Hydration Shells of DNA from the Point of View of Terahertz Time-Domain Spectroscopy
Abstract
:1. Introduction
2. Results
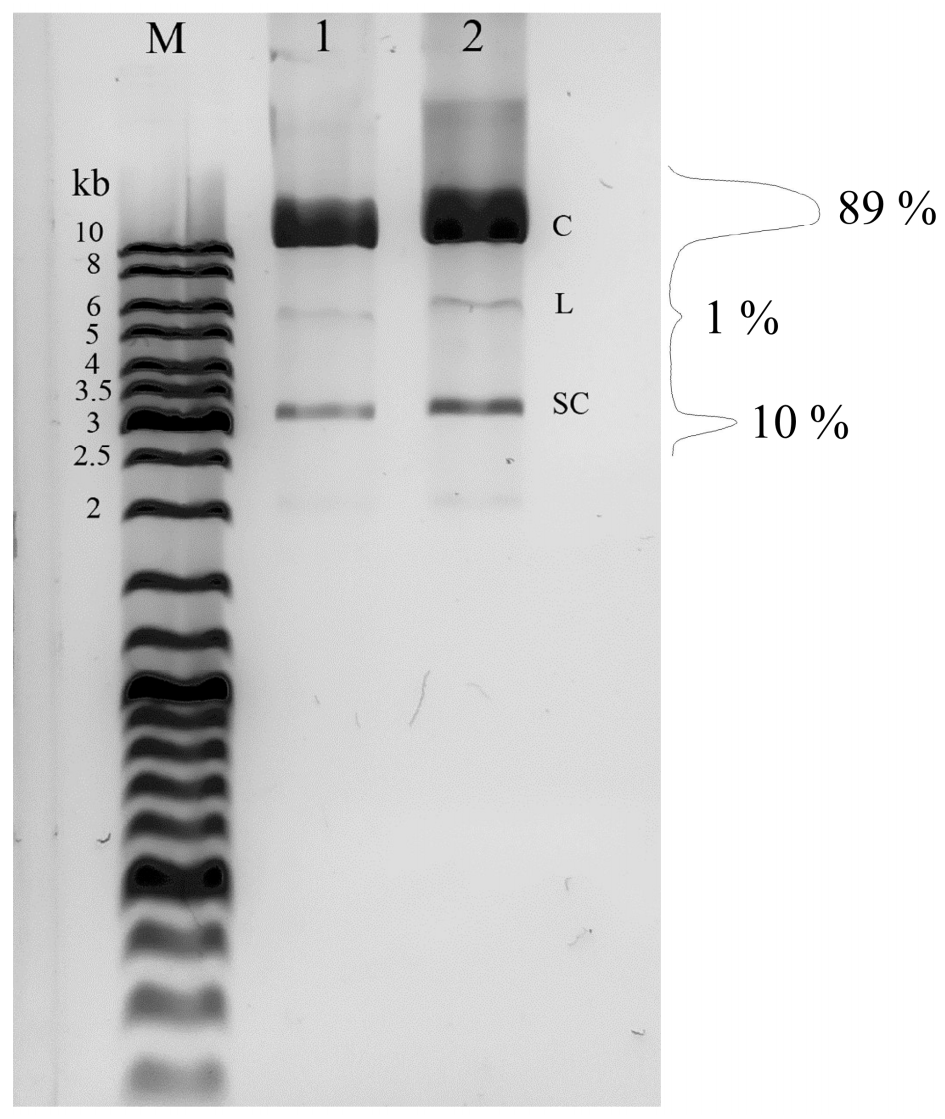
2.1. The Form of the Studied Plasmid DNA
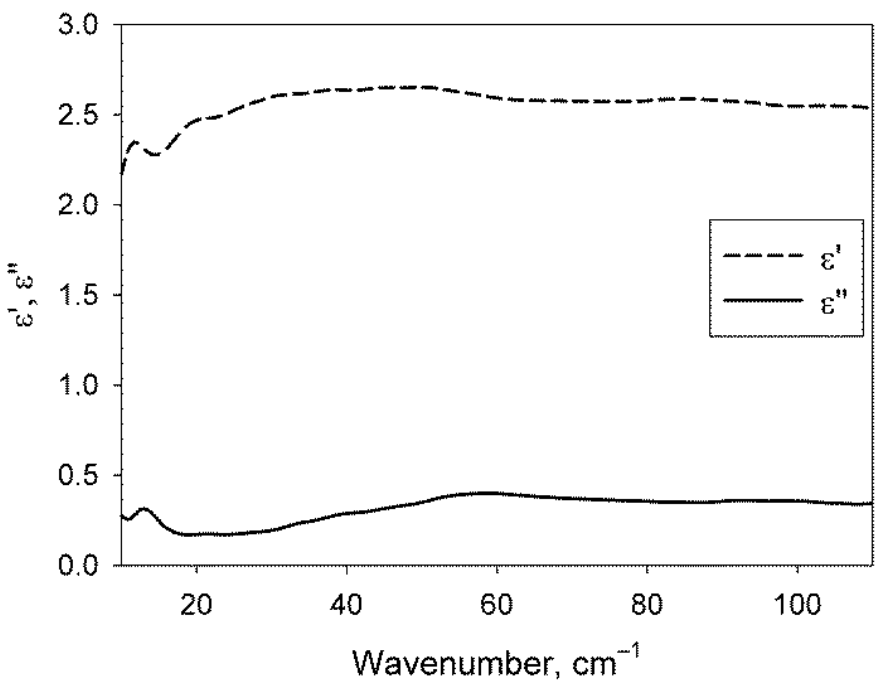
2.2. Dielectric Permittivities of the Water Phase of the Studied Solutions
2.3. Parameters of the Model Dielectric Permittivity and the Percentage of Free Water Molecules in the Studied Solutions
2.4. Dielectric Permittivity of Dry DNA
2.5. Conductivities of the Studied Solutions
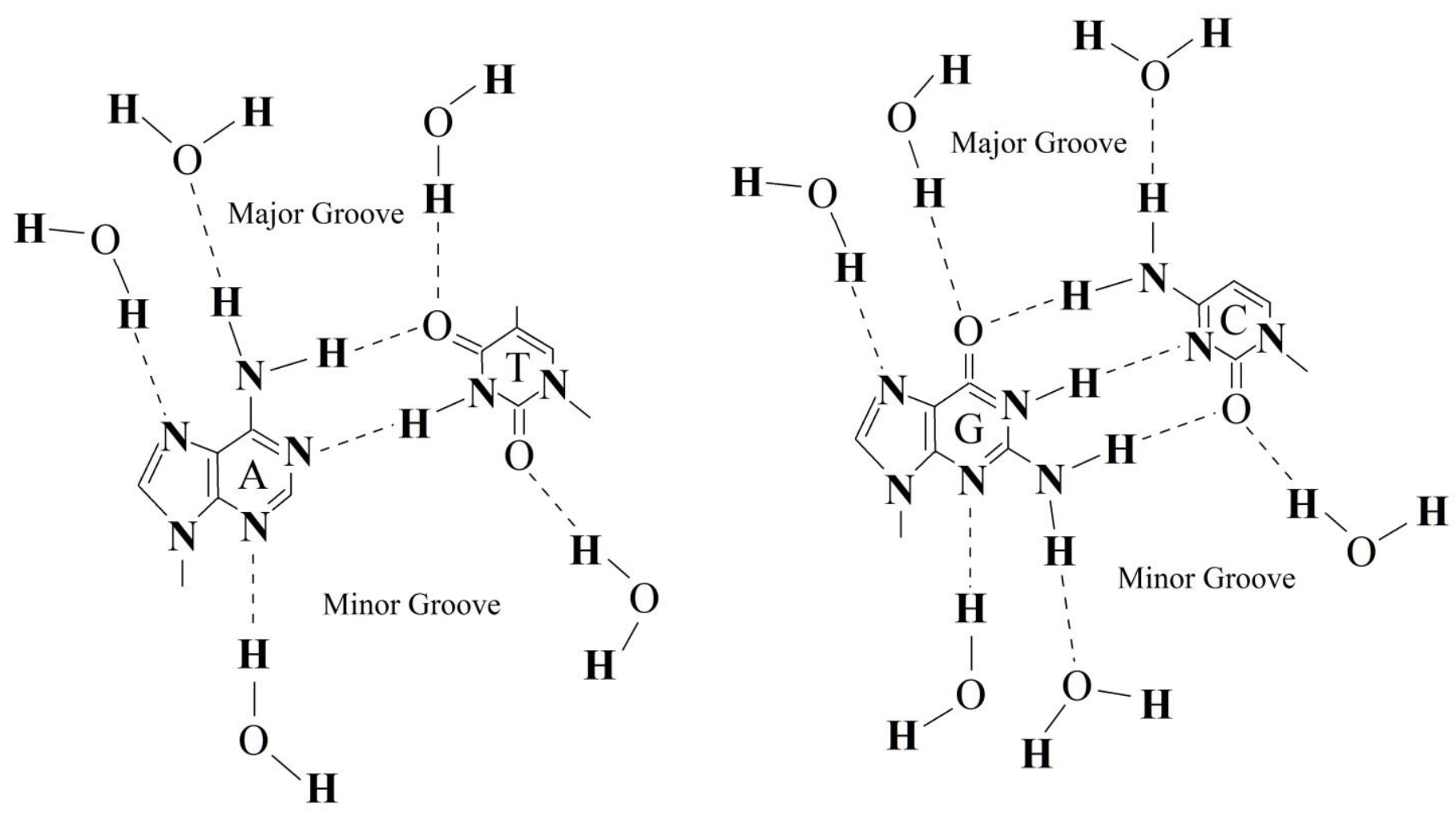
3. Discussion
4. Materials and Methods
4.1. Preparation of Samples
4.2. THz-TDS
4.3. Calculation of DP of DNA Solution Water Phase
4.4. Analysis of Water Solution DPs
4.5. Preparation of Dry DNA Films and Measurement of Their DPs
4.6. Measurement of Solutions Conductivity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Falk, M.; Hartman, K.A.; Lord, R.C. Hydration of Deoxyribonucleic Acid. III. A Spectroscopic Study of the Effect of Hydration on the Structure of Deoxyribonucleic Acid. J. Am. Chem. Soc. 1963, 85, 391–394. [Google Scholar] [CrossRef]
- Lindsay, S.M.; Lee, S.A.; Powell, J.W.; Weidlich, T.; DeMarco, C.; Lewen, G.D.; Tao, N.J.; Rupprecht, A. The origin of the A to B transition in DNA fibers and films. Biopolymers 1988, 27, 1015–1043. [Google Scholar] [CrossRef] [PubMed]
- Lavalle, N.; Lee, S.A.; Rupprecht, A. Counterion effects on the physical properties and the A to B transition of calf-thymus DNA films. Biopolymers 1990, 30, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, Y.; Kamiya, N.; Iwasaki, H.; Ito, T.; Satow, Y. Humidity-controlled reversible structure transition of disodium adenosine 5’-triphosphate between dihydrate and trihydrate in a single crystal state. J. Am. Chem. Soc. 1991, 113, 5440–5445. [Google Scholar] [CrossRef]
- Saenger, W.; Hunter, W.N.; Kennard, O. DNA conformation is determined by economics in the hydration of phosphate groups. Nature 1986, 324, 385–388. [Google Scholar] [CrossRef]
- Falk, M.; Hartman, K.A.; Lord, R.C. Hydration of Deoxyribonucleic Acid. I. a Gravimetric Study. J. Am. Chem. Soc. 1962, 84, 3843–3846. [Google Scholar] [CrossRef]
- Falk, M.; Hartman, K.A.; Lord, R.C. Hydration of Deoxyribonucleic Acid. II. An Infrared Study. J. Am. Chem. Soc. 1963, 85, 387–391. [Google Scholar] [CrossRef]
