Atorvastatin Ester Regulates Lipid Metabolism in Hyperlipidemia Rats via the PPAR-signaling Pathway and HMGCR Expression in the Liver
Abstract
1. Introduction
2. Results
2.1. Characterization of Ate
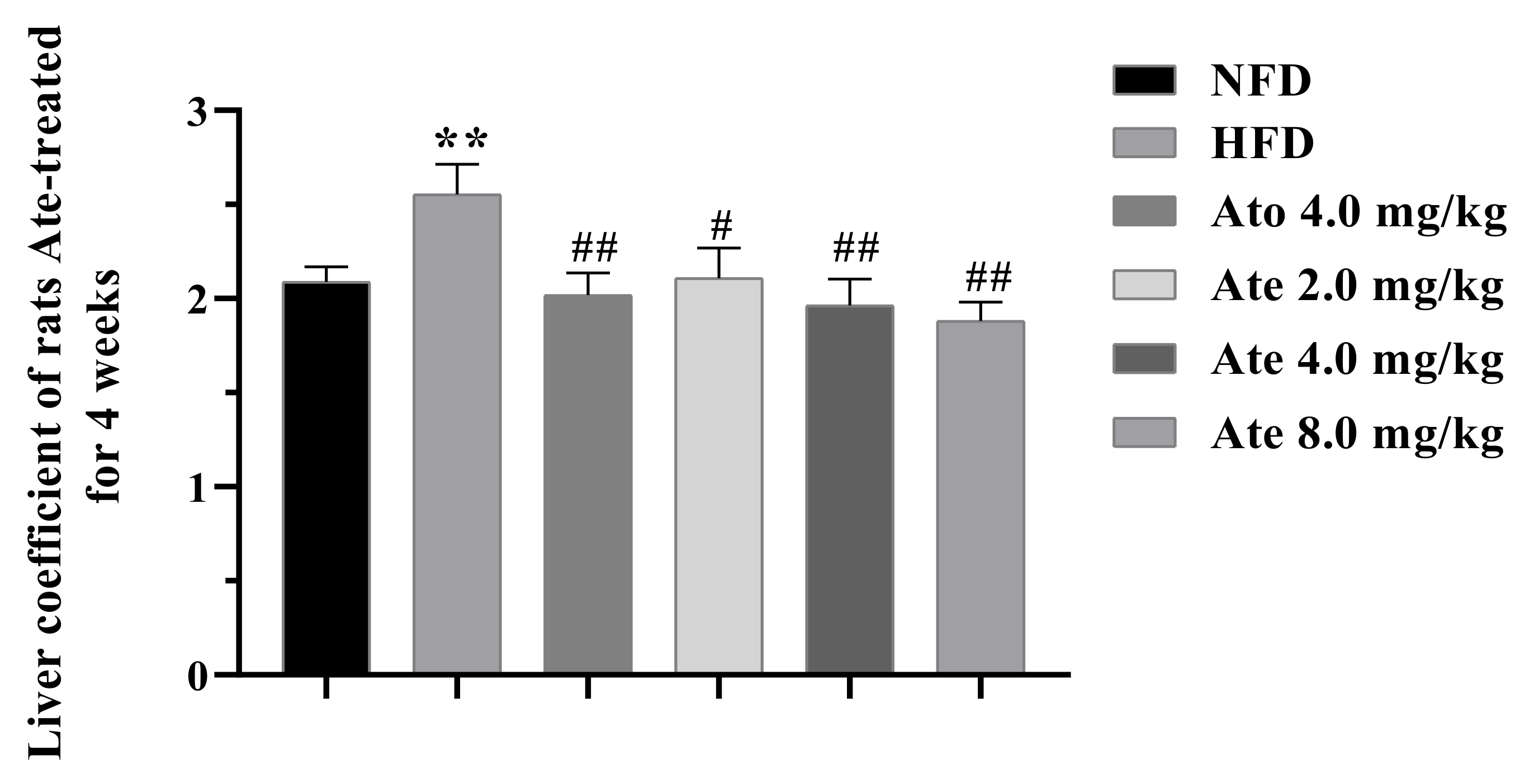
2.2. Ate Can Significantly Reduce the Body Weight and Liver Index of the Hyperlipidemia Rats
2.3. Biochemical Analysis
2.4. Histopathological Analysis
2.5. RNA-Sequencing Analysis
2.5.1. Number of DEGs
2.5.2. GO Enrichment and KEGG Enrichment Analysis
2.6. Real-Time PCR Analysis
2.7. Western Blot
3. Discussion
4. Materials and Methods
4.1. Animals and Treatments
4.2. The Synthesis of Atorvastatin Ester (Ate)
4.3. Liver Index
4.4. Biochemical Analysis
4.5. Histopathological Analysis
4.6. RNA-sequencing Analysis
4.7. Real-Time PCR Analysis
4.8. Western Blot
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALT | Glutamate-pyruvate transaminase |
| AST | Glutamic-oxalacetic transaminase |
| Ate | Atorvastatin ester |
| Ato | Atorvastatin |
| CD36 | Fatty acid translocase |
| DEGs | Differently expressed genes |
| GADPH | Glyceraldehyde-3-phosphate dehydrogenase |
| GO | Gene ontology |
| HDL | High-density lipoprotein |
| HE | Hematoxylin-eosin-staining |
| HFD | High-fat diet |
| HMGCR | 3-hydroxy-3-methylglutaryl coenzyme A reductase |
| HMGCS1 | 3-hydroxy-3-methylglutaryl- coenzyme A synthase 1 |
| KEGG | Kyoto Encyclopedia of genes and genomes |
| LDL | Low-density lipoprotein |
| LPL | Lipoprotein lipase |
| mRNA | Messenger RNA |
| NHF | Non-high-fat diet |
| ORO | Oil Red O |
| PPARα | Peroxisome proliferators-activated receptor α |
| PPARγ | Peroxisome proliferators-activated receptor γ |
| RNA seq | RNA sequence |
| TC | Total cholesterol |
| TG | Triglycerides |
References
- Li, M.; Shu, X.; Xu, H.; Zhang, C.; Yang, L.; Zhang, L.; Ji, G. Integrative analysis of metabolome and gut microbiota in diet-induced hyperlipidemic rats treated with berberine compounds. J. Transl. Med. 2016, 14, 237. [Google Scholar] [CrossRef] [PubMed]
- Goode, G.K.; Miller, J.P.; Heagerty, A.M. Hyperlipidaemia, hypertension, and coronary heart disease. Lancet 1995, 345, 362–364. [Google Scholar] [CrossRef]
- Valdivielso, P.; Rioja, J.; García-Arias, C.; Sánchez-Chaparro, M.A.; González-Santos, P. Omega 3 fatty acids induce a marked reduction of apolipoprotein B48 when added to fluvastatin in patients with type 2 diabetes and mixed hyperlipidemia: A preliminary report. Cardiovasc. Diabetol. 2009, 8, 1. [Google Scholar] [CrossRef]
