Transcriptomic and Metabolic Analyses Reveal the Mechanism of Ethylene Production in Stony Hard Peach Fruit during Cold Storage
Abstract
:1. Introduction
2. Results and Discussion
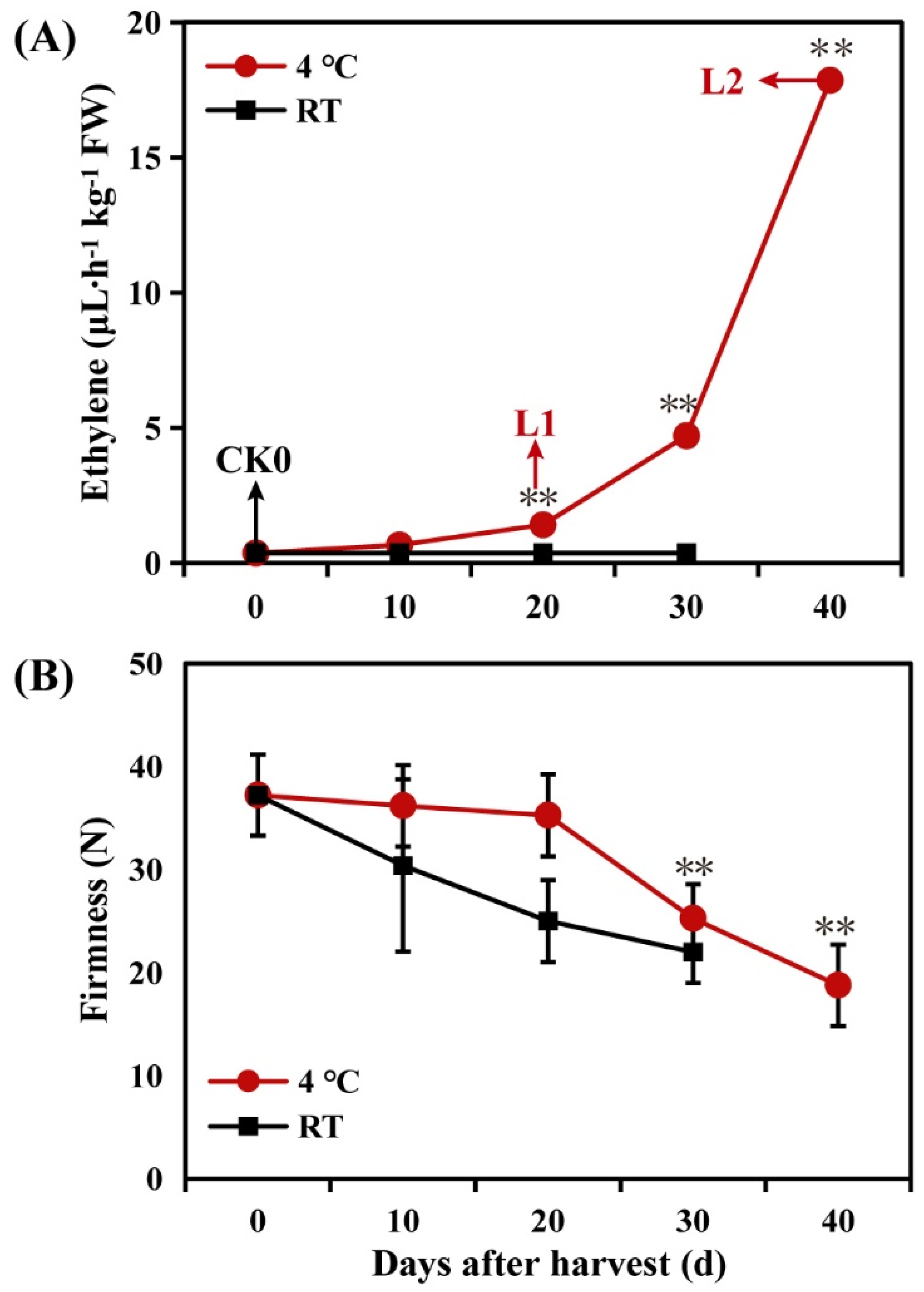
2.1. Changes in Ethylene Release during Storage
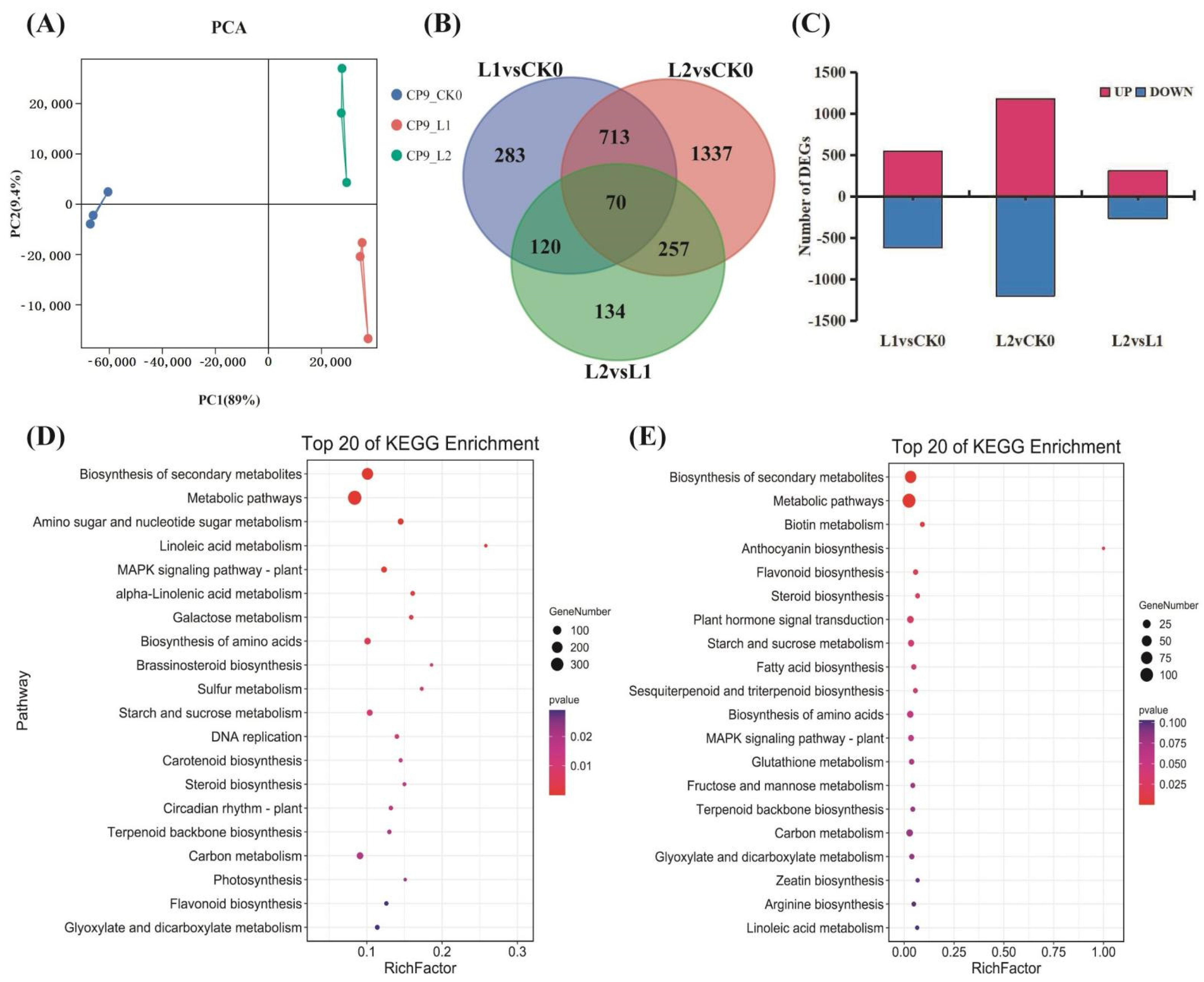
2.2. Changes in Transcript Abundance during Cold Storage
