carcinogenesis 2019, 41, 1294–1305. [Google Scholar] [CrossRef]
- Wang, H.; Huang, R.; Guo, W.; Qin, X.; Yang, Z.; Yuan, Z.; Wei, Y.; Mo, C.; Zeng, Z.; Luo, J.; et al. RNA-binding protein CELF1 enhances cell migration, invasion, and chemoresistance by targeting ETS2 in colorectal cancer. Clin. Sci. 2020, 134, 1973–1990. [Google Scholar] [CrossRef]
- Tang, M.; Yang, M.; He, K.; Li, R.; Chen, X.; Wang, Y.; Zhang, X.; Qiu, T. Glycyrrhetinic acid remodels the tumor microenvironment and synergizes with doxorubicin for breast cancer treatment in a murine model. Nanotechnology 2021, 32, 185702. [Google Scholar] [CrossRef]
- Wang, Z.-L.; Li, B.; Luo, Y.-X.; Lin, Q.; Liu, S.-R.; Zhang, X.-Q.; Zhou, H.; Yang, J.-H.; Qu, L.-H. Comprehensive Genomic Characterization of RNA-Binding Proteins across Human Cancers. Cell Rep. 2018, 22, 286–298. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Liu, Z.; Liu, L.; Guo, C.; Jiao, D.; Li, L.; Zhao, J.; Han, X.; Sun, Y. CELF2 is a candidate prognostic and immunotherapy biomarker in triple-negative breast cancer and lung squamous cell carcinoma: A pan-cancer analysis. J. Cell. Mol. Med. 2021, 25, 7559–7574. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Xie, X. RNA-binding protein CELF2 inhibits breast cancer cell invasion and angiogenesis by downregulating NFATc. Exp. Ther. Med. 2021, 22, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Piqué, L.; de Paz, A.M.; Piñeyro, D.; Martínez-Cardús, A.; de Moura, M.C.; Llinàs-Arias, P.; Setien, F.; Gomez-Miragaya, J.; Gonzalez-Suarez, E.; Sigurdsson, S.; et al. Epigenetic inactivation of the splicing RNA-binding protein CELF2 in human breast cancer. Oncogene 2019, 38, 7106–7112. [Google Scholar] [CrossRef] [PubMed]
- Yeung, Y.T.; Fan, S.; Lu, B.; Yin, S.; Yang, S.; Nie, W.; Wang, M.; Zhou, L.; Li, T.; Li, X.; et al. CELF2 suppresses non-small cell lung carcinoma growth by inhibiting the PREX2-PTEN interaction. Carcinogenesis 2019, 41, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-S.; Tu, S.-J.; Chiang, H.-S.; Yen, J.-C.; Lee, Y.-T.; Fang, H.-Y.; Chang, J.-G. Genome-Wide Analysis of Prognostic Alternative Splicing Signature and Splicing Factors in Lung Adenocarcinoma. Genes 2020, 11, 1300. [Google Scholar] [CrossRef]
- Guo, Q.; Wu, Y.; Guo, X.; Cao, L.; Xu, F.; Zhao, H.; Zhu, J.; Wen, H.; Ju, X.; Wu, X. The RNA-Binding Protein CELF2 Inhibits Ovarian Cancer Progression by Stabilizing FAM198B. Mol. Ther.-Nucleic Acids 2020, 23, 169–184. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, X.; Zhang, X.; Chen, R. Identification of important long non-coding RNAs and highly recurrent aberrant alternative splicing events in hepatocellular carcinoma through integrative analysis of multiple RNA-Seq datasets. Mol. Genet. Genom. 2015, 291, 1035–1051. [Google Scholar] [CrossRef]
- Wu, J.-Z.; Jiang, N.; Lin, J.-M.; Liu, X. STYXL1 promotes malignant progression of hepatocellular carcinoma via downregulating CELF2 through the PI3K/Akt pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2977–2985. [Google Scholar]
- Li, Y.; Ren, Z.; Peng, Y.; Li, K.; Wang, X.; Huang, G.; Qi, S.; Liu, Y. Classification of glioma based on prognostic alternative splicing. BMC Med Genom. 2019, 12, 165. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Zhang, J.; Jiang, L.; Wang, Y.; Ren, X.; Cheng, B.; Xia, J. Comprehensive Analysis of Prognostic Alternative Splicing Signatures in Oral Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 1740. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hu, Z.; Lan, Y.; Long, J.; Wang, Y.; Chen, X.; Xu, X.; Zeng, Z.; Ouyang, Y. Prognostic significance of survival-associated alternative splicing events in gastric cancer. Aging 2020, 12, 21923–21941. [Google Scholar] [CrossRef] [PubMed]
- Schultz, A.-S.; Preussner, M.; Bunse, M.; Karni, R.; Heyd, F. Activation-Dependent TRAF3 Exon 8 Alternative Splicing Is Controlled by CELF2 and hnRNP C Binding to an Upstream Intronic Element. Mol. Cell. Biol. 2017, 37. [Google Scholar] [CrossRef] [Green Version]
