Structure-Based Functional Analysis of a Hormone Belonging to an Ecdysozoan Peptide Superfamily: Revelation of a Common Molecular Architecture and Residues Possibly for Receptor Interaction
Abstract
:1. Introduction
2. Results
2.1. Sco-CHH-L and Sco-CHH Transcript, and Protein Expression Levels in the Pericardial Organs
2.2. Pattern of Changes in Sco-CHH-L and Sco-CHH Transcript Levels under Osmotic Stresses
2.3. Characterization of Wild-Type and Mutated Sco-CHH-L Peptides
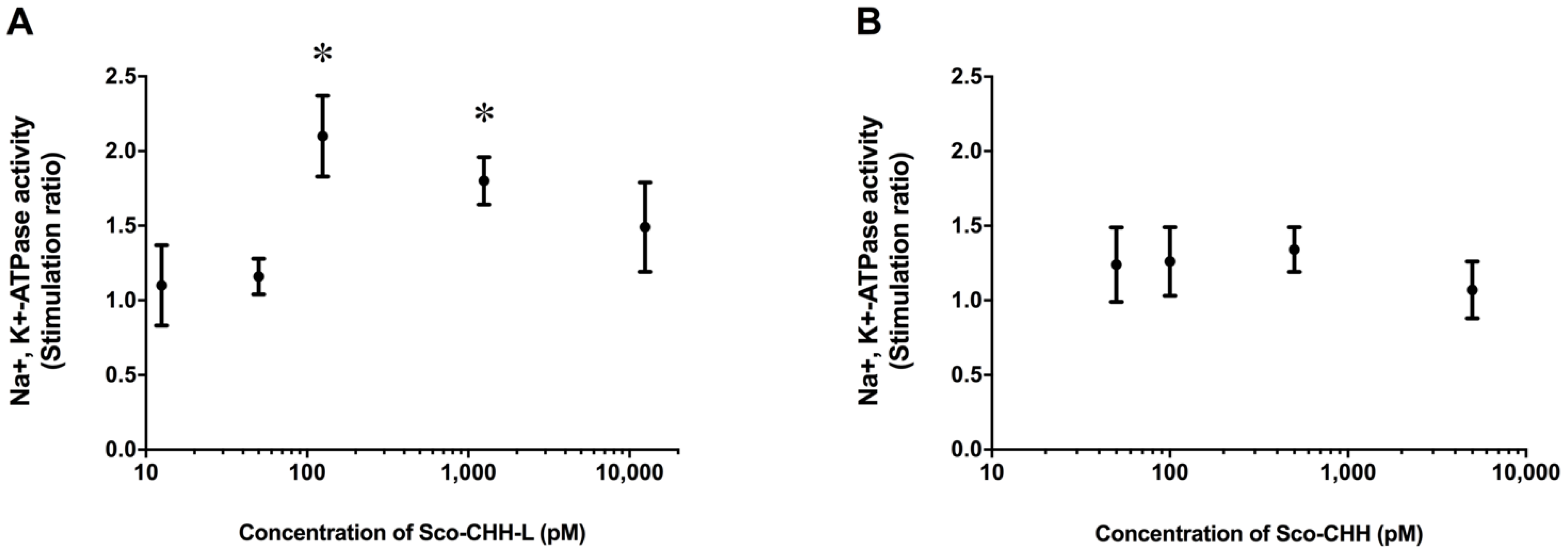
2.4. Effects of Sco-CHH and Sco-CHH-L on Na+, K+-ATPase Activity in Posterior Gills
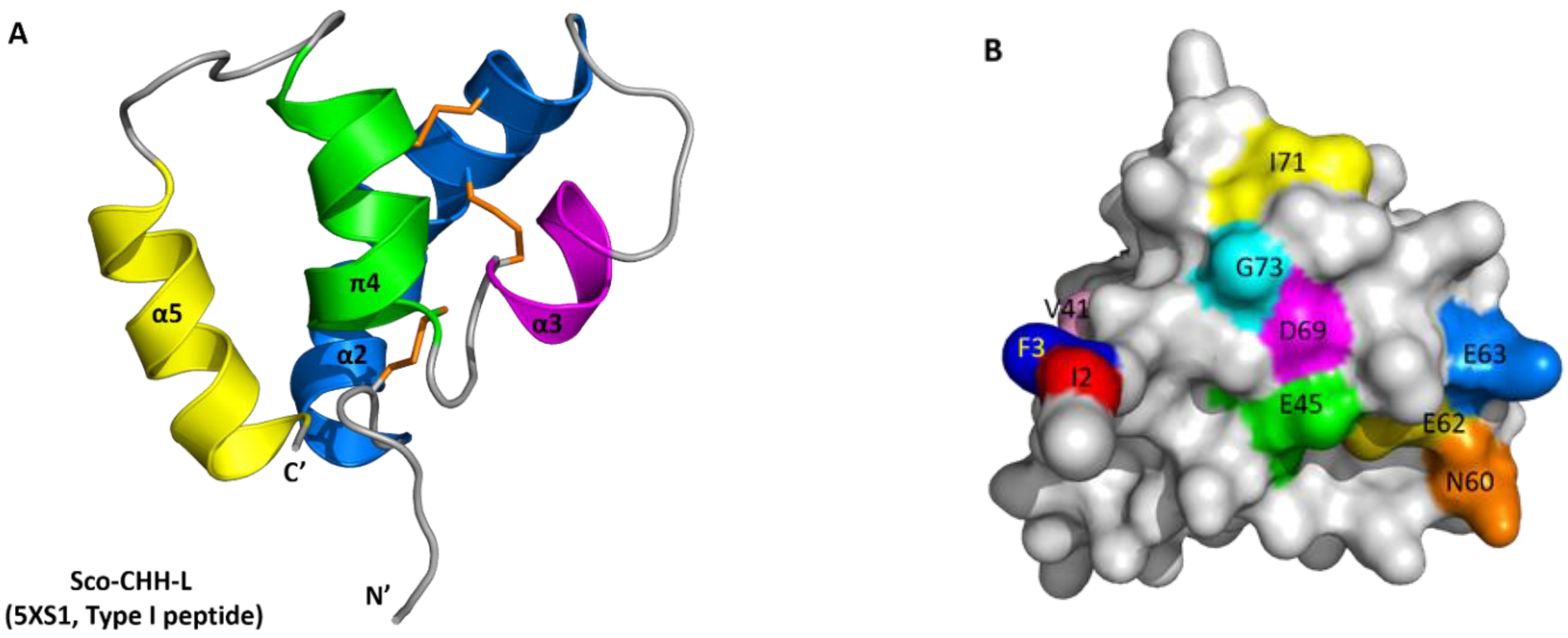
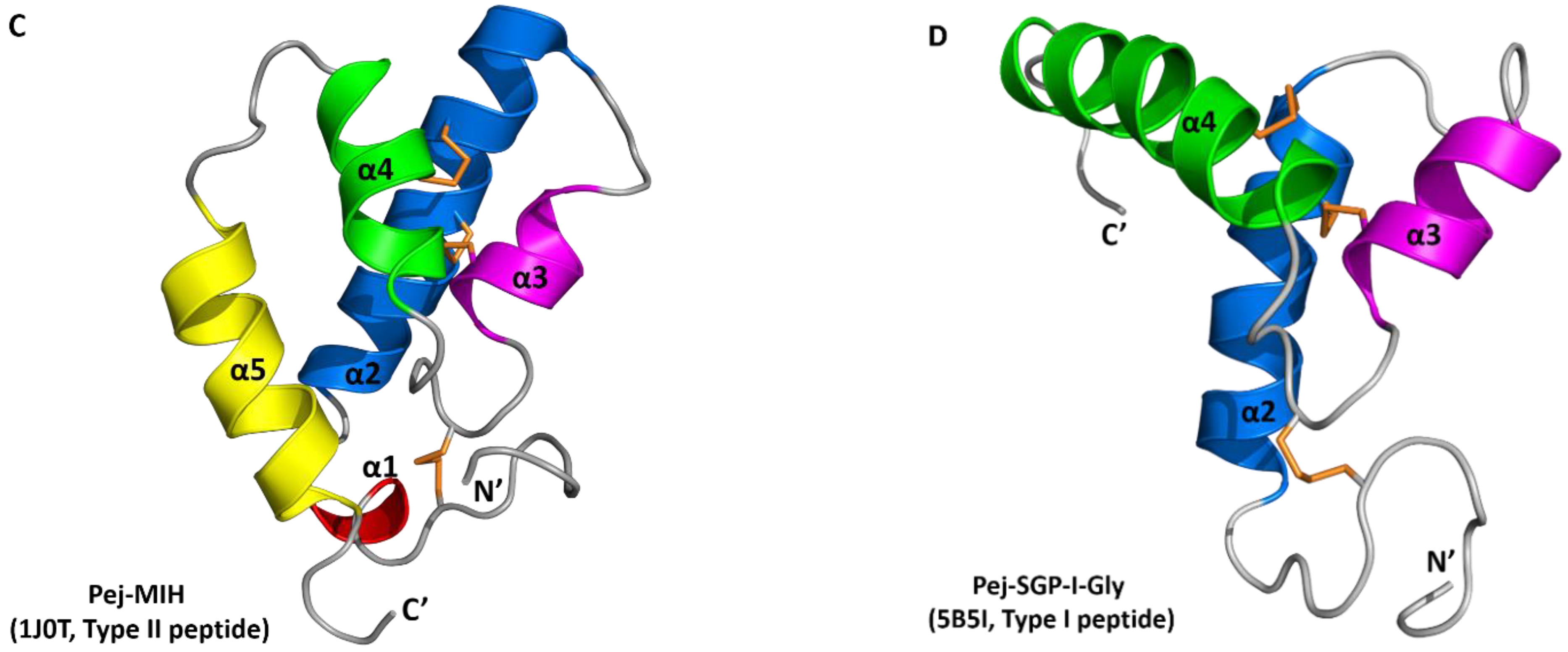
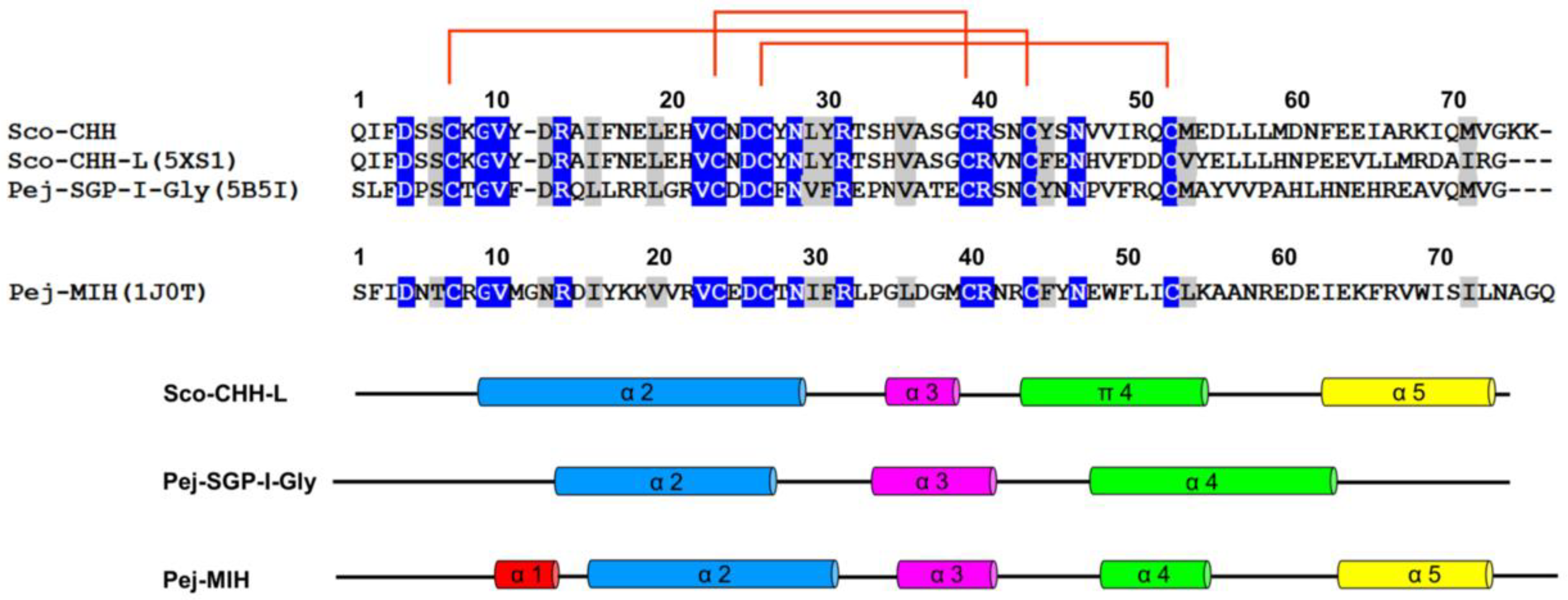
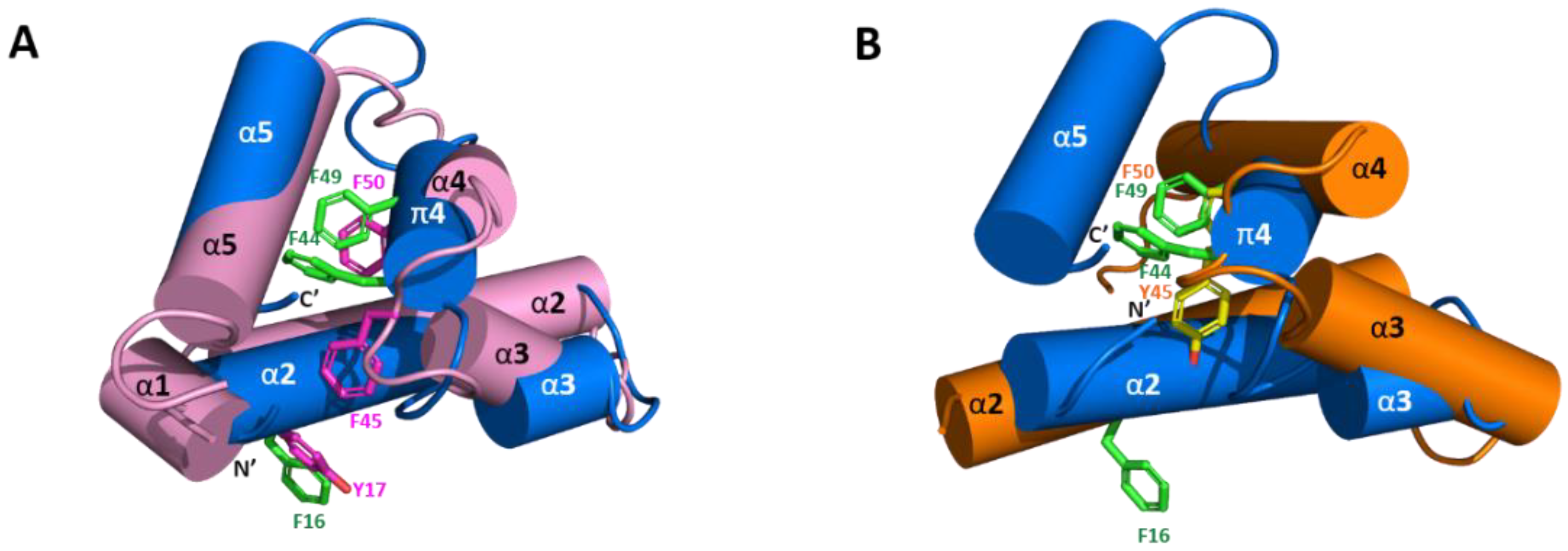
2.5. Solution Structure of Sco-CHH-L
2.6. Functionally Important Residues of Sco-CHH-L
3. Discussion
4. Materials and Methods
4.1. Animal Maintenance
4.2. ELISA for the Quantitative Analysis of Sco-CHH-L and Sco-CHH in Pericardial Organs
4.3. Gene Expression under Osmotic Stresses
4.4. Construction of Expression Plasmids and Production of Recombinant Peptides
4.5. Circular Dichroism Spectral Analysis of Recombinant Wild-Type and Alanine-Substituted Sco-CHH-L
4.6. Effects of Wild-Type Peptides and Mutated Sco-CHH-L Peptides on Na+, K+-ATPase Activity
4.7. Nuclear Magnetic Resonance Spectroscopy and Structure Determination
4.8. Data and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lacombe, C.; Greve, P.; Martin, G. Overview on the sub-grouping of the crustacean hyperglycemic hormone family. Neuropeptides 1999, 33, 71–80. [Google Scholar] [CrossRef]
- Wang, Y.J.; Zhao, Y.; Meredith, J.; Phillips, J.E.; Theilmann, D.A.; Brock, H.W. Mutational analysis of the C-terminus in ion transport peptide (ITP) expressed in Drosophila Kc1 cells. Arch. Insect Biochem. Physiol. 2000, 45, 129–138. [Google Scholar] [CrossRef]
- Chan, S.M.; Gu, P.L.; Chu, K.H.; Tobe, S.S. Crustacean neuropeptide genes of the CHH/MIH/GIH family: Implications from molecular studies. Gen. Comp. Endocrinol. 2003, 134, 214–219. [Google Scholar] [CrossRef]