- Falk, M.; Poole, A.G.; Goymour, C.G. Infrared study of the state of water in the hydration shell of DNA. Can. J. Chem. 1970, 48, 1536–1542. [Google Scholar] [CrossRef]
- Qian, Y.Q.; Otting, G.; Wuethrich, K. NMR detection of hydration water in the intermolecular interface of a protein-DNA complex. J. Am. Chem. Soc. 1993, 115, 1189–1190. [Google Scholar] [CrossRef]
- Michalarias, I.; Gao, X.; Ford, R.C.; Li, J. Recent progress on our understanding of water around biomolecules. J. Mol. Liq. 2005, 117, 107–116. [Google Scholar] [CrossRef]
- Chalikian, T.V.; Sarvazyan, A.P.; Plum, G.E.; Breslauer, K.J. Influence of base composition, base sequence, and duplex structure on DNA hydration: Apparent molar volumes and apparent molar adiabatic compressibilities of synthetic and natural DNA duplexes at 25 °C. Biochemistry 1994, 33, 2394–2401. [Google Scholar] [CrossRef]
- Duboué-Dijon, E.; Fogarty, A.C.; Hynes, J.T.; Laage, D. Dynamical Disorder in the DNA Hydration Shell. J. Am. Chem. Soc. 2016, 138, 7610–7620. [Google Scholar] [CrossRef]
- Schneider, B.; Patel, K.; Berman, H.M. Hydration of the phosphate group in double-helical DNA. Biophys. J. 1998, 75, 2422–2434. [Google Scholar] [CrossRef] [Green Version]
- Mashimo, S.; Umehara, T.; Kuwabara, S.; Yagihara, S. Dielectric study on dynamics and structure of water bound to DNA using a frequency range 107–1010 Hz. J. Phys. Chem. 1989, 93, 4963–4967. [Google Scholar] [CrossRef]
- Takashima, S. Dielectric dispersion of deoxyribonucleic acid. II. J. Phys. Chem. 1966, 70, 1372–1380. [Google Scholar] [CrossRef]
- Takashima, S. Effect of ions on the dielectric relaxation of DNA. Biopolymers 1967, 5, 899–913. [Google Scholar] [CrossRef]
- Takashima, S.; Gabriel, C.; Sheppard, R.J.; Grant, E.H. Dielectric behavior of DNA solution at radio and microwave frequencies (at 20 °C). Biophys. J. 1984, 46, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Kuwabara, S.; Umehara, T.; Mashimo, S.; Yagihara, S. Dynamics and structure of water bound to DNA. J. Phys. Chem. 1988, 92, 4839–4841. [Google Scholar] [CrossRef]
- Lee, Y.-S. Principles of Terahertz Science and Technology; Springer: New York, NY, USA, 2009. [Google Scholar]
- Theuer, M.; Harsha, S.S.; Molter, D.; Torosyan, G.; Beigang, R. Terahertz Time-Domain Spectroscopy of Gases, Liquids and Solids. Chem. Phys. Chem. 2011, 12, 2695–2705. [Google Scholar] [CrossRef]