- Mallory, M.J.; Allon, S.J.; Qiu, J.; Gazzara, M.R.; Tapescu, I.; Martinez, N.M.; Fu, X.-D.; Lynch, K.W. Induced transcription and stability of CELF2 mRNA drives widespread alternative splicing during T-cell signaling. Proc. Natl. Acad. Sci. USA 2015, 112, E2139–E2148. [Google Scholar] [CrossRef] [Green Version]
- Martinez, N.; Agosto, L.; Qiu, J.; Mallory, M.J.; Gazzara, M.R.; Barash, Y.; Fu, X.-D.; Lynch, K.W. Widespread JNK-dependent alternative splicing induces a positive feedback loop through CELF2-mediated regulation of MKK7 during T-cell activation. Genes Dev. 2015, 29, 2054–2066. [Google Scholar] [CrossRef] [Green Version]
- Van Der Leun, A.M.; Thommen, D.S.; Schumacher, T.N. CD8+ T cell states in human cancer: Insights from single-cell analysis. Nat. Rev. Cancer 2020, 20, 218–232. [Google Scholar] [CrossRef]
- Subramaniam, D.; Ramalingam, S.; Linehan, D.C.; Dieckgraefe, B.K.; Postier, R.G.; Houchen, C.W.; Jensen, R.A.; Anant, S. RNA Binding Protein CUGBP2/CELF2 Mediates Curcumin-Induced Mitotic Catastrophe of Pancreatic Cancer Cells. PLoS ONE 2011, 6, e16958. [Google Scholar] [CrossRef] [Green Version]
- Jakstaite, A.; Maziukiene, A.; Silkuniene, G.; Kmieliute, K.; Dauksa, A.; Paskauskas, S.; Gulbinas, A.; Dambrauskas, Z. Upregulation of cugbp2 increases response of pancreatic cancer cells to chemotherapy. Langenbeck’s Arch. Surg. 2015, 401, 99–111. [Google Scholar] [CrossRef]
- New, J.; Subramaniam, D.; Ramalingam, S.; Enders, J.; Sayed, A.A.A.; Ponnurangam, S.; Standing, D.; Ramamoorthy, P.; O’Neil, M.; Dixon, D.A.; et al. Pleotropic role of RNA binding protein CELF2 in autophagy induction. Mol. Carcinog. 2019, 58, 1400–1409. [Google Scholar] [CrossRef]
- Xu, J.; Fang, Y.; Qin, J.; Chen, X.; Liang, X.; Xie, X.; Lu, W. A transcriptomic landscape of human papillomavirus 16 E6-regulated gene expression and splicing events. FEBS Lett. 2016, 590, 4594–4605. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Guo, R. Integrative analysis of significant RNA-binding proteins in colorectal cancer metastasis. J. Cell. Biochem. 2018, 119, 9730–9741. [Google Scholar] [CrossRef]
- Wang, X.; Sun, C.-L.; Lombraña, A.Q.; Singh, P.; Landier, W.; Hageman, L.; Mather, M.; Rotter, J.I.; Taylor, K.D.; Chen, Y.-D.I.; et al. CELF4 Variant and Anthracycline-Related Cardiomyopathy: A Children’s Oncology Group Genome-Wide Association Study. J. Clin. Oncol. 2016, 34, 863–870. [Google Scholar] [CrossRef] [Green Version]
- Linschoten, M.; Teske, A.J.; Cramer, M.J.; Van Der Wall, E.; Asselbergs, F.W. Chemotherapy-Related Cardiac Dysfunction. Circ. Genom. Precis. Med. 2018, 11, e001753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, R.L.; Su, P.-H.; Liao, Y.-P.; Wu, T.-I.; Hsu, Y.-T.; Lin, W.-Y.; Wang, H.-C.; Weng, Y.-C.; Ou, Y.-C.; Huang, T.H.-M.; et al. Integrated Epigenomics Analysis Reveals a DNA Methylation Panel for Endometrial Cancer Detection Using Cervical Scrapings. Clin. Cancer Res. 2016, 23, 263–272. [Google Scholar] [CrossRef] [Green Version]
- Teerlink, C.C.; Stevens, J.; Hernandez, R.; Facelli, J.C.; Cannon-Albright, L.A. An intronic variant in the CELF4 gene is associated with risk for colorectal cancer. Cancer Epidemiol. 2021, 72, 101941. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Liu, L.; Shi, Y.; Guo, F.; Wang, H.; Zhao, X.; Zhong, D.; Li, G. Integrated analysis of RNA-binding proteins in human colorectal cancer. World J. Surg. Oncol. 2020, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Yuan, C.; Liu, X. A New RBPs-Related Signature Predicts the Prognosis of Colon Adenocarcinoma Patients. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef]
- Miao, Y.; Zhang, H.; Su, B.; Wang, J.; Quan, W.; Li, Q.; Mi, D. Construction and validation of an RNA-binding protein-associated prognostic model for colorectal cancer. PeerJ 2021, 9, e11219. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Hui, W.; Halike, H.; Gao, F. RNA Binding Protein-Based Model for Prognostic Prediction of Colorectal Cancer. Technol. Cancer Res. Treat. 2021, 20. [Google Scholar] [CrossRef] [PubMed]