- Soh, J.; Iqbal, J.; Queiroz, J.; Fernandez-Hernando, C.; Hussain, M.M. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat. Med. 2013, 19, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Forouzanfar, M.H.; Afshin, A.; Alexander, L.T.; Anderson, H.R.; Bhutta, Z.A.; Biryukov, S.; Brauer, M. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1659–1724. [Google Scholar] [CrossRef]
- Broeders, E.N.; Knoop, C.; Abramowicz, D. Drug treatment of lipid disorders. N. Engl. J. Med. 1999, 341, 2020. [Google Scholar]
- Kobashigawa, J.; Kasiske, B.J.T. Hyperlipidemia in solid organ transplantation. Liver Transplant. Surg. 1997, 63, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Goli, A.K.; Goli, S.A.; Byrd, R.P., Jr.; Roy, T.M. Simvastatin-induced lactic acidosis: A rare adverse reaction? Clin. Pharmacol. Ther. 2002, 72, 461–464. [Google Scholar] [CrossRef]
- Wang, L.; Fan, W.; Zhang, M.; Zhang, Q.; Li, L.; Wang, J.; Zhu, L.; Wei, D.; Peng, W.; Wu, C. Antiobesity, Regulation of Lipid Metabolism, and Attenuation of Liver Oxidative Stress Effects of Hydroxy-alpha-sanshool Isolated from Zanthoxylum bungeanum on High-Fat Diet-Induced Hyperlipidemic Rats. Oxid. Med. Cell Longev. 2019, 2019, 5852494. [Google Scholar] [CrossRef]
- Liu, A.; Wu, Q.; Guo, J.; Ares, I.; Rodriguez, J.L.; Martinez-Larranaga, M.R.; Yuan, Z.; Anadon, A.; Wang, X.; Martinez, M.A. Statins: Adverse reactions, oxidative stress and metabolic interactions. Pharmacol. Ther. 2019, 195, 54–84. [Google Scholar] [CrossRef]
- Okopien, B.; Buldak, L.; Boldys, A. Current and future trends in the lipid lowering therapy. Pharmacol. Rep. 2016, 68, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Istvan, E.; Deisenhofer, J.J.S. Structural mechanism for statin inhibition of HMG-CoA reductase. Science 2001, 292, 1160–1164. [Google Scholar] [CrossRef]
- Dehnavi, S.; Sohrabi, N.; Sadeghi, M.; Lansberg, P.; Banach, M.; Al-Rasadi, K.; Johnston, T.P.; Sahebkar, A. Statins and autoimmunity: State-of-the-art. Pharmacol. Ther. 2020, 214, 107614. [Google Scholar] [CrossRef]
- Cote, D.J.; Rosner, B.A.; Smith-Warner, S.A.; Egan, K.M.; Stampfer, M.J. Statin use, hyperlipidemia, and risk of glioma. Eur. J. Epidemiol. 2019, 34, 997–1011. [Google Scholar] [CrossRef]
- Oesterle, A.; Laufs, U.; Liao, J.K. Pleiotropic Effects of Statins on the Cardiovascular System. Circ. Res. 2017, 120, 229–243. [Google Scholar] [CrossRef]
- Holmes, M.V.; Ala-Korpela, M. What is ‘LDL cholesterol’? Nat. Rev. Cardiol. 2019, 16, 197–198. [Google Scholar] [CrossRef]
- Lewington, S.; Whitlock, G.; Clarke, R.; Sherliker, P.; Emberson, J.; Halsey, J.; Qizilbash, N.; Peto, R.; Collins, R.J.L. Blood cholesterol and vascular mortality by age, sex, and blood pressure: A meta-analysis of individual data from 61 prospective studies with 55,000 vascular deaths. NIH Public Access 2007, 370, 1829–1839. [Google Scholar]