- Asaki, M.L.T.; Redondo, A.; Zawodzinski, T.A.; Taylor, A.J. Dielectric relaxation of electrolyte solutions using terahertz transmission spectroscopy. J. Chem. Phys. 2002, 116, 8469. [Google Scholar] [CrossRef] [Green Version]
- Glancy, P.; Beyermann, W.P. Dielectric properties of fully hydrated nucleotides in the terahertz frequency range. J. Chem. Phys. 2010, 132, 245102. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.H.; Jung, S.; Park, J.; Park, W.Y.; Kwon, O.S.; Park, G.S. Determination of relaxation time of DNA hydration water by THz-TDS. In Proceedings of the 37th International Conference on Infrared, Millimeter, and Terahertz Waves, Wollongong, NSW, Australia, 23–28 September 2012; pp. 1–2. [Google Scholar] [CrossRef]
- Glancy, P. Concentration-dependent effects on fully hydrated DNA at terahertz frequencies. J. Biol. Phys. 2015, 41, 247–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraszewski, A.; Kulinski, S.; Matuszewski, M. Dielectric properties and a model of biphase water suspension at 9.4 GHz. J. Appl. Phys. 1976, 47, 1275. [Google Scholar] [CrossRef]
- Polley, D.; Patra, A.; Mitra, R.K. Dielectric relaxation of the extended hydration sheathe of DNA in the THz frequency region. Chem. Phys. Lett. 2013, 586, 143–147. [Google Scholar] [CrossRef]
- Penkov, N.; Shvirst, N.; Yashin, V.; Fesenko, E., Jr.; Fesenko, E. Terahertz Spectroscopy Applied for Investigation of Water Structure. J. Phys. Chem. B 2015, 119, 12664–12670. [Google Scholar] [CrossRef]
- Penkov, N.; Yashin, V.; Fesenko, E., Jr.; Manokhin, A.; Fesenko, E. A Study of the Effect of a Protein on the Structure of Water in Solution Using Terahertz Time-Domain Spectroscopy. Appl. Spectrosc. 2018, 72, 257–267. [Google Scholar] [CrossRef]
- Penkov, N.V.; Yashin, V.; Belosludtsev, K.N. Hydration Shells of DPPC Liposomes from the Point of View of Terahertz Time-Domain Spectroscopy. Appl. Spectrosc. 2021, 75, 189–198. [Google Scholar] [CrossRef]
- Penkov, N.V.; Penkova, N. Key Differences of the Hydrate Shell Structures of ATP and Mg·ATP Revealed by Terahertz Time-Domain Spectroscopy and Dynamic Light Scattering. J. Phys. Chem. B. 2021, 125, 4375–4382. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V. A virocentric perspective on the evolution of life. Curr. Opin. Virol. 2013, 3, 546–557. [Google Scholar] [CrossRef]