- Ladd, A.N.; Charlet, N.; Cooper, T.A. The CELF Family of RNA Binding Proteins Is Implicated in Cell-Specific and Developmentally Regulated Alternative Splicing. Mol. Cell. Biol. 2001, 21, 1285–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loria, P.M.; Duke, A.; Rand, J.B.; Hobert, O. Two Neuronal, Nuclear-Localized RNA Binding Proteins Involved in Synaptic Transmission. Curr. Biol. 2003, 13, 1317–1323. [Google Scholar] [CrossRef] [Green Version]
- Kline, R.A.; Kaifer, K.A.; Osman, E.; Carella, F.; Tiberi, A.; Ross, J.; Pennetta, G.; Lorson, C.L.; Murray, L.M. Comparison of independent screens on differentially vulnerable motor neurons reveals alpha-synuclein as a common modifier in motor neuron diseases. PLoS Genet. 2017, 13, e1006680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, F.; Lu, Z.; Wang, S.; Wu, S.; Wu, Y.; Sun, Z. Human cytomegalovirus UL141 protein interacts with CELF5 and affects viral DNA replication. Mol. Med. Rep. 2018, 17, 4657–4664. [Google Scholar] [CrossRef]
- Manicklal, S.; Emery, V.; Lazzarotto, T.; Boppana, S.B.; Gupta, R. The “Silent” Global Burden of Congenital Cytomegalovirus. Clin. Microbiol. Rev. 2013, 26, 86–102. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.; Li, Z.; Li, C.; Wang, G.; Yan, P.; Peng, L.; Wang, J.; Zhu, X.; Hu, P.; Zhang, J.; et al. Germ Cell-Specific Gene 1-Like Protein Regulated by Splicing Factor CUGBP Elav-Like Family Member 5 and Primary Bile Acid Biosynthesis are Prognostic in Glioblastoma Multiforme. Front. Genet. 2020, 10, 1380. [Google Scholar] [CrossRef]
- Ladd, A.; Nguyen, N.H.; Malhotra, K.; Cooper, T.A. CELF6, a Member of the CELF Family of RNA-binding Proteins, Regulates Muscle-specific Splicing Enhancer-dependent Alternative Splicing. J. Biol. Chem. 2004, 279, 17756–17764. [Google Scholar] [CrossRef] [Green Version]
- Sutandy, F.R.; Ebersberger, S.; Huang, L.; Busch, A.; Bach, M.; Kang, H.-S.; Fallmann, J.; Maticzka, D.; Backofen, R.; Stadler, P.F.; et al. In vitro iCLIP-based modeling uncovers how the splicing factor U2AF2 relies on regulation by cofactors. Genome Res. 2018, 28, 699–713. [Google Scholar] [CrossRef] [Green Version]
- Mackereth, C.; Madl, T.; Bonnal, S.; Simon, B.; Zanier, K.; Gasch, A.; Rybin, V.; Valcarcel, J.; Sattler, M. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature 2011, 475, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Berglund, J.A.; Abovich, N.; Rosbash, M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 1998, 12, 858–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maji, D.; Glasser, E.; Henderson, S.; Galardi, J.; Pulvino, M.J.; Jenkins, J.L.; Kielkopf, C.L. Representative cancer-associated U2AF2 mutations alter RNA interactions and splicing. J. Biol. Chem. 2020, 295, 17148–17157. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cheng, D.; Zhu, M.; Yu, H.; Pan, Z.; Liu, L.; Geng, Q.; Pan, H.; Yan, M.; Yao, M. OTUB2 stabilizes U2AF2 to promote the Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in non-small cell lung cancer. Theranostics 2019, 9, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhou, J.; Zhao, J.; Zhang, H.; Li, L.; Li, H.; Chen, L.; Hu, J.; Zheng, W.; Jing, Z. The U2AF2 /circRNA ARF1/miR-342–3p/ISL2 feedback loop regulates angiogenesis in glioma stem cells. J. Exp. Clin. Cancer Res. 2020, 39, 1–22. [Google Scholar] [CrossRef]
- Fang, J.; Li, Y.; Zhang, J.; Yan, M.; Li, J.; Bao, S.; Jin, T. Correlation between polymorphisms in microRNA-regulated genes and cervical cancer susceptibility in a Xinjiang Uygur population. Oncotarget 2017, 8, 31758–31764. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Zhang, Q.; Xia, L.; Shi, M.; Cai, J.; Zhang, H.; Li, J.; Lin, G.; Xie, W.; Zhang, Y.; et al. RNA-binding protein CELF6 is cell cycle regulated and controls cancer cell proliferation by stabilizing p21. Cell Death Dis. 2019, 10, 688–714. [Google Scholar] [CrossRef]