- Katayama, H.; Ohira, T.; Nagata, S.; Nagasawa, H. Structure-activity relationship of crustacean molt-inhibiting hormone from the kuruma prawn Marsupenaeus japonicus. Biochemistry 2004, 43, 9629–9635. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Meredith, J.; Brock, H.W.; Phillips, J.E. Mutational analysis of the N-terminus in Schistocerca gregaria ion-transport peptide expressed in Drosophila Kc1 cells. Arch. Insect Biochem. Physiol. 2005, 58, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Padhi, A.; Verghese, B.; Otta, S.K.; Varghese, B.; Ramu, K. Positive Darwinian selection on crustacean hyperglycemic hormone (CHH) of the green shore crab, Carcinus maenas. In Silico Biol. 2007, 7, 355–367. [Google Scholar] [PubMed]
- Montagné, N.; Desdevises, Y.; Soyez, D.; Toullec, J.-Y. Molecular evolution of the crustacean hyperglycemic hormone family in ecdysozoans. BMC Evol. Biol. 2010, 10, 62. [Google Scholar] [CrossRef] [Green Version]
- McCowan, C.; Garb, J.E. Recruitment and diversification of an ecdysozoan family of neuropeptide hormones for black widow spider venom expression. Gene 2014, 536, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Huang, S.S.; Toullec, J.Y.; Chang, C.Y.; Chen, Y.R.; Huang, W.S.; Lee, C.Y. Functional Assessment of Residues in the Amino- and Carboxyl-Termini of Crustacean Hyperglycemic Hormone (CHH) in the Mud Crab Scylla olivacea Using Point-Mutated Peptides. PLoS ONE 2015, 10, e0134983. [Google Scholar] [CrossRef] [Green Version]
- Undheim, E.A.; Grimm, L.L.; Low, C.F.; Morgenstern, D.; Herzig, V.; Zobel-Thropp, P.; Pineda, S.S.; Habib, R.; Dziemborowicz, S.; Fry, B.G.; et al. Weaponization of a Hormone: Convergent Recruitment of Hyperglycemic Hormone into the Venom of Arthropod Predators. Structure 2015, 23, 1283–1292. [Google Scholar] [CrossRef] [Green Version]
- Webster, S.G.; Keller, R.; Dircksen, H. The CHH-superfamily of multifunctional peptide hormones controlling crustacean metabolism, osmoregulation, moulting, and reproduction. Gen. Comp. Endocrinol. 2012, 175, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Kleinholz, L.H. Crustacean Neurosecretory Hormones and Physiological Specificity. Am. Zool. 1976, 16, 151–166. [Google Scholar] [CrossRef] [Green Version]
- Kegel, G.; Reichwein, B.; Tensen, C.P.; Keller, R. Amino acid sequence of crustacean hyperglycemic hormone (CHH) from the crayfish, Orconectes limosus: Emergence of a novel neuropeptide family. Peptides 1991, 12, 909–913. [Google Scholar] [CrossRef]
- Keller, R. Crustacean neuropeptides: Structures, functions and comparative aspects. Experientia 1992, 48, 439–448. [Google Scholar] [CrossRef]