- Haberland, M.; Mottino, G.; Le, M.; Frank, J.S. Sequestration of aggregated LDL by macrophages studied with freeze-etch electron microscopy. J. Lipid Res. 2001, 42, 605–619. [Google Scholar] [CrossRef]
- Hofmann, A.; Brunssen, C.; Morawietz, H. Contribution of lectin-like oxidized low-density lipoprotein receptor-1 and LOX-1 modulating compounds to vascular diseases. Vasc. Pharmacol. 2017, 107, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Younis, N.; Charlton-Menys, V.; Sharma, R.; Soran, H.; Durrington, P.J.A. Glycation of LDL in non-diabetic people: Small dense LDL is preferentially glycated both in vivo and in vitro. Atherosclerosis 2009, 202, 162–168. [Google Scholar] [CrossRef]
- Borén, J.; Williams, K.J. The central role of arterial retention of cholesterol-rich apolipoprotein-B-containing lipoproteins in the pathogenesis of atherosclerosis: A triumph of simplicity. Curr. Opin. Lipidol. 2016, 27, 473–483. [Google Scholar] [CrossRef]
- Voight, B.; Peloso, G.; Orho-Melander, M.; Frikke-Schmidt, R.; Barbalic, M.; Jensen, M.; Hindy, G. Plasma HDL cholesterol and risk of myocardial infarction: A mendelian randomisation study. Lancet 2012, 380, 572–580. [Google Scholar] [CrossRef]
- Adams, S.P.; Tsang, M.; Wright, J.M. Lipid-lowering efficacy of atorvastatin. Cochrane Database Syst. Rev. 2015, 12, 1–460. [Google Scholar] [CrossRef]
- Ward, N.C.; Watts, G.F.; Eckel, R.H. Statin Toxicity. Circ. Res. 2019, 124, 328–350. [Google Scholar] [CrossRef]
- Ward, N.C.; Pang, J.; Ryan, J.D.M.; Watts, G.F. Nutraceuticals in the management of patients with statin-associated muscle symptoms, with a note on real-world experience. Clin. Cardiol. 2018, 41, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Du Souich, P.; Roederer, G.; Dufour, R. Myotoxicity of statins: Mechanism of action. Pharmacol. Ther. 2017, 175, 1–16. [Google Scholar] [CrossRef]
- Chang, C.; Chang, Y.; Lee, Y.; Liu, Y.; Chuang, L.; Lin, J.W. Severe hepatic injury associated with different statins in patients with chronic liver disease: A nationwide population-based cohort study. J. Gastroenterol. Hepatol. 2015, 30, 155–162. [Google Scholar] [CrossRef]
- Stapleton, P.; Goodwill, A.; James, M.; Brock, R.; Frisbee, J.C. Hypercholesterolemia and microvascular dysfunction: Interventional strategies. J. Inflamm. 2010, 7, 54. [Google Scholar] [CrossRef]