- Karimian, A.; Ahmadi, Y.; Yousefi, B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair 2016, 42, 63–71. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, L.; Pei, J.; Wang, Z.; Zhang, J.; Wang, B. CELF6 modulates triple-negative breast cancer progression by regulating the stability of FBP1 mRNA. Breast Cancer Res. Treat. 2020, 183, 71–82. [Google Scholar] [CrossRef]
- Dong, C.; Yuan, T.; Wu, Y.; Wang, Y.; Fan, T.W.; Miriyala, S.; Lin, Y.; Yao, J.; Shi, J.; Kang, T.; et al. Loss of FBP1 by Snail-Mediated Repression Provides Metabolic Advantages in Basal-like Breast Cancer. Cancer Cell 2013, 23, 316–331. [Google Scholar] [CrossRef] [Green Version]
- Sundar, I.K.; Mullapudi, N.; Yao, H.; Spivack, S.D.; Rahman, I. Lung cancer and its association with chronic obstructive pulmonary disease: Update on nexus of epigenetics. Curr. Opin. Pulm. Med. 2011, 17, 279–285. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Amitay, E.; Harrison, T.A.; Banbury, B.L.; Berndt, S.I.; Brenner, H.; Buchanan, D.D.; Campbell, P.T.; Cao, Y.; Chan, A.T.; et al. Association between Smoking and Molecular Subtypes of Colorectal Cancer. JNCI Cancer Spectr. 2021, 5. [Google Scholar] [CrossRef]
- Lee, J.-E.; Kim, H.-R.; Lee, M.-H.; Kim, N.-H.; Wang, K.-M.; Lee, S.-H.; Park, O.; Hong, E.-J.; Youn, J.-W.; Kim, Y.-Y. Smoking-Related DNA Methylation is Differentially Associated with Cadmium Concentration in Blood. Biochem. Genet. 2020, 58, 617–630. [Google Scholar] [CrossRef]
- Yang, I.V.; Schwartz, D.A. Epigenetic Control of Gene Expression in the Lung. Am. J. Respir. Crit. Care Med. 2011, 183, 1295–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2013, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuyama, H.; Suzuki, H.I. Systems and Synthetic microRNA Biology: From Biogenesis to Disease Pathogenesis. Int. J. Mol. Sci. 2019, 21, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, G.; Wu, X. Hsa_circ_0032463 acts as the tumor promoter in osteosarcoma by regulating the miR-330-3p/PNN axis. Int. J. Mol. Med. 2021, 47, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, G.; Li, T.; Wang, N.; Wu, J.; Zhi, H. MiR-330-3p functions as a tumor suppressor that regulates glioma cell proliferation and migration by targeting CELF. Arch. Med Sci. 2020, 16, 1166–1175. [Google Scholar] [CrossRef]
- Saul, M.J.; Baumann, I.; Bruno, A.; Emmerich, A.C.; Wellstein, J.; Ottinger, S.M.; Contursi, A.; Dovizio, M.; Donnini, S.; Tacconelli, S.; et al. miR-574-5p as RNA decoy for CUGBP1 stimulates human lung tumor growth by mPGES-1 induction. FASEB J. 2019, 33, 6933–6947. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, L.; Sun, Y.; Xue, Y.; Qu, J.; Pan, S.; Li, H.; Qu, H.; Wang, J.; Zhang, J. miR-615-3p promotes proliferation and migration and inhibits apoptosis through its potential target CELF2 in gastric cancer. Biomed. Pharmacother. 2018, 101, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wei, H.; Chen, Q. Long noncoding RNA HOTTIP promotes the metastatic potential of ovarian cancer through the regulation of the miR -615-3p/ SMARCE1 pathway. Kaohsiung J. Med Sci. 2020, 36, 973–982. [Google Scholar] [CrossRef]
- Liao, C.; Chen, W.; Wang, J. MicroRNA-20a Regulates Glioma Cell Proliferation, Invasion, and Apoptosis by Targeting CUGBP Elav-Like Family Member. World Neurosurg. 2019, 121, e519–e527. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Jiao, B.-H.; Fan, F.-S.; Lu, S.-K.; Song, J.; Guo, C.-Y.; Yang, J.-K.; Yang, L. Downregulation of miR-95-3p inhibits proliferation, and invasion promoting apoptosis of glioma cells by targeting CELF. Int. J. Oncol. 2015, 47, 1025–1033. [Google Scholar] [CrossRef]
- Xu, H.; Wang, F.; Wang, L. Suppression of miR-106a-5p expression inhibits tumorigenesis via increasing CELF-2 expression in spinal cord glioma. Oncol. Lett. 2021, 22, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Zhou, F.; Nie, J.; Wang, X.; Zhao, Q. Hypoxic colorectal cancer-secreted exosomes deliver miR-210-3p to normoxic tumor cells to elicit a protumoral effect. Exp. Biol. Med. 2021. [Google Scholar] [CrossRef]