- Soyez, D. Occurrence and diversity of neuropeptides from the crustacean hyperglycemic hormone family in arthropods. A short review. Ann. N. Y. Acad. Sci. 1997, 814, 319–323. [Google Scholar] [CrossRef]
- Meredith, J.; Ring, M.; Macins, A.; Marschall, J.; Cheng, N.N.; Theilmann, D.; Brock, H.W.; Phillips, J.E. Locust ion transport peptide (ITP): Primary structure, cDNA and expression in a baculovirus system. J. Exp. Biol. 1996, 199, 1053–1061. [Google Scholar] [CrossRef]
- Christie, A.E. Neuropeptide discovery in Ixodoidea: An in silico investigation using publicly accessible expressed sequence tags. Gen. Comp. Endocrinol. 2008, 157, 174–185. [Google Scholar] [CrossRef]
- Christie, A.E.; Nolan, D.H.; Garcia, Z.A.; McCoole, M.D.; Harmon, S.M.; Congdon-Jones, B.; Ohno, P.; Hartline, N.; Congdon, C.B.; Baer, K.N.; et al. Bioinformatic prediction of arthropod/nematode-like peptides in non-arthropod, non-nematode members of the Ecdysozoa. Gen. Comp. Endocrinol. 2011, 170, 480–486. [Google Scholar] [CrossRef]
- Santos, E.A.; Keller, R. Crustacean hyperglycemic hormone (CHH) and the regulation of carbohydrate metabolism: Current perspectives. Comp. Biochem. Physiol. Part A Physiol. 1993, 106, 405–411. [Google Scholar] [CrossRef]
- Li, W.; Chiu, K.H.; Tien, Y.C.; Tsai, S.F.; Shih, L.J.; Lee, C.H.; Toullec, J.Y.; Lee, C.Y. Differential effects of silencing crustacean hyperglycemic hormone gene expression on the metabolic profiles of the muscle and hepatopancreas in the crayfish Procambarus clarkii. PLoS ONE 2017, 12, e0172557. [Google Scholar] [CrossRef]
- Li, W.; Chiu, K.H.; Lee, C.Y. Regulation of amino acid and nucleotide metabolism by crustacean hyperglycemic hormone in the muscle and hepatopancreas of the crayfish Procambarus clarkia. PLoS ONE 2019, 14, e0221745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dircksen, H.; Bocking, D.; Heyn, U.; Mandel, C.; Chung, J.S.; Baggerman, G.; Verhaert, P.; Daufeldt, S.; Plosch, T.; Jaros, P.P.; et al. Crustacean hyperglycaemic hormone (CHH)-like peptides and CHH-precursor-related peptides from pericardial organ neurosecretory cells in the shore crab, Carcinus maenas, are putatively spliced and modified products of multiple genes. Biochem. J. 2001, 356, 159–170. [Google Scholar] [CrossRef]