- Yao, N.Y.; O’Donnell, M.E. Evolution of replication machines. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 135–149. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Turner, K.M.; Nguyen, N.; Raviram, R.; Erb, M.; Santini, J.; Luebeck, J.; Rajkumar, U.; Diao, Y.; Li, B.; et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 2019, 575, 699–703. [Google Scholar] [CrossRef]
- Nortemann, K.; Hilland, J.; Kaatze, U. Dielectric Properties of Aqueous NaCl Solutions at Microwave Frequencies. J. Phys. Chem. A 1997, 101, 6864–6869. [Google Scholar] [CrossRef]
- Barthel, J.; Kleebauer, M.; Buchner, R. Dielectric Relaxation of Electrolyte Solutions in Acetonitrile. J. Solut. Chem. 1995, 24, 1–17. [Google Scholar] [CrossRef]
- Penkov, N.V.; Shvirst, N.E.; Yashin, V.A.; Fesenko, E.E. On singularities of molecular relaxation in water solutions. Biophysics 2013, 58, 731–738. [Google Scholar] [CrossRef]
- Yada, H.; Nagai, M.; Tanaka, K. Origin of the fast relaxation component of water and heavy water revealed by terahertz time domain attenuated total reflection spectroscopy. Chem. Phys. Lett. 2008, 464, 166–170. [Google Scholar] [CrossRef]
- Zasetsky, A.Y. Dielectric relaxation in liquid water: Two fractions or two dynamics? Phys. Rev. Lett. 2011, 107, 117601. [Google Scholar] [CrossRef]
- Shiraga, K.; Suzuki, T.; Kondo, N.; Baerdemaeker, J.; Ogawa, Y. Quantitative characterization of hydration state and destructuring effect of monosaccharides and disaccharides on water hydrogen bond network. Carbohydr. Res. 2015, 406, 46–54. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M. What is “hypermobile” water?: Detected in alkali halide, adenosine phosphate, and F-actin solutions by highresolution microwave dielectric spectroscopy. Pure Appl. Chem. 2014, 86, 181–189. [Google Scholar] [CrossRef]
- Ebbinghaus, S.; Kim, S.J.; Heyden, M.; Xu, Y.; Heugen, U.; Gruebele, M.; Leitner, D.M.; Havenith, M. An Extended Dynamical Hydration Shell Around Proteins. Proc. Nat. Acad. Sci. USA 2007, 104, 20749–20752. [Google Scholar] [CrossRef] [Green Version]
- Bye, J.W.; Meliga, S.; Ferachou, D.; Cinque, G.; Zeitler, J.A.; Falconer, R.J. Analysis of the Hydration Water around Bovine Serum Albumin Using Terahertz Coherent Synchrotron Radiation. J. Phys. Chem. A 2014, 118, 83–88. [Google Scholar] [CrossRef] [Green Version]
- Heyden, M.; Tobias, D.J.; Matyushov, D.V. Terahertz Absorption of Dilute Aqueous Solutions. J. Chem. Phys. 2012, 137, 235103. [Google Scholar] [CrossRef]