- Thompson, G.; Catapano, A.; Saheb, S.; Atassi-Dumont, M.; Barbir, M.; Eriksson, M.; Paulweber, B.; Sijbrands, E.; Stalenhoef, A.; Parhofer, K.G. Severe hypercholesterolaemia: Therapeutic goals and eligibility criteria for LDL apheresis in Europe. Curr. Opin. Lipidol. 2010, 21, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Reiner, Ž. Hypertriglyceridaemia and risk of coronary artery disease. Nat. Rev. Cardiol. 2017, 14, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, R.; Hu, N.; Yu, C.; Song, H.; Li, Y.; Dai, Y.; Guo, Z.; Li, M.; Zheng, Y.; et al. Baihe Wuyao decoction ameliorates CCl-induced chronic liver injury and liver fibrosis in mice through blocking TGF-β1/Smad2/3 signaling, anti-inflammation and anti-oxidation effects. J. Ethnopharmacol. 2020, 263, 113227. [Google Scholar] [CrossRef] [PubMed]
- Derosa, G.; Sahebkar, A.; Maffioli, P. The role of various peroxisome proliferator-activated receptors and their ligands in clinical practice. J. Cell. Physiol. 2018, 233, 153–161. [Google Scholar] [CrossRef]
- Kim, S.; Brown, D.; Jester, J.V. Transcriptome analysis after PPARγ activation in human meibomian gland epithelial cells (hMGEC). Ocul. Surf. 2019, 17, 809–816. [Google Scholar] [CrossRef]
- Wang, Y.X. PPARs: Diverse regulators in energy metabolism and metabolic diseases. Cell Res. 2010, 20, 124–137. [Google Scholar] [CrossRef]
- Juge-Aubry, C.; Gorla-Bajszczak, A.; Pernin, A.; Lemberger, T.; Wahli, W.; Burger, A.; Meier, C.A. Peroxisome proliferator-activated receptor mediates cross-talk with thyroid hormone receptor by competition for retinoid X receptor. Possible role of a leucine zipper-like heptad repeat. J. Biol. Chem. 1995, 270, 18117–18122. [Google Scholar] [CrossRef]
- Braissant, O.; Foufelle, F.; Scotto, C.; Dauça, M.; Wahli, W.J.E. Differential expression of peroxisome proliferator-activated receptors (PPARs): Tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinplogy 1996, 137, 354–366. [Google Scholar] [CrossRef]
- Robinson-Rechavi, M.; Carpentier, A.; Duffraisse, M.; Laudet, V. How many nuclear hormone receptors are there in the human genome? Trends Genet. 2001, 17, 554–556. [Google Scholar] [CrossRef]
- Robinson-Rechavi, M.; Marchand, O.; Escriva, H.; Bardet, P.; Zelus, D.; Hughes, S.; Laudet, V. Euteleost fish genomes are characterized by expansion of gene families. Genome Res. 2001, 11, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Baumrucker, C.; Shirk, E.; Vanden Heuvel, J.; Block, E.; Varga, G.A. Characterization of Madin-Darby bovine kidney cell line for peroxisome proliferator activated receptors: Temporal response and sensitivity to fatty acids. J. Dairy Sci. 2008, 91, 2808–2813. [Google Scholar] [CrossRef] [PubMed]