- Ohira, T.; Tsutsui, N.; Nagasawa, H.; Wilder, M.N. Preparation of two recombinant crustacean hyperglycemic hormones from the giant freshwater prawn, Macrobrachium rosenbergii, and their hyperglycemic activities. Zool. Sci. 2006, 23, 383–391. [Google Scholar] [CrossRef]
- Chang, C.C.; Tsai, K.W.; Hsiao, N.W.; Chang, C.Y.; Lin, C.L.; Watson, R.D.; Lee, C.Y. Structural and functional comparisons and production of recombinant crustacean hyperglycemic hormone (CHH) and CHH-like peptides from the mud crab Scylla olivacea. Gen. Comp. Endocrinol. 2010, 167, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Katayama, H.; Nagata, K.; Ohira, T.; Yumoto, F.; Tanokura, M.; Nagasawa, H. The solution structure of molt-inhibiting hormone from the Kuruma prawn Marsupenaeus japonicus. J. Biol. Chem. 2003, 278, 9620–9623. [Google Scholar] [CrossRef] [Green Version]
- Tsutsui, N.; Sakamoto, T.; Arisaka, F.; Tanokura, M.; Nagasawa, H.; Nagata, K. Crystal structure of a crustacean hyperglycemic hormone (CHH) precursor suggests structural variety in the C-terminal regions of CHH superfamily members. FEBS J. 2016, 283, 4325–4339. [Google Scholar] [CrossRef] [PubMed]
- Carlisle, D.B.; Knowles, F.G.W. Neurohæmal Organs in Crustaceans. Nature 1953, 172, 404–405. [Google Scholar] [CrossRef]
- Cooke, I.M.; Sullivan, R.E. Hormones and neurosecretion. In The Biology of Crustacea; Atwood, H.L., Sandeman, D.C., Eds.; Academic Press: New York, NY, USA, 1982; Volume 3. [Google Scholar]
- Christie, A.E.; Skiebe, P.; Marder, E. Matrix of neuromodulators in neurosecretory structures of the crab Cancer borealis. J. Exp. Biol. 1995, 198, 2431–2439. [Google Scholar] [CrossRef]
- Kamemoto, F.I. Neuroendocrinology of Osmoregulation in Decapod Crustacea. Am. Zool. 1976, 16, 141–150. [Google Scholar] [CrossRef]
- Tsai, K.W.; Chang, S.J.; Wu, H.J.; Shih, H.Y.; Chen, C.H.; Lee, C.Y. Molecular cloning and differential expression pattern of two structural variants of the crustacean hyperglycemic hormone family from the mud crab Scylla olivacea. Gen. Comp. Endocrinol. 2008, 159, 16–25. [Google Scholar] [CrossRef]
- Aurora, R.; Srinivasan, R.; Rose, G.D. Rules for alpha-helix termination by glycine. Science 1994, 264, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-Y.; Tsai, K.-W.; Tsai, W.-S.; Jiang, J.-Y.; Chen, Y.-J. Crustacean hyperglycemic hormone: Structural variants, physiological function, and cellular mechanism of action. J. Mar. Sci. Technol. 2014, 22, 75–81. [Google Scholar] [CrossRef]