- Shiraga, K.; Ogawa, Y.; Kondo, N.; Irisawa, A.; Imamura, M. Evaluation of the hydration state of saccharides using terahertztime-domain attenuated total reflection spectroscopy. Food Chem. 2013, 140, 315–320. [Google Scholar] [CrossRef]
- Djikaev, Y.S.; Ruckenstein, E. Dependence of the number of hydrogen bonds per water molecule on its distance to a hydrophobic surface and a thereupon-based model for hydrophobic attraction. J. Chem. Phys. 2010, 133, 194105. [Google Scholar] [CrossRef]
- Bartha, F.; Kapuy, O.; Kozmutza, C.; Alsenoy, C.V. Analysis of weakly bound structures: Hydrogen bond and the electron density in a water dimmer. J. Mol. Struct. Theochem. 2003, 666–667, 117–122. [Google Scholar] [CrossRef]
- Duguid, J.; Bloomfield, V.A.; Benevides, J.; Thomas, G.J., Jr. Raman spectroscopy of DNA-metal complexes. I. Interactions and conformational effects of the divalent cations: Mg, Ca, Sr, Ba, Mn, Co, Ni, Cu, Pd, and Cd. Biophys. J. 1993, 65, 1916–1928. [Google Scholar] [CrossRef] [Green Version]
- Serec, K.; Babic, S.D.; Podgornik, R.; Tomic, S. Effect of magnesium ions on the structure of DNA thin films: An infrared spectroscopy study. Nucleic Acids Res. 2016, 44, 8456–8464. [Google Scholar] [CrossRef] [Green Version]
- Bugaenko, L.T.; Ryabykh, S.M.; Bugaenko, A.L. A Near Total System of the Average Ionic Crystallo-Graphic Radius and Their Usage for Ionization Potentials Determination. Vestn. Mosc. State Univ. 2008, 49, 363–384. (In Russian) [Google Scholar]
- Koneshan, S.; Rasaiah, J.C.; Lynden-Bell, R.M.; Lee, S.H. Solvent Structure, Dynamics, and Ion Mobility in Aqueous Solutions at 25 °C. J. Phys. Chem. B 1998, 102, 4193–4204. [Google Scholar] [CrossRef]
- Yang, Z. Hofmeister effects: An explanation for the impact of ionic liquids on biocatalysis. J. Biotechnol. 2009, 144, 12–22. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Lee, M.P.H.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Zinchenko, A.A.; Yoshikawa, K. Na+ Shows a Markedly Higher Potential than K+ in DNA Compaction in a Crowded Environment. Biophys. J. 2005, 88, 4118–4123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savelyev, A.; Papoian, G.A. Electrostatic, Steric, and Hydration Interactions Favor Na+ Condensation around DNA Compared with K+. J. Am. Chem. Soc. 2006, 128, 14506–14518. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Aaij, C.; Borst, P. The gel electrophoresis of DNA. Biochim. Biophys. Acta 1972, 269, 192–200. [Google Scholar] [CrossRef]