- Harano, Y.; Yasui, K.; Toyama, T.; Nakajima, T.; Mitsuyoshi, H.; Mimani, M.; Hirasawa, T.; Itoh, Y.; Okanoue, T. Fenofibrate, a peroxisome proliferator-activated receptor alpha agonist, reduces hepatic steatosis and lipid peroxidation in fatty liver Shionogi mice with hereditary fatty liver. Liver Int. 2006, 26, 613–620. [Google Scholar] [CrossRef]
- Mancini, F.; Lanni, A.; Sabatino, L.; Moreno, M.; Giannino, A.; Contaldo, F.; Colantuoni, V.; Goglia, F. Fenofibrate prevents and reduces body weight gain and adiposity in diet-induced obese rats. Fed. Eur. Biochem. Soc. 2001, 491, 154–158. [Google Scholar] [CrossRef]
- Staels, B.; Dallongeville, J.; Auwerx, J.; Schoonjans, K.; Leitersdorf, E.; Fruchart, J.J.C. Mechanism of action of fibrates on lipid and lipoprotein metabolism. Cardiovasc. Drugs 1998, 98, 2088–2093. [Google Scholar] [CrossRef]
- Kersten, S. Integrated physiology and systems biology of PPARα. Mol. Metab. 2014, 3, 354–371. [Google Scholar] [CrossRef]
- Forest, C.; Tordjman, J.; Glorian, M.; Duplus, E.; Chauvet, G.; Quette, J.; Beale, E.G.; Antoine, B. Fatty acid recycling in adipocytes: A role for glyceroneogenesis and phosphoenolpyruvate carboxykinase. Biochem. Soc. Trans. 2003, 31, 1125–1129. [Google Scholar] [CrossRef]
- Medina-Gomez, G.; Gray, S.; Vidal-Puig, A. Adipogenesis and lipotoxicity: Role of peroxisome proliferator-activated receptor gamma (PPARgamma) and PPARgammacoactivator-1 (PGC1). Public Health Nutr. 2007, 10, 1132–1137. [Google Scholar] [CrossRef]
- Lavoie, B.; Nausch, B.; Zane, E.; Leonard, M.; Balemba, O.; Bartoo, A.; Wilcox, R.; Nelson, M.; Carey, M.; Mawe, G.M. Disruption of gallbladder smooth muscle function is an early feature in the development of cholesterol gallstone disease. Neurogastroenterol. Motil. 2012, 24, e313–e324. [Google Scholar] [CrossRef]
- Assy, N.; Kaita, K.; Mymin, D.; Levy, C.; Rosser, B.; Minuk, G. Fatty infiltration of liver in hyperlipidemic patients. Dig. Dis. Sci. 2000, 45, 1929–1934. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.P. Serum triglycerides, the liver and the pancreas. Hyperlipid. Cardiovasc. Dis. 2000, 11, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Balazs, Z.; Panzenboeck, U.; Hammer, A.; Sovic, A.; Quehenberger, O.; Malle, E.; Sattler, W. Uptake and transport of high-density lipoprotein (HDL) and HDL-associated alpha-tocopherol by an in vitro blood-brain barrier model. J. Neurochem. 2004, 89, 939–950. [Google Scholar] [CrossRef] [PubMed]