- Yasuda, A.; Yasuda, Y.; Fujita, T.; Naya, Y. Characterization of crustacean hyperglycemic hormone from the crayfish (Procambarus clarkii): Multiplicity of molecular forms by stereoinversion and diverse functions. Gen. Comp. Endocrinol. 1994, 95, 387–398. [Google Scholar] [CrossRef]
- Chung, J.S.; Webster, S.G. Moult cycle-related changes in biological activity of moult-inhibiting hormone (MIH) and crustacean hyperglycaemic hormone (CHH) in the crab, Carcinus maenas. From target to transcript. Eur. J. Biochem. 2003, 270, 3280–3288. [Google Scholar] [CrossRef]
- Chang, E.S.; Prestwich, G.D.; Bruce, M.J. Amino acid sequence of a peptide with both molt-inhibiting and hyperglycemic activities in the lobster, Homarus americanus. Biochem. Biophys. Res. Commun. 1990, 171, 818–826. [Google Scholar] [CrossRef]
- Sefiani, M.; Le Caer, J.P.; Soyez, D. Characterization of hyperglycemic and molt-inhibiting activity from sinus glands of the penaeid shrimp Penaeus vannamei. Gen. Comp. Endocrinol. 1996, 103, 41–53. [Google Scholar] [CrossRef]
- Sommer, M.J.; Mantel, L.H. Effect of dopamine, cyclic AMP, and pericardial organs on sodium uptake and Na/K-ATPase activity in gills of the green crab Carcinus maenas (L). J. Exp. Zool. Part A Ecol. Genet. Physiol. 1988, 248, 272–277. [Google Scholar] [CrossRef]
- Serrano, L.; Blanvillain, G.; Soyez, D.; Charmantier, G.; Grousset, E.; Aujoulat, F.; Spanings-Pierrot, C. Putative involvement of crustacean hyperglycemic hormone isoforms in the neuroendocrine mediation of osmoregulation in the crayfish Astacus leptodactylus. J. Exp. Biol. 2003, 206, 979–988. [Google Scholar] [CrossRef] [Green Version]
- Charmantier-Daures, M.; Charmantier, G.; Janssen, K.P.; Aiken, D.E.; van Herp, F. Involvement of eyestalk factors in the neuroendocrine control of osmoregulation in adult American lobster Homarus americanus. Gen. Comp. Endocrinol. 1994, 94, 281–293. [Google Scholar] [CrossRef]
- Spanings-Pierrot, C.; Soyez, D.; Van Herp, F.; Gompel, M.; Skaret, G.; Grousset, E.; Charmantier, G. Involvement of crustacean hyperglycemic hormone in the control of gill ion transport in the crab Pachygrapsus marmoratus. Gen. Comp. Endocrinol. 