2.3. Changes in Transcript Abundance during Cold Storage
2.4. Changes in Transcript Abundance during Cold Storage
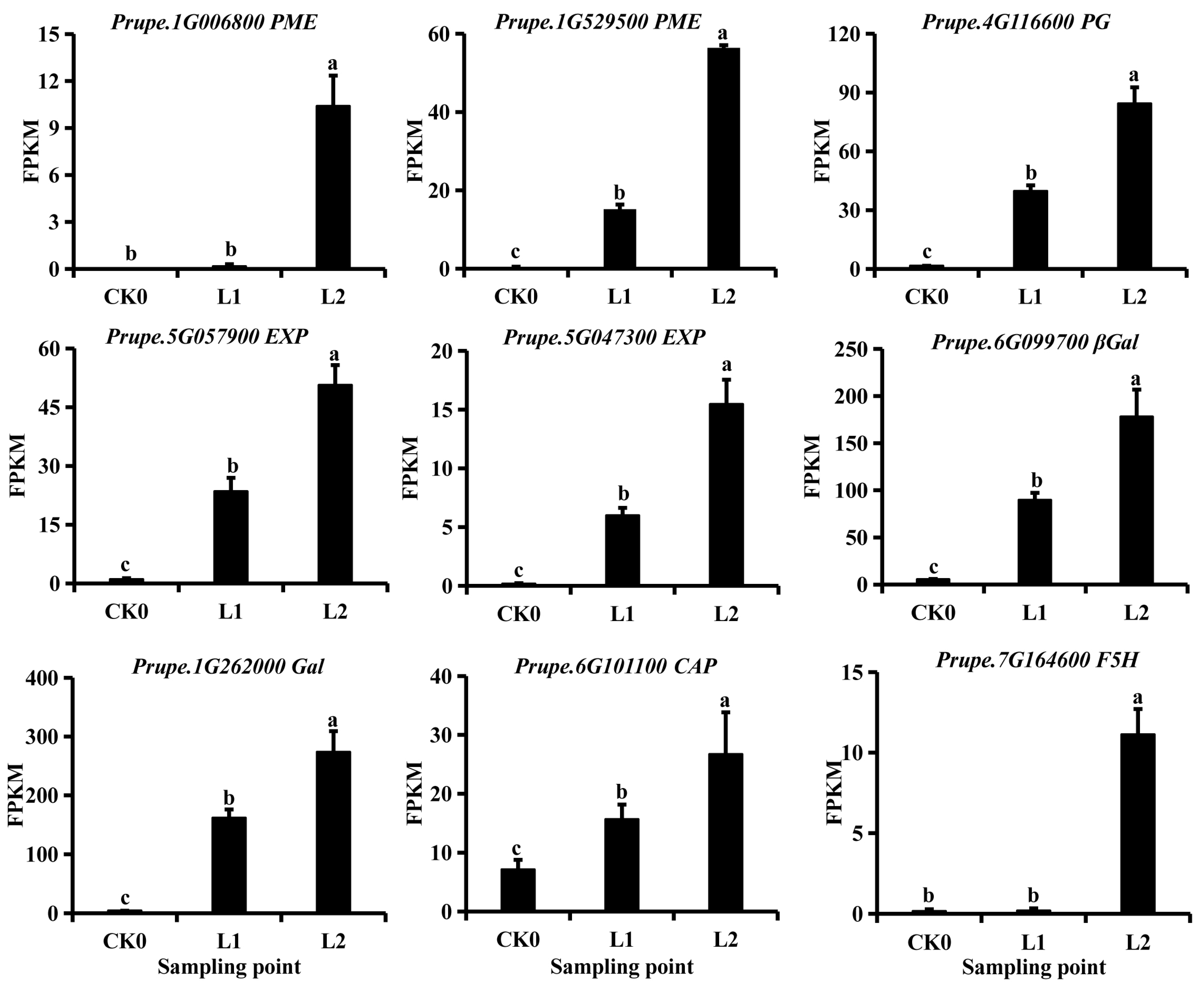
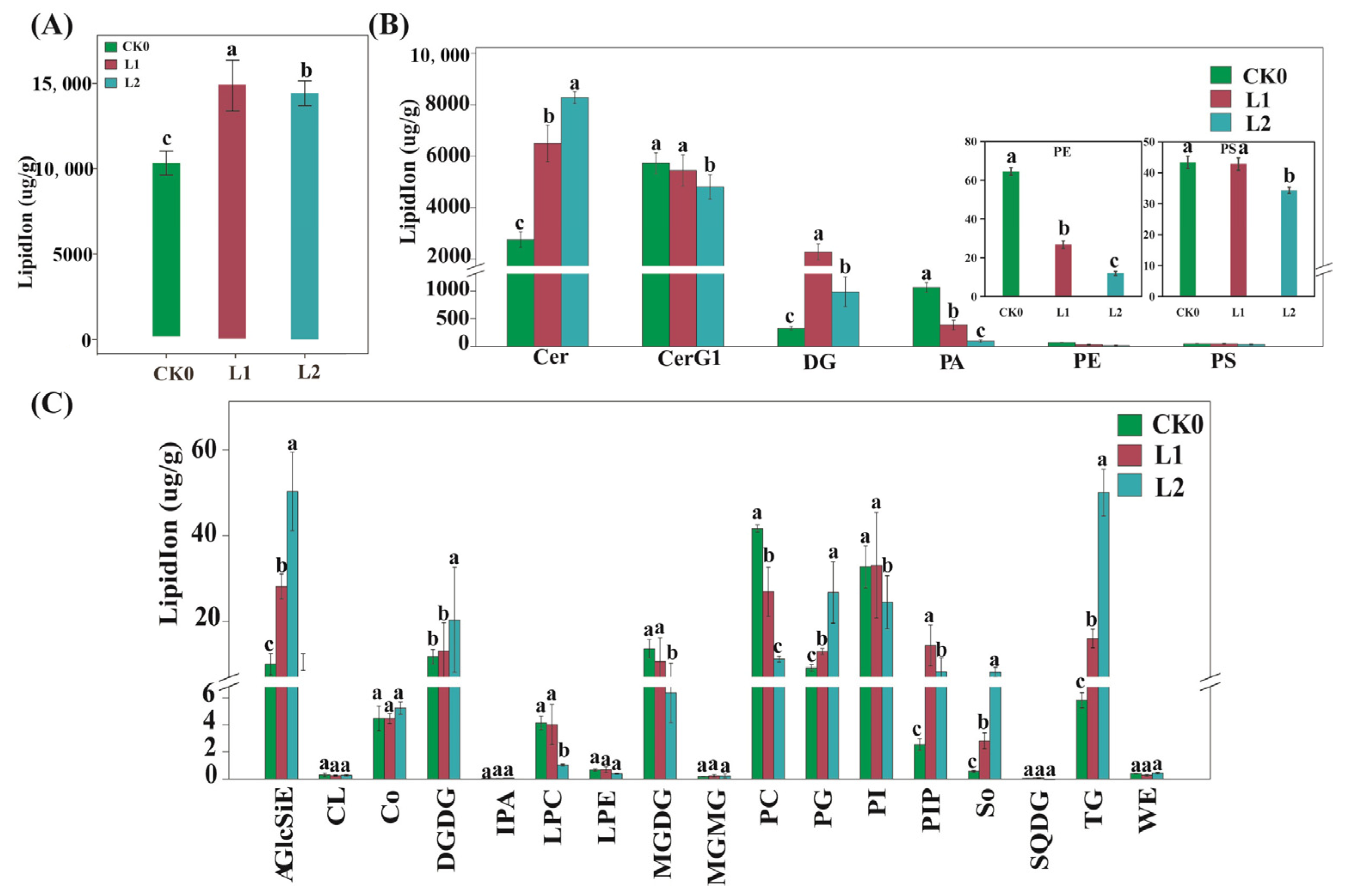
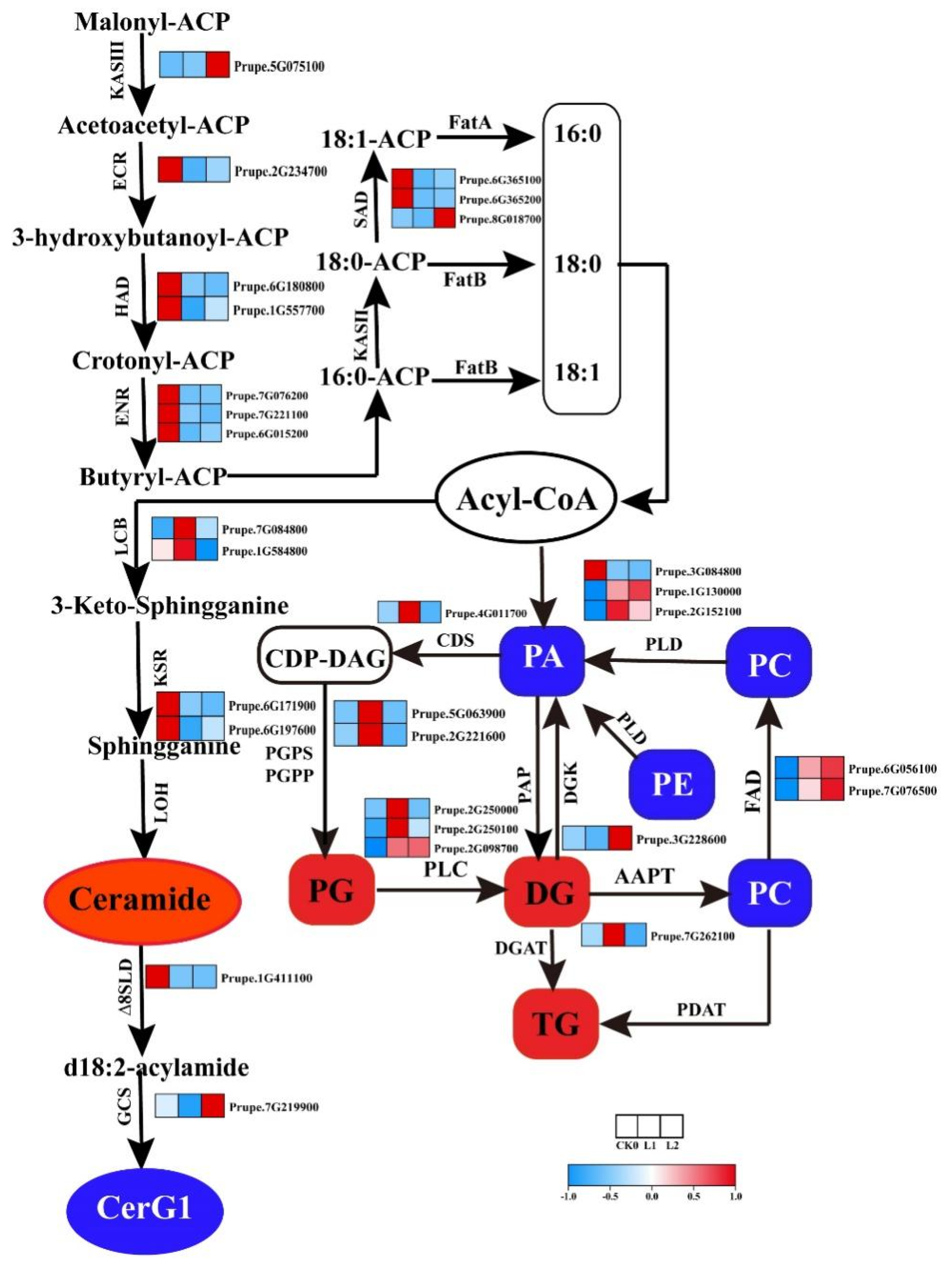
2.5. Analysis of the Expression of Lipid Metabolic Genes and Changes in Lipid Content
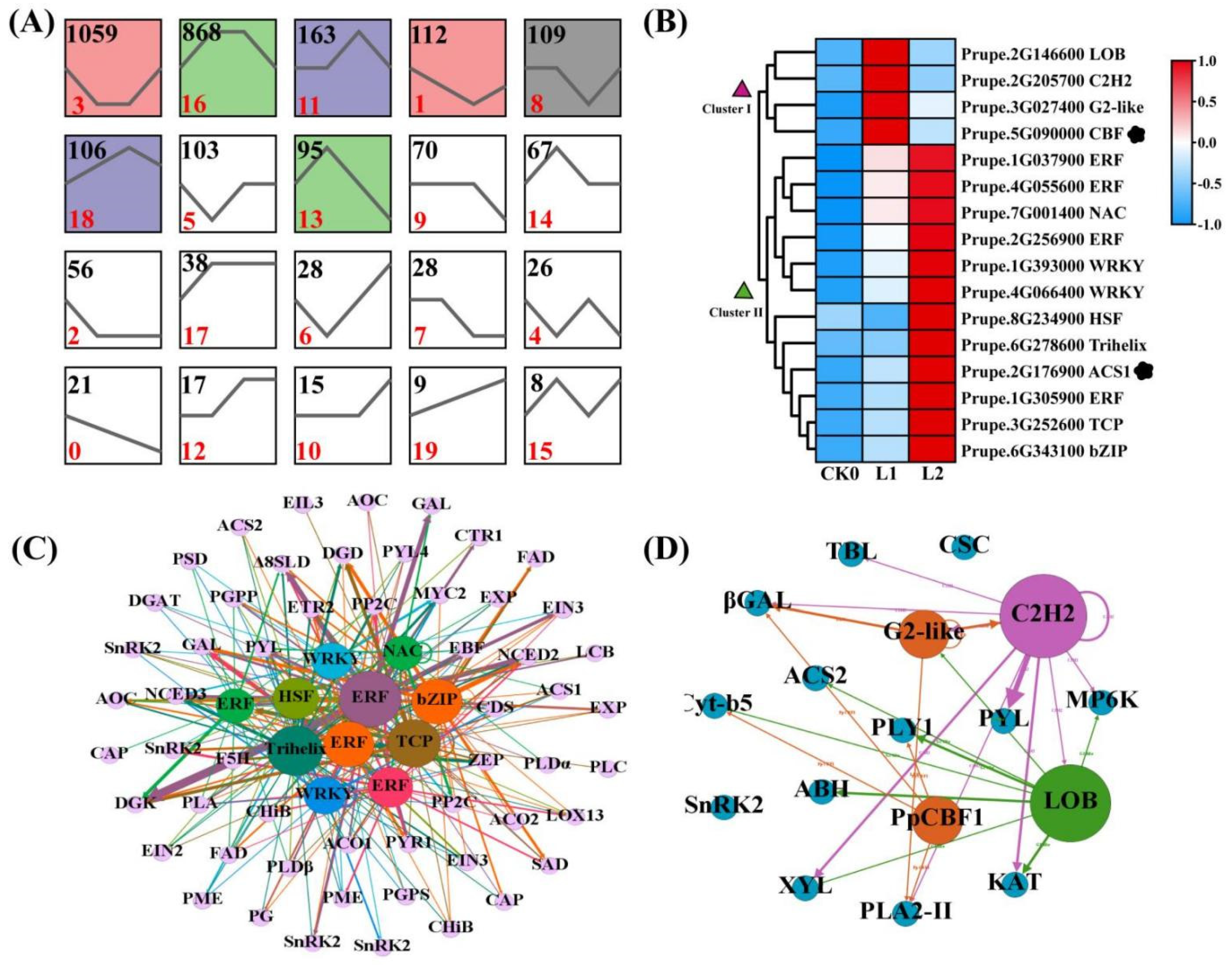
2.6. Key TFs and Their Related Co-Expression Regulatory Network during Long-Term Cold Storage