- Beckstead, J.; Oda, M.; Martin, D.; Forte, T.; Bielicki, J.; Berger, T.; Luty, R.; Kay, C.; Ryan, R.J.B. Structure-function studies of human apolipoprotein A-V: A regulator of plasma lipid homeostasis. Biochemistry 2003, 42, 9416–9423. [Google Scholar] [CrossRef][Green Version]
- Yang, Y.; Thyagarajan, N.; Coady, B.; Brown, R.J. Cholesterol efflux from THP-1 macrophages is impaired by the fatty acid component from lipoprotein hydrolysis by lipoprotein lipase. Biochem. Biophys. Res. Commun. 2014, 451, 632–636. [Google Scholar] [CrossRef]
- Brun, R.; Tontonoz, P.; Forman, B.; Ellis, R.; Chen, J.; Evans, R.; Spiegelman, B.M. Differential activation of adipogenesis by multiple PPAR isoforms. Genes Dev. 1996, 10, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Jay, A.; Brunaldi, K.; Huang, N.; Hamilton, J.J.B. CD36 enhances fatty acid uptake by increasing the rate of intracellular esterification but not transport across the plasma membrane. Biochemistry 2013, 52, 7254–7261. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, I.J.; Eckel, R.H.; Abumrad, N.A. Regulation of fatty acid uptake into tissues: Lipoprotein lipase- and CD36-mediated pathways. J. Lipid Res. 2009, 50, S86–S90. [Google Scholar] [CrossRef]
- Greenwalt, D.E.; Lipsky, R.H.; Ockenhouse, C.F.; Ikeda, H.; Tandon, N.N.; Jamieson, G.A. Membrane glycoprotein CD36: A review of its roles in adherence, signal transduction, and transfusion medicine. Blood 1992, 80, 1105–1115. [Google Scholar] [CrossRef]
- Han, J.; Hajjar, D.P.; Zhou, X.; Gotto, A.M.; Nicholson, A.C. Regulation of PPARγ-Mediated Gene Expression:A New Mechanism of Action for High Density Lipoprotein. J. Biol. Chem. 2002, 277, 23582–23586. [Google Scholar] [CrossRef]
- Kimak, E.; Bylina, J.; Solski, J.; Hałabiś, M.; Baranowicz-Gąszczyk, I.; Książek, A. Association between lipids, lipoproteins composition of HDL particles and triglyceride-rich lipoproteins, and LCAT and CETP activity in post-renal transplant patients. Cell Biochem. Biophys. 2013, 67, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Niño, H.; García-Pintos, I.; Rodríguez-Borges, J.; Escobar-Cubiella, M.; García-Mera, X.; Prado-Prado, F. Review of synthesis, biological assay and QSAR studies of β-secretase inhibitors. Curr. Comput. Aided Drug Des. 2011, 7, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Vock, C.; Döring, F.; Nitz, I. Transcriptional Regulation of HMG-CoA Synthase and HMG-CoA Reductase Genes by Human ACBP. Cell. Physiol. Biochem. 2008, 22, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yang, Q.; Li, J.; Tian, Y.; Chen, F.; Lu, L. Molecular Cloning and Expression of Goose(Anser anser) 3-hydroxy-3-methylglutaryl Coenzyme A Synthase Gene(HMGCS1) in Overfeeding Landes Goose Liver. J. Agric. Biotechnol. 2014, 22, 8. [Google Scholar]