- Glasel, J.A. Validity of nucleic acid purities monitored by 260 nm/280 nm absorbance ratios. Biotechniques 1995, 18, 62–63. [Google Scholar]
- Penkov, N.V. Peculiarities of the Perturbation of Water Structure by Ions with Various Hydration in Concentrated Solutions of CaCl2, CsCl, KBr, and KI Physics of Wave Phenomena. Phys. Wave Phenom. 2019, 27, 128–134. [Google Scholar] [CrossRef]
- Taschin, A.; Bartolini, P.; Tasseva, J.; Torre, R. THz time-domain spectroscopic investigations of thin films. Measurement 2018, 118, 282–288. [Google Scholar] [CrossRef] [Green Version]
- Kindt, J.T.; Schmuttenmaer, C.A. Far-Infrared Dielectric Properties of Polar Liquids Probed by Femtosecond Terahertz Pulse Spectroscopy. Phys. Chem. 1996, 100, 10373–10379. [Google Scholar] [CrossRef]
- Penkov, N.V.; Penkova, N.A. Effective Medium Model Applied to Biopolymer Solutions. Appl. Spectrosc. 2021, in press. [Google Scholar] [CrossRef]
- Durchschlag, H. Specific Volumes of Biological Macromolecules and Some Other Molecules of Biological Interest. In Thermodynamic Data for Biochemistry and Biotechnology; Hinz, H.J., Ed.; Springer: Berlin/Heidelberg, Germany, 1986. [Google Scholar]
- Hippel, A.R. The Dielectric Relaxation Spectra of Water, Ice, and Aqueous Solutions, and Their Interpretation. 2. Tentative Interpretation of the Relaxation Spectrum of Water in the Time and Frequency Domain. IEEE Trans. Electr. Insul. 1988, 23, 817–823. [Google Scholar] [CrossRef]
- Laage, D.; Hynes, J.T. A Molecular Jump Mechanism of Water Reorientation. Science 2006, 311, 832–835. [Google Scholar] [CrossRef]
- Nielsen, O.F. Low-frequency spectroscopic studies of interactions in liquids. Annu. Rep. Prog. Chem. Sect. C Phys. Chem. 1993, 90, 3–44. [Google Scholar] [CrossRef]
- Nielsen, O.F. Low-frequency spectroscopic studies and intermolecular vibrational energy transfer in liquids. Annu. Rep. Prog. Chem. Sect. C Phys. Chem. 1996, 93, 57–99. [Google Scholar] [CrossRef]
- Ellison, W.J. Permittivity of pure water, at standard atmospheric pressure, over the frequency range 0–25 THz and the temperature range 0–100 °C. J. Phys. Chem. Ref. Data 2007, 36, 1–18. [Google Scholar] [CrossRef]