- Xiao, Z.; Chow, S.C.; Li, C.H.; Tang, S.C.; Tsui, S.K.; Lin, Z.; Chen, Y. Role of microRNA-95 in the anticancer activity of Brucein D in hepatocellular carcinoma. Eur. J. Pharmacol. 2014, 728, 141–150. [Google Scholar] [CrossRef]
- Lee, K.H.; Lee, J.K.; Choi, D.W.; Do, I.-G.; Sohn, I.; Jang, K.-T.; Jung, S.-H.; Heo, J.S.; Choi, S.H.; Lee, K.T. Postoperative Prognosis Prediction of Pancreatic Cancer With Seven MicroRNAs. Pancreas 2015, 44, 764–768. [Google Scholar] [CrossRef]
- Zehentmayr, F.; Hauser-Kronberger, C.; Zellinger, B.; Hlubek, F.; Schuster, C.; Bodenhofer, U.; Fastner, G.; Deutschmann, H.; Steininger, P.; Reitsamer, R.; et al. Hsa-miR-375 is a predictor of local control in early stage breast cancer. Clin. Epigenetics 2016, 8, 28. [Google Scholar] [CrossRef] [Green Version]
- Zellinger, B.; Bodenhofer, U.; Engländer, I.A.; Kronberger, C.; Strasser, P.; Grambozov, B.; Fastner, G.; Stana, M.; Reitsamer, R.; Sotlar, K.; et al. Hsa-miR-375/RASD1 Signaling May Predict Local Control in Early Breast Cancer. Genes 2020, 11, 1404. [Google Scholar] [CrossRef]
- Tang, W.; Li, G.-S.; Li, J.-D.; Pan, W.-Y.; Shi, Q.; Xiong, D.-D.; Mo, C.-H.; Zeng, J.-J.; Chen, G.; Feng, Z.-B.; et al. The role of upregulated miR-375 expression in breast cancer: An in vitro and in silico study. Pathol.-Res. Pr. 2019, 216, 152754. [Google Scholar] [CrossRef]
- Jonsdottir, K.; Janssen, S.R.; Da Rosa, F.C.; Gudlaugsson, E.; Skaland, I.; Baak, J.P.A.; Janssen, E.A.M. Validation of Expression Patterns for Nine miRNAs in 204 Lymph-Node Negative Breast Cancers. PLoS ONE 2012, 7, e48692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, Y.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Wang, H.; Tang, L.; Huang, H.; Xu, M.; Lin, Y.; Zhou, L.; Ho, L.; Lu, J.; Ai, X. LncRNA BACE1-AS enhances the invasive and metastatic capacity of hepatocellular carcinoma cells through mediating miR-377-3p/CELF1 axis. Life Sci. 2021, 275, 119288. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Liang, G.; Yang, L.; Liu, L.; Wang, B.; Yan, F. SP1-induced AFAP1-AS1 contributes to proliferation and invasion by regulating miR-497-5p/CELF1 pathway in nasopharyngeal carcinoma. Hum. Cell 2021, 34, 491–501. [Google Scholar] [CrossRef]
- Shi, M.; Yang, R.; Lin, J.; Wei, Q.; Chen, L.; Gong, W.; Guo, X. LncRNA-SNHG16 promotes proliferation and migration of acute myeloid leukemia cells via PTEN/PI3K/AKT axis through suppressing CELF2 protein. J. Biosci. 2021, 46, 1–13. [Google Scholar] [CrossRef]
- Xie, S.-C.; Zhang, J.-Q.; Jiang, X.-L.; Hua, Y.-Y.; Xie, S.-W.; Qin, Y.-A.; Yang, Y.-J. LncRNA CRNDE facilitates epigenetic suppression of CELF2 and LATS2 to promote proliferation, migration and chemoresistance in hepatocellular carcinoma. Cell Death Dis. 2020, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhou, H.; Dong, W. LncRNA RHPN1-AS1 promotes the progression of nasopharyngeal carcinoma by targeting CELF2 expression. Exp. Mol. Pathol. 2021, 122, 104671. [Google Scholar] [CrossRef]
- Li, C.; Mu, J.; Shi, Y.; Xin, H. LncRNA CCDC26 Interacts with CELF2 Protein to Enhance Myeloid Leukemia Cell Proliferation and Invasion via the circRNA_ANKIB1/miR-195-5p/PRR11 Axis. Cell Transplant. 2021, 30. [Google Scholar] [CrossRef]
- Jacobsen, A.; Wen, J.; Marks, D.S.; Krogh, A. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 2010, 20, 1010–1019. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zubovic, L.; Yang, F.; Godin, K.; Pavelitz, T.; Castellanos, J.; Macchi, P.; Varani, G. Rbfox proteins regulate microRNA biogenesis by sequence-specific binding to their precursors and target downstream Dicer. Nucleic Acids Res. 2016, 44, 4381–4395. [Google Scholar] [CrossRef] [PubMed]
- Nussbacher, J.K.; Yeo, G.W. Systematic Discovery of RNA Binding Proteins that Regulate MicroRNA Levels. Mol. Cell 2018, 69, 1005–1016.e7. [Google Scholar] [CrossRef] [Green Version]