2000, 119, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Prymaczok, N.C.; Pasqualino, V.M.; Viau, V.E.; Rodriguez, E.M.; Medesani, D.A. Involvement of the crustacean hyperglycemic hormone (CHH) in the physiological compensation of the freshwater crayfish Cherax quadricarinatus to low temperature and high salinity stress. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2016, 186, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.S.; Dircksen, H.; Webster, S.G. A remarkable, precisely timed release of hyperglycemic hormone from endocrine cells in the gut is associated with ecdysis in the crab Carcinus maenas. Proc. Natl. Acad. Sci. USA 1999, 96, 13103–13107. [Google Scholar] [CrossRef] [Green Version]
- Chung, J.S.; Webster, S.G. Binding sites of crustacean hyperglycemic hormone and its second messengers on gills and hindgut of the green shore crab, Carcinus maenas: A possible osmoregulatory role. Gen. Comp. Endocrinol. 2006, 147, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Turner, L.M.; Webster, S.G.; Morris, S. Roles of crustacean hyperglycaemic hormone in ionic and metabolic homeostasis in the Christmas Island blue crab, Discoplax celeste. J. Exp. Biol. 2013, 216, 1191–1201. [Google Scholar] [CrossRef] [Green Version]
- Sun, D.; Lv, J.; Gao, B.; Liu, P.; Li, J. Crustacean hyperglycemic hormone of Portunus trituberculatus: Evidence of alternative splicing and potential roles in osmoregulation. Cell Stress Chaperones 2019, 24, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Mantel, L.H.; Farmer, L.L. Osmotic and ionic regulation. Intern. Anat. Physiol. Regul. 1983, 5, 53–161. [Google Scholar]
- Davenport, J.; Wong, T. Responses of adult mud crabs (Scylla serrata) (Forskal) to salinity and low oxygen tension. Comp. Biochem. Physiol. Part A Physiol. 1987, 86, 43–47. [Google Scholar] [CrossRef]
- Chen, J.-C.; Chia, P.-G. Osmotic and ionic concentrations of Scylla serrata (Forskål) subjected to different salinity levels. Comp. Biochem. Physiol. Part A Physiol. 1997, 117, 239–244. [Google Scholar] [CrossRef]
- Chou, P.Y. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. Relat. Areas Mol. Biol. 1978, 47, 45–148. [Google Scholar]
- Katayama, H.; Ohira, T.; Aida, K.; Nagasawa, H. Significance of a carboxyl-terminal amide moiety in the folding and biological activity of crustacean hyperglycemic hormone. Peptides 2002, 23, 1537–1546. [Google Scholar] [CrossRef]