- Penkov, N.V.; Yashin, V.A.; Fesenko, E.E., Jr.; Fesenko, E.E. Calculation of the Amount of Free Water Molecules in Aqueous Solutions by Means of Spectral Parameters from the Terahertz Frequency Domain Taking into Account Processes of Screening. Biophysics 2014, 59, 347–350. [Google Scholar] [CrossRef]

| Title 1 | Δε1 | Δε2 | τ2, ps | ω, cm−1 | A/ω2 | γ, cm−1 | n, % |
|---|---|---|---|---|---|---|---|
| Water | 68.86 ± 0.81 | 2.691 ± 0.042 | 0.316 ± 0.006 | 207.2 ± 4.3 | 1.702 ± 0.019 | 196.5 ± 11.1 | 3.78 ± 0.04 |
| Water with DNA | 61.70 ± 0.97 | 3.088 ± 0.095 | 0.346 ± 0.009 | 217.3 ± 7.5 | 1.831 ± 0.037 | 215.4 ± 16.2 | 4.01 ± 0.07 |
| MgCl2 40 mM | 67.59 ± 0.63 | 2.709 ± 0.036 | 0.318 ± 0.004 | 209.2 ± 4.8 | 1.710 ± 0.016 | 201.9 ± 11.1 | 3.79 ± 0.04 |
| MgCl2 40 mM with DNA | 60.55 ± 0.71 | 2.992 ± 0.046 | 0.337 ± 0.005 | 218.1 ± 6.3 | 1.809 ± 0.026 | 216.9 ± 15.0 | 3.95 ± 0.04 |
| KCl 150 mM | 63.11 ± 0.93 | 2.953 ± 0.066 | 0.329 ± 0.007 | 211.6 ± 5.3 | 1.771 ± 0.025 | 201.2 ± 11.8 | 3.95 ± 0.06 |
| KCl 150 mM with DNA | 59.44 ± 0.66 | 3.105 ± 0.040 | 0.335 ± 0.005 | 217.3 ± 4.0 | 1.807 ± 0.025 | 215.5 ± 9.2 | 4.05 ± 0.03 |
| Title 2 | ||||||
|---|---|---|---|---|---|---|
| Solution | Water | Water with DNA | MgCl2 40 mM | MgCl2 40 mM with DNA | KCl 150 mM | KCl 150 mM with DNA |
| Conductivity, S/m | 0 | 0.502 | 0.825 | 1.466 | 1.740 | 3.325 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Penkova, N.A.; Sharapov, M.G.; Penkov, N.V. Hydration Shells of DNA from the Point of View of Terahertz Time-Domain Spectroscopy. Int. J. Mol. Sci. 2021, 22, 11089. https://doi.org/10.3390/ijms222011089
Penkova NA, Sharapov MG, Penkov NV. Hydration Shells of DNA from the Point of View of Terahertz Time-Domain Spectroscopy. International Journal of Molecular Sciences. 2021; 22(20):11089. https://doi.org/10.3390/ijms222011089
Chicago/Turabian StylePenkova, Nadezda A., Mars G. Sharapov, and Nikita V. Penkov. 2021. "Hydration Shells of DNA from the Point of View of Terahertz Time-Domain Spectroscopy" International Journal of Molecular Sciences 22, no. 20: 11089. https://doi.org/10.3390/ijms222011089
APA StylePenkova, N. A., Sharapov, M. G., & Penkov, N. V. (2021). Hydration Shells of DNA from the Point of View of Terahertz Time-Domain Spectroscopy. International Journal of Molecular Sciences, 22(20), 11089. https://doi.org/10.3390/ijms222011089