- Ruggiero, T.; Trabucchi, M.; De Santa, F.; Zupo, S.; Harfe, B.D.; McManus, M.T.; Rosenfeld, M.G.; Briata, P.; Gherzi, R. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J. 2009, 23, 2898–2908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahuja, D.; Goyal, A.; Ray, P.S. Interplay between RNA-binding protein HuR and microRNA-125b regulates p53 mRNA translation in response to genotoxic stress. RNA Biol. 2016, 13, 1152–1165. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Ouyang, M.; Rao, J.N.; Zou, T.; Xiao, L.; Chung, H.K.; Wu, J.; Donahue, J.M.; Gorospe, M.; Wang, J.-Y. Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol. Biol. Cell 2015, 26, 1797–1810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebert, M.S.; Sharp, P.A. Emerging Roles for Natural MicroRNA Sponges. Curr. Biol. 2010, 20, R858–R861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumbovic, G.; Biayna, J.; Banús, J.; Samuelsson, J.; Roth, A.; Diederichs, S.; Alonso, S.; Buschbeck, M.; Perucho, M.; Forcales, S.-V. A novel long non-coding RNA from NBL2 pericentromeric macrosatellite forms a perinucleolar aggregate structure in colon cancer. Nucleic Acids Res. 2018, 46, 5504–5524. [Google Scholar] [CrossRef]
- Govardhanagiri, S.; Bethi, S.; Nagaraju, G.P. Small Molecules and Pancreatic Cancer Trials and Troubles. In Breaking Tolerance to Pancreatic Cancer Unresponsiveness to Chemotherapy; Academic Press: Cambridge, MA, USA, 2019; pp. 117–131. [Google Scholar] [CrossRef]
- D’Souza, A.M.; Cast, A.; Kumbaji, M.; Rivas, M.; Gulati, R.; Johnston, M.; Smithrud, D.; Geller, J.; Timchenko, N. Small Molecule Cjoc42 Improves Chemo-Sensitivity and Increases Levels of Tumor Suppressor Proteins in Hepatoblastoma Cells and in Mice by Inhibiting Oncogene Gankyrin. Front. Pharmacol. 2021, 12, 146. [Google Scholar] [CrossRef]
- Zheng, B.; Yuan, M.; Wang, S.; Tan, Y.; Xu, Y.; Ye, J.; Gao, Y.; Sun, X.; Wang, T.; Kong, L.; et al. Fraxinellone alleviates kidney fibrosis by inhibiting CUG-binding protein 1-mediated fibroblast activation. Toxicol. Appl. Pharmacol. 2021, 420, 115530. [Google Scholar] [CrossRef]
- Chandler, C.; Liu, T.; Buckanovich, R.; Coffman, L.G. The double edge sword of fibrosis in cancer. Transl. Res. 2019, 209, 55–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-H.; Kwon, H.Y.; Ryu, D.H.; Nam, M.-H.; Shim, B.S.; Kim, J.H.; Lee, J.Y.; Kim, S.-H. Inhibition of CUG-binding protein 1 and activation of caspases are critically involved in piperazine derivative BK10007S induced apoptosis in hepatocellular carcinoma cells. PLoS ONE 2017, 12, e0186490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, M.Y.; Pan, X.; Fix, L.N.; Farwell, M.A.; Zhang, B. 5-fluorouracil drug alters the microrna expression profiles in MCF-7 breast cancer cells. J. Cell. Physiol. 2010, 226, 1868–1878. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-H.; Lotterman, C.; Karikari, C.; Omura, N.; Feldmann, G.; Habbe, N.; Goggins, M.G.; Mendell, J.T.; Maitra, A. Epigenetic Silencing of MicroRNA miR-107 Regulates Cyclin-Dependent Kinase 6 Expression in Pancreatic Cancer. Pancreatology 2009, 9, 293–301. [Google Scholar] [CrossRef] [Green Version]
- Boren, T.; Xiong, Y.; Hakam, A.; Wenham, R.; Apte, S.; Chan, G.; Kamath, S.G.; Chen, D.-T.; Dressman, H.; Lancaster, J.M. MicroRNAs and their target messenger RNAs associated with ovarian cancer response to chemotherapy. Gynecol. Oncol. 2009, 113, 249–255. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Lee, E.-M.; Shin, S.; Cha, H.J.; Yoon, Y.; Bae, S.; Jung, J.H.; Lee, S.-M.; Lee, S.-J.; Park, I.-C.; et al. Suberoylanilide hydroxamic acid (SAHA) changes microRNA expression profiles in A549 human non-small cell lung cancer cells. Int. J. Mol. Med. 2009, 24, 45–50. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zhou, Y.; Yin, B.; Hao, W.; Zhao, L.; Ju, W.; Bai, C. 5-Fluorouracil and oxaliplatin modify the expression profiles of microRNAs in human colon cancer cells in vitro. Oncol. Rep. 2009, 23, 121–128. [Google Scholar] [CrossRef] [Green Version]