- Mosco, A.; Edomi, P.; Guarnaccia, C.; Lorenzon, S.; Pongor, S.; Ferrero, E.A.; Giulianini, P.G. Functional aspects of cHH C-terminal amidation in crayfish species. Regul. Pept. 2008, 147, 88–95. [Google Scholar] [CrossRef]
- Webster, S. High-affinity binding of putative moult-inhibiting hormone (MIH) and crustacean hyperglycaemic hormone (CHH) to membrane-bound receptors on the Y-organ of the shore crab Carcinus maenus. Proc. R. Soc. London Ser. B Biol. Sci. 1993, 251, 53–59. [Google Scholar]
- Katayama, H.; Chung, J.S. The specific binding sites of eyestalk- and pericardial organ-crustacean hyperglycaemic hormones (CHHs) in multiple tissues of the blue crab, Callinectes sapidus. J. Exp. Biol. 2009, 212, 542–549. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-Y.; Toullec, J.-Y.; Lee, C.-Y. The Crustacean Hyperglycemic Hormone Superfamily: Progress Made in the Past Decade. Front. Endocrinol. 2020, 11, 578958. [Google Scholar] [CrossRef] [PubMed]
- Schägger, H.; Von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- Chen, H.Y.; Watson, R.D.; Chen, J.C.; Liu, H.F.; Lee, C.Y. Molecular characterization and gene expression pattern of two putative molt-inhibiting hormones from Litopenaeus vannamei. Gen. Comp. Endocrinol. 2007, 151, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.X.; Lyu, P.; Wemmer, D.E.; Kallenbach, N.R. Alpha helix capping in synthetic model peptides by reciprocal side chain–main chain interactions: Evidence for an N terminal “capping box”. Proteins Struct. Funct. Bioinform. 1994, 18, 1–7. [Google Scholar] [CrossRef]
- Lobley, A.; Whitmore, L.; Wallace, B. DICHROWEB: An interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics 2002, 18, 211–212. [Google Scholar] [CrossRef] [Green Version]
- Chung, K.F.; Lin, H.C. Osmoregulation and Na, K-ATPase expression in osmoregulatory organs of Scylla paramamosain. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2006, 144, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Peterson, G.L. A simplified method for analysis of inorganic phosphate in the presence of interfering substances. Anal. Biochem. 1978, 84, 164–172. [Google Scholar] [CrossRef]
- Holliday, C.W. Salinity-induced changes in gill Na, K-ATPase activity in the mud fiddler crab, Uca pugnax. J. Exp. Zool. Part A Ecol. Genet. Physiol. 1985, 233, 199–208. [Google Scholar] [CrossRef]