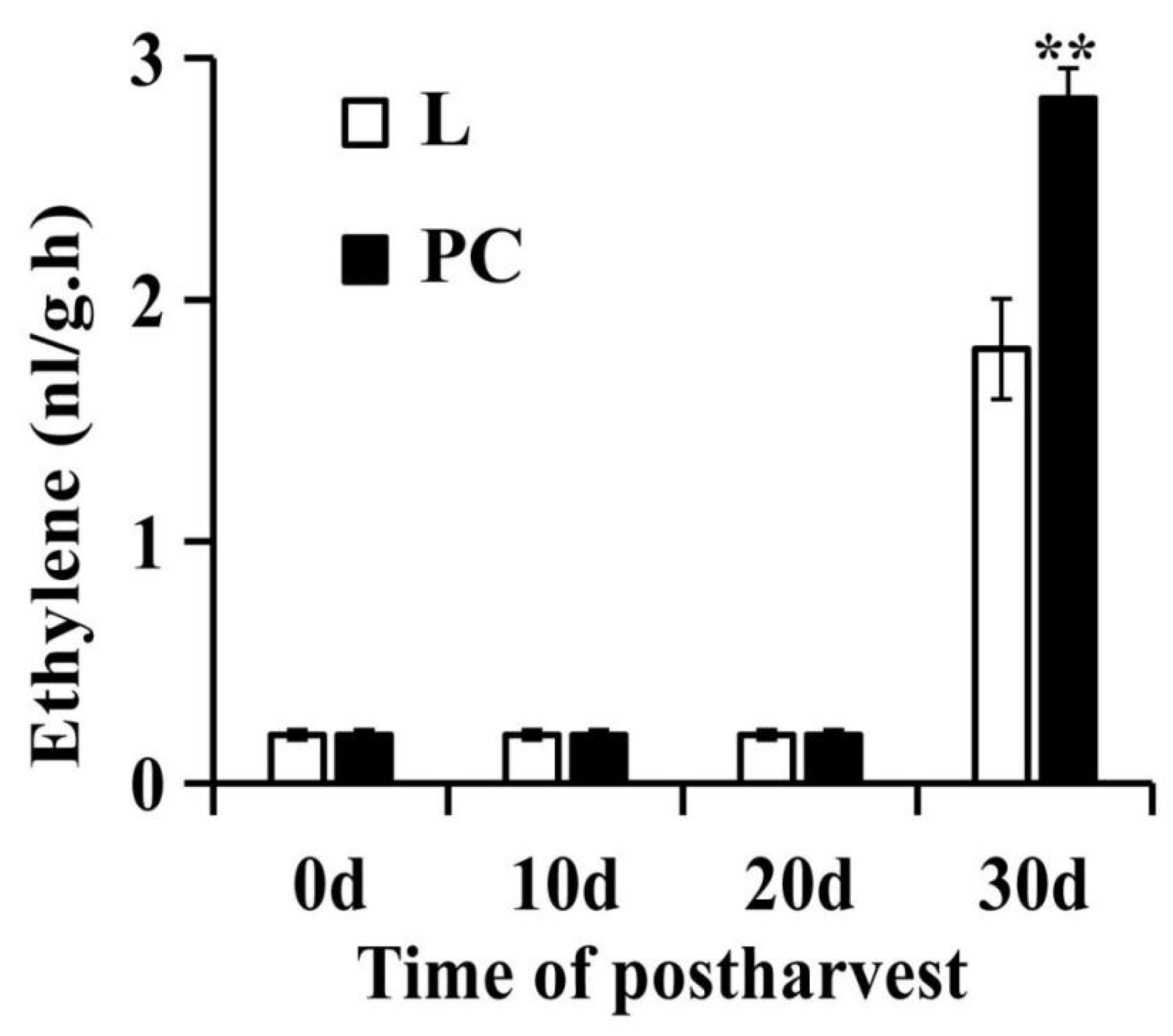
2.7. PC Promoted Ethylene Production in SH Peach Fruit under Low Temperature
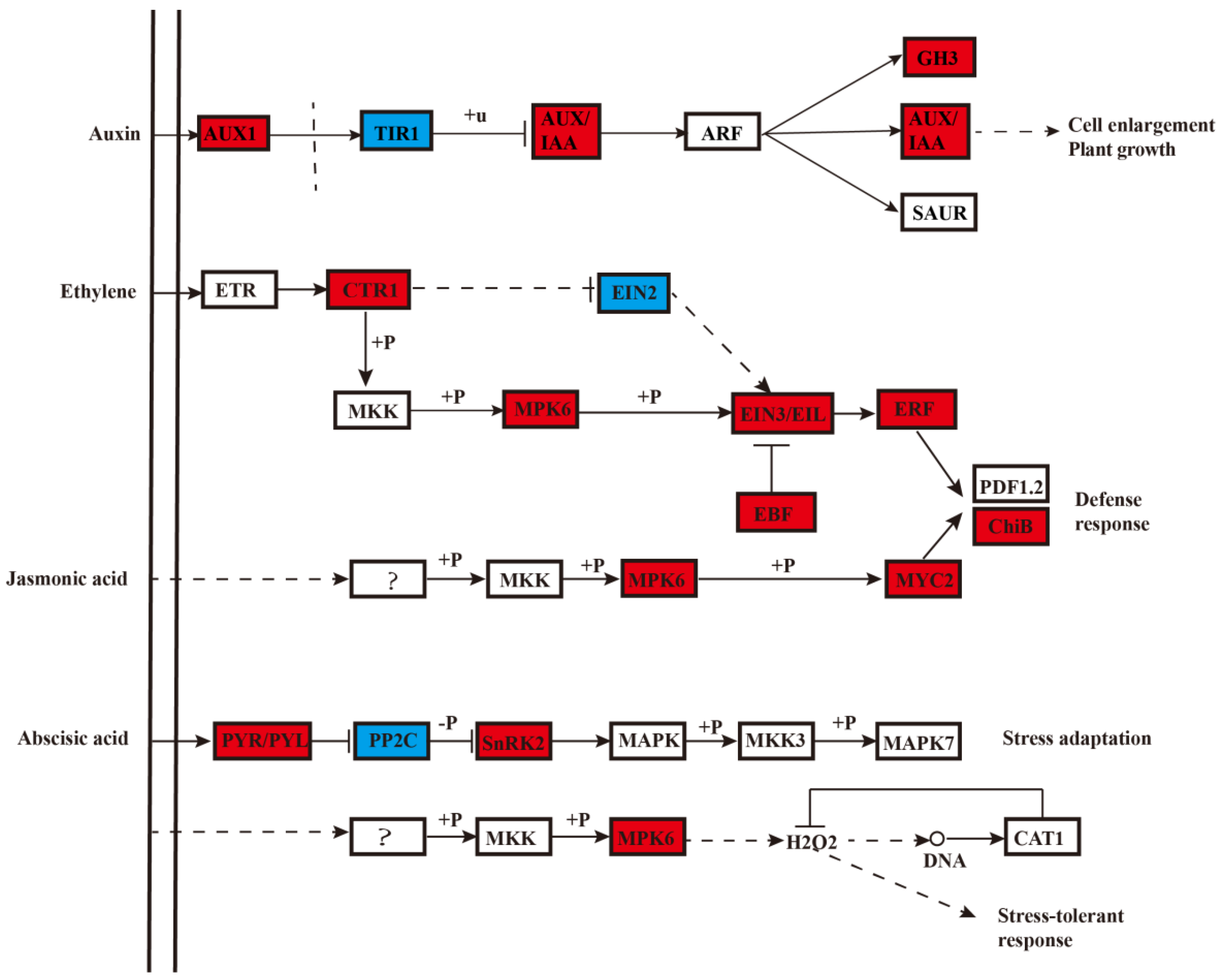
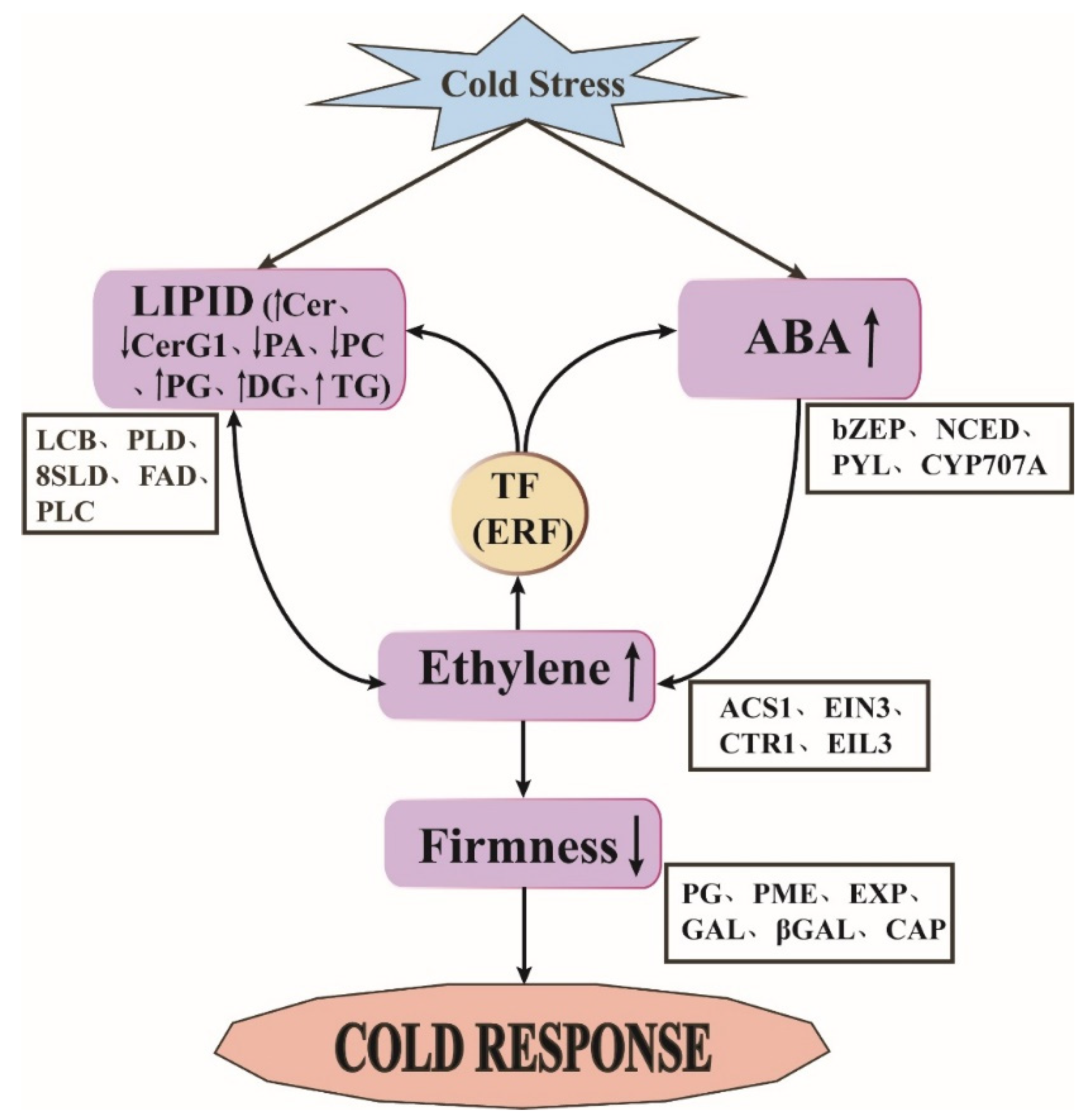
2.8. Mechanism and Transcriptional Regulation of of Peach Fruit during Long-Term Cold Storage
3. Materials and Methods
3.1. Plant Material and Treatments