- Lehmusvaara, S.; Erkkilä, T.; Urbanucci, A.; Jalava, S.; Seppälä, J.; Kaipia, A.; Kujala, P.; Lähdesmäki, H.; Tammela, T.L.; Visakorpi, T. Goserelin and bicalutamide treatments alter the expression of microRNAs in the prostate. Prostate 2012, 73, 101–112. [Google Scholar] [CrossRef]
- Ramaiah, J.; Pushpavalli, S.N.; Lavanya, A.; Bhadra, K.; Haritha, V.; Patel, N.; Tamboli, J.R.; Kamal, A.; Bhadra, U.; Pal-Bhadra, M. Novel anthranilamide-pyrazolo[1,5-a]pyrimidine conjugates modulate the expression of p53-MYCN associated micro RNAs in neuroblastoma cells and cause cell cycle arrest and apoptosis. Bioorganic Med. Chem. Lett. 2013, 23, 5699–5706. [Google Scholar] [CrossRef]
- Wu, N.; Wu, G.-C.; Hu, R.; Li, M.; Feng, H. Ginsenoside Rh2 inhibits glioma cell proliferation by targeting microRNA-128. Acta Pharmacol. Sin. 2011, 32, 345–353. [Google Scholar] [CrossRef] [Green Version]
- Ichikawa, T.; Sato, F.; Terasawa, K.; Tsuchiya, S.; Toi, M.; Tsujimoto, G.; Shimizu, K. Trastuzumab Produces Therapeutic Actions by Upregulating miR-26a and miR-30b in Breast Cancer Cells. PLoS ONE 2012, 7, e31422. [Google Scholar] [CrossRef] [Green Version]
- Yin, W.; Wang, P.; Wang, X.; Song, W.; Cui, X.; Yu, H.; Zhu, W. Identification of microRNAs and mRNAs associated with multidrug resistance of human laryngeal cancer Hep-2 cells. Braz. J. Med Biol. Res. 2013, 46, 546–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radpour, R.; Barekati, Z.; Kohler, C.; Schumacher, M.M.; Grussenmeyer, T.; Jenoe, P.; Hartmann, N.; Moes, S.; Letzkus, M.; Bitzer, J.; et al. Integrated Epigenetics of Human Breast Cancer: Synoptic Investigation of Targeted Genes, MicroRNAs and Proteins upon Demethylation Treatment. PLoS ONE 2011, 6, e27355. [Google Scholar] [CrossRef]
- Seol, H.S.; Akiyama, Y.; Shimada, S.; Lee, H.J.; Kim, T.I.; Chun, S.M.; Singh, S.R.; Jang, S.J. Epigenetic silencing of microRNA-373 to epithelial-mesenchymal transition in non-small cell lung cancer through IRAK2 and LAMP1 axes. Cancer Lett. 2014, 353, 232–241. [Google Scholar] [CrossRef]
- Qiu, Y.-Y.; Hu, Q.; Tang, Q.-F.; Feng, W.; Hu, S.-J.; Liang, B.; Peng, W.; Yin, P.-H. MicroRNA-497 and bufalin act synergistically to inhibit colorectal cancer metastasis. Tumor Biol. 2013, 35, 2599–2606. [Google Scholar] [CrossRef] [PubMed]
- An, I.-S.; An, S.; Kwon, K.J.; Kim, Y.J.; Bae, S. Ginsenoside Rh2 mediates changes in the microRNA expression profile of human non-small cell lung cancer A549 cells. Oncol. Rep. 2012, 29, 523–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.-N.; Zhang, H.-D.; Zhi, R.; Yuan, S.-J. Down-regulation of some miRNAs by degrading their precursors contributes to anti-cancer effect of mistletoe lectin-I. Br. J. Pharmacol. 2010, 162, 349–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaghoobi, H.; Azizi, M.; Banitalebi-Dehkordi, F.; Mohammad Rezaei, S.; Arsang-Jnag, M.; Taheri, S. Ghafouri-Fard, Be-ta-Secretase 1 (BACE1) Is Down-Regulated in Invasive Ductal Carcinoma of Breast. Rep. Biochem. Mol. Biol. 2019, 8, 200–207. [Google Scholar]
- Yang, M.; Wei, W. SNHG16: A Novel Long-Non Coding RNA in Human Cancers. OncoTargets Ther. 2019, 12, 11679–11690. [Google Scholar] [CrossRef] [Green Version]
- Ghidini, A.; Cléry, A.; Halloy, F.; Allain, F.H.T.; Hall, J. RNA-PROTACs: Degraders of RNA-Binding Proteins. Angew. Chem. Int. Ed. 2020, 60, 3163–3169. [Google Scholar] [CrossRef]
- Khan, A.A.; Allemailem, K.S.; Almatroudi, A.; Almatroodi, S.A.; Alsahli, M.A.; Rahmani, A.H. Novel strategies of third level (Organelle-specific) drug targeting: An innovative approach of modern therapeutics. J. Drug Deliv. Sci. Technol. 2020, 61, 102315. [Google Scholar] [CrossRef]
- Ho, J.J.D.; Marsden, P.A. Competition and collaboration between RNA-binding proteins and microRNAs. Wiley Interdiscip. Rev. RNA 2013, 5, 69–86. [Google Scholar] [CrossRef]