- Tsai, J.R.; Lin, H.C. V-type H+-ATPase and Na+,K+-ATPase in the gills of 13 euryhaline crabs during salinity acclimation. J. Exp. Biol. 2007, 210, 620–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goddard, T.D.; Kneller, D.G. SPARKY 3; University of California: San Francisco, CA, USA, 2008. [Google Scholar]
- Brunger, A.T. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007, 2, 2728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oostenbrink, C.; Villa, A.; Mark, A.E.; van Gunsteren, W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004, 25, 1656–1676. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]

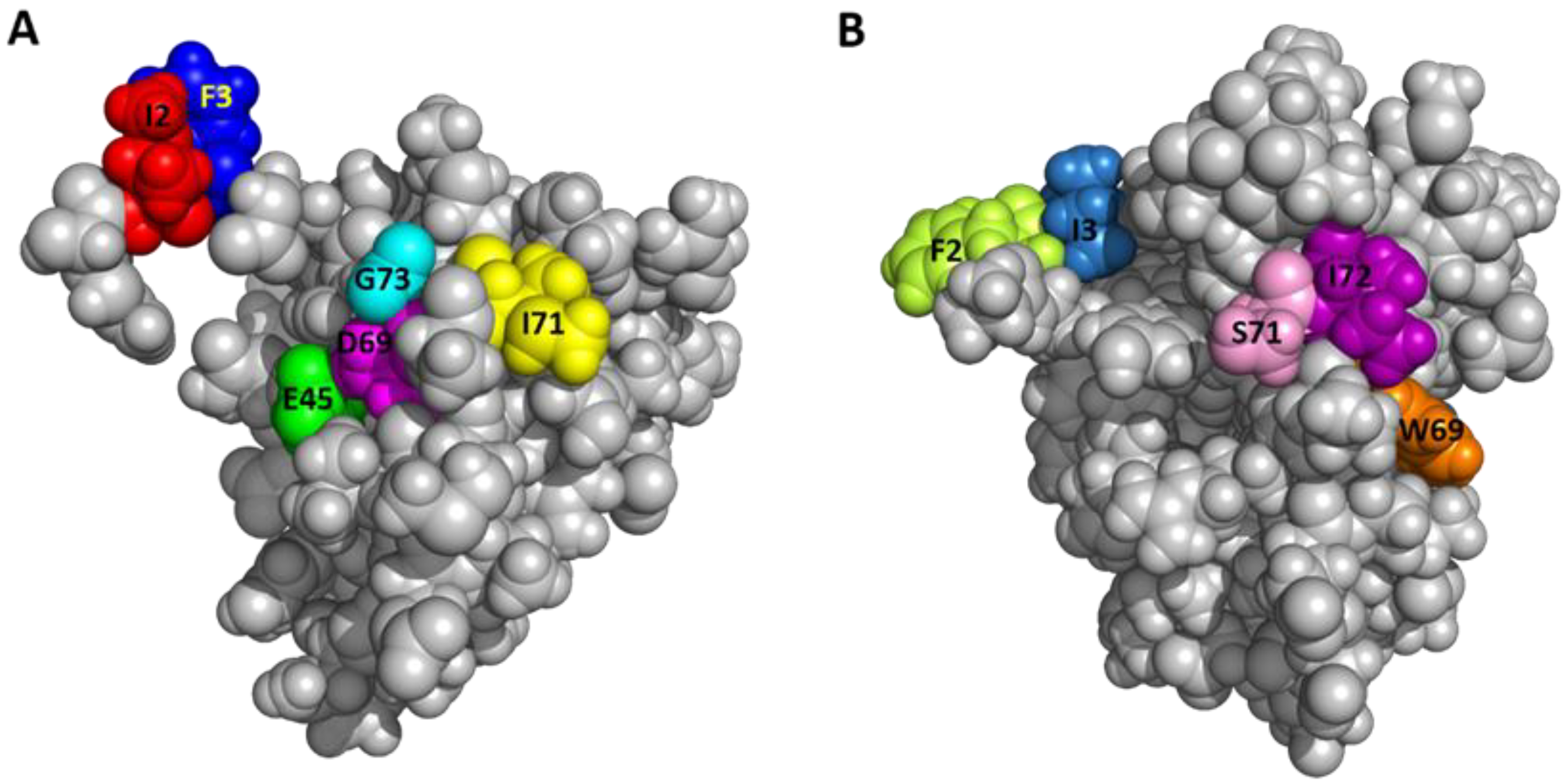
| Peptide | Theoretical Mass (Da) | Observed Mass (Da) | α-Helix (%) |
|---|---|---|---|
| Sco-CHH | 8527.9 | 8528.0 | 47.0 |
| Sco-CHH-L | 8626.8 | 8626.0 | 40.0 |
| I2A | 8584.7 | 8584.0 | 36.1 |
| F3A | 8550.7 | 8551.0 | 31 |
| V41A | 8598.8 | 8598.3 | 38.5 |
| E45A | 8568.8 | 8568.0 | 38.0 |
| N60A | 8583.8 | 8584.5 | 28.1 |
| E62A | 8568.8 | 8568.5 | 37.5 |
| E63A | 8568.8 | 8568.5 | 37.1 |
| D69A | 8582.8 | 8583.0 | 39.1 |
| I71A | 8584.7 | 8584.0 | 35 |
| G73A | 8640.9 | 8642.0 | 26.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-R.; Hsiao, N.-W.; Lee, Y.-Z.; Huang, S.-S.; Chang, C.-C.; Tsai, J.-R.; Lin, H.-C.; Toullec, J.-Y.; Lee, C.-Y.; Lyu, P.-C. Structure-Based Functional Analysis of a Hormone Belonging to an Ecdysozoan Peptide Superfamily: Revelation of a Common Molecular Architecture and Residues Possibly for Receptor Interaction. Int. J. Mol. Sci. 2021, 22, 11142. https://doi.org/10.3390/ijms222011142
Chen Y-R, Hsiao N-W, Lee Y-Z, Huang S-S, Chang C-C, Tsai J-R, Lin H-C, Toullec J-Y, Lee C-Y, Lyu P-C. Structure-Based Functional Analysis of a Hormone Belonging to an Ecdysozoan Peptide Superfamily: Revelation of a Common Molecular Architecture and Residues Possibly for Receptor Interaction. International Journal of Molecular Sciences. 2021; 22(20):11142. https://doi.org/10.3390/ijms222011142
Chicago/Turabian StyleChen, Yun-Ru, Nai-Wan Hsiao, Yi-Zong Lee, Shiau-Shan Huang, Chih-Chun Chang, Jyuan-Ru Tsai, Hui-Chen Lin, Jean-Yves Toullec, Chi-Ying Lee, and Ping-Chiang Lyu. 2021. "Structure-Based Functional Analysis of a Hormone Belonging to an Ecdysozoan Peptide Superfamily: Revelation of a Common Molecular Architecture and Residues Possibly for Receptor Interaction" International Journal of Molecular Sciences 22, no. 20: 11142. https://doi.org/10.3390/ijms222011142