| Nutrient Composition | Per 1001.54 g (g) | Nutrient Reference Value |
|---|---|---|
| Casein, 80 mesh | 195.00 | 19.47 |
| DL Methionine | 3.00 | 0.30% |
| Corn starch | 50.00 | 4.99% |
| Maltodextrin | 100.00 | 9.98% |
| Sucrose | 341.00 | 34.05% |
| Cellulose | 50.00 | 4.99% |
| Anhydrous milk fat | 200.00 | 19.97% |
| Mineral mixture s10001 | 35.0 | 3.49% |
| Calcium carbonate | 4.00 | 0.40% |
| Vitamin mixture v10001 | 10.00 | 1.00% |
| Choline tartrate | 2.00 | 0.20% |
| Cholesterol | 1.50 | 0.15% |
| Ethoxyquine | 0.04 | 0.00% |
| Nutrient Composition | Per 1000 g (g) | Nutrient Reference Value |
|---|---|---|
| Water content | ≤100 | ≤10% |
| Crude protein | ≥180 | ≥18% |
| Crude fat | ≥40 | ≥4% |
| Crude fiber | ≤50 | ≤5% |
| Coarse ash | ≤80 | ≤8% |
| Calcium | 10–18 | 1–1.8% |
| Phosphorus | 6–12 | 0.6–1.2% |
| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| CD36 | AACATCGAGTGTCGAATATGTGG | CCGAATAGTTCGCCGAAAGAA |
| HMGCS1 | TGAACTGGGTCGAATCCAGC | CCTGTAGGTCTGGCATTTCCT |
| PPARα | AACATCGAGTGTCGAATATGTGG | CCGAATAGTTCGCCGAAAGAA |
| PPARγ | TCGCTGATGCACTGCCTATG | GAGAGGTCCACAGAGCTGATT |
| LPL | GGGAGTTTGGCTCCAGAGTTT | TGTGTCTTCAGGGGTCCTTAG |
| GAPDH | AGGTCGGTGTGAACGGATTTG | GGGGTCGTTGATGGCAACA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, N.; Chen, C.; Wang, J.; Huang, J.; Yao, D.; Li, C. Atorvastatin Ester Regulates Lipid Metabolism in Hyperlipidemia Rats via the PPAR-signaling Pathway and HMGCR Expression in the Liver. Int. J. Mol. Sci. 2021, 22, 11107. https://doi.org/10.3390/ijms222011107
Hu N, Chen C, Wang J, Huang J, Yao D, Li C. Atorvastatin Ester Regulates Lipid Metabolism in Hyperlipidemia Rats via the PPAR-signaling Pathway and HMGCR Expression in the Liver. International Journal of Molecular Sciences. 2021; 22(20):11107. https://doi.org/10.3390/ijms222011107
Chicago/Turabian StyleHu, Nan, Chunyun Chen, Jinhui Wang, Jian Huang, Dahong Yao, and Chunli Li. 2021. "Atorvastatin Ester Regulates Lipid Metabolism in Hyperlipidemia Rats via the PPAR-signaling Pathway and HMGCR Expression in the Liver" International Journal of Molecular Sciences 22, no. 20: 11107. https://doi.org/10.3390/ijms222011107
APA StyleHu, N., Chen, C., Wang, J., Huang, J., Yao, D., & Li, C. (2021). Atorvastatin Ester Regulates Lipid Metabolism in Hyperlipidemia Rats via the PPAR-signaling Pathway and HMGCR Expression in the Liver. International Journal of Molecular Sciences, 22(20), 11107. https://doi.org/10.3390/ijms222011107