- Cazzalini, O.; Scovassi, A.I.; Savio, M.; Stivala, L.A.; Prosperi, E. Multiple roles of the cell cycle inhibitor p21CDKN1A in the DNA damage response. Mutat. Res. Mutat. Res. 2010, 704, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Wang, N.; Lin, G.; Zhang, H.; Xie, W.; Zhang, Y.; Xu, N. MBNL2 Regulates DNA Damage Response via Stabilizing p21. Int. J. Mol. Sci. 2021, 22, 783. [Google Scholar] [CrossRef] [PubMed]


| miRNA | Molecule/Drug | Effect on miRNA Expression | Cancer Cell Line |
|---|---|---|---|
| mir-20b | 5-Fluorouracil | Downregulated | Breast cancer cells [163] |
| 5-aza-2′-deoxycytidine | Upregulated | Human pancreatic cancer cell lines [164] | |
| miR-106a | Cisplatin | Downregulated | Ovarian cancer cell lines [165] |
| Suberoylanilide hydroxamic acid | Downregulated | Human lung carcinoma cell line [166] | |
| 5-Fluorouracil | Downregulated | Colon cancer cell line [167] | |
| Bicalutamide | Downregulated | Prostate cancer cell line [168] | |
| miR-107 | 5-Fluorouracil | Upregulated | Breast cancer cells [163] |
| Anthranilamide-pyrazolo [1,5-a] pyrimidine | Upregulated | Neuroblastoma cell [169] | |
| miR-140 | 5-Fluorouracil | Upregulated | Breast cancer cells [163] |
| miR-140-5p | Suberoylanilide hydroxamic acid | Upregulated | Human lung carcinoma cell line [166] |
| mir-155 | Suberoylanilide hydroxamic acid | Downregulated | Human lung carcinoma cell line [166] |
| Ginsenoside Rh2 | Downregulated | Human glioma cells [170] | |
| miR-195 | 5-Fluorouracil | Downregulated | Breast cancer cells [163] |
| Trastuzumab | Upregulated | Breast cancer cells [171] | |
| miR-210 | 5-Fluorouracil | Upregulated | Breast cancer cell line [163] |
| Vincristine | Upregulated | Human laryngeal cancer Hep-2 cells [172] | |
| Ginsenoside Rh2 | Downregulated | Human glioma cells [170] | |
| 5-aza-2′-deoxycytidine | Downregulated | Human breast cancer cell line [173] | |
| miR-330 | Gemcitabine | Downregulated | Ovarian cancer cell lines [165] |
| miR-375 | 5-Fluorouracil | Downregulated | Breast cancer cells [163] |
| Suberoylanilide hydroxamic acid | Upregulated | Non-small cell lung cancer cell line [174] | |
| Trichostatin A | Downregulated | Non-small cell lung cancer cell line [174] | |
| miR-497 | 5-Fluorouracil | Diferentially expressed on breast cancer cells in treatment group and not in control group | Breast cancer cells [163] |
| Bufalin | Upregulated | Human colorectal cancer cell line [175] | |
| miR-574-3p | Suberoylanilide hydroxamic acid (SAHA) | Upregulated | Human lung carcinoma cell line [166] |
| miR-574-5p | 5-Fluorouracil | Upregulated | Breast cancer cells [163] |
| Ginsenoside Rh2 | Upregulated | Non-small-cell lung cancer cell line [176] | |
| miR-615-3p | Mistletoe lectin-I | Downregulated | CRC cell line CLY (established from liver metastases of a CRC patient) [177] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasiri-Aghdam, M.; Garcia-Garduño, T.C.; Jave-Suárez, L.F. CELF Family Proteins in Cancer: Highlights on the RNA-Binding Protein/Noncoding RNA Regulatory Axis. Int. J. Mol. Sci. 2021, 22, 11056. https://doi.org/10.3390/ijms222011056
Nasiri-Aghdam M, Garcia-Garduño TC, Jave-Suárez LF. CELF Family Proteins in Cancer: Highlights on the RNA-Binding Protein/Noncoding RNA Regulatory Axis. International Journal of Molecular Sciences. 2021; 22(20):11056. https://doi.org/10.3390/ijms222011056
Chicago/Turabian StyleNasiri-Aghdam, Maryam, Texali C. Garcia-Garduño, and Luis Felipe Jave-Suárez. 2021. "CELF Family Proteins in Cancer: Highlights on the RNA-Binding Protein/Noncoding RNA Regulatory Axis" International Journal of Molecular Sciences 22, no. 20: 11056. https://doi.org/10.3390/ijms222011056
APA StyleNasiri-Aghdam, M., Garcia-Garduño, T. C., & Jave-Suárez, L. F. (2021). CELF Family Proteins in Cancer: Highlights on the RNA-Binding Protein/Noncoding RNA Regulatory Axis. International Journal of Molecular Sciences, 22(20), 11056. https://doi.org/10.3390/ijms222011056