3.2. Measurement of Ethylene and ABA Contents and Firmness of Peach Fruit
3.3. Library Construction and Transcriptome Sequencing
3.4. RNA-Seq Data Analysis
3.5. Lipidome Analysis
3.6. Transcription Factor (TF) Binding Site Prediction and Coexpression Network Construction
3.7. Treatment with PC in SH Peach
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Padilla, M.N.; Hernandez, M.L.; Sanz, C.; Martinez-Rivas, J.M. Molecular cloning, functional characterization and transcriptional regulation of a 9-lipoxygenase gene from olive. Phytochemistry 2012, 74, 58–68. [Google Scholar] [CrossRef]
- Wang, K.; Yin, X.R.; Zhang, B.; Grierson, D.; Xu, C.J.; Chen, K.S. Transcriptomic and metabolic analyses provide new insights into chilling injury in peach fruit. Plant Cell Environ. 2017, 40, 1531–1551. [Google Scholar] [CrossRef]
- Lee, H.G.; Seo, P.J. The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 2015, 82, 962–977. [Google Scholar] [CrossRef] [PubMed]
- Brovelli, E.A.; Brecht, J.K.; Sherman, W.B.; Sims, C.A. Anatomical and physiological responses of melting- and nonmelting-flesh peaches to postharvest chilling. J. Am. Soc. Hortic. Sci. 1998, 123, 668–674. [Google Scholar] [CrossRef] [Green Version]
- Brummell, D.A.; Dal Cin, V.; Lurie, S.; Crisosto, C.H.; Labavitch, J.M. Cell wall metabolism during the development of chilling injury in cold-stored peach fruit: Association of mealiness with arrested disassembly of cell wall pectins. J. Exp. Bot. 2004, 55, 2041–2052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Quan, P.K.; Liu, H.K.; Li, L.; Qi, S.Y.; Zhang, M.S.; Zhang, B.; Ma, B.; Han, M.; Zhang, H.; et al. Transcriptomic and Metabolic Analyses Provide New Insights into the Apple Fruit Quality Decline during Long-Term Cold Storage. J. Agric. Food Chem. 2020, 68, 4699–4716. [Google Scholar] [CrossRef]
- Sun, H.J.; Zhou, X.; Zhou, Q.; Zhao, Y.B.; Kong, X.M.; Luo, M.L.; Ji, S.J. Disorder of membrane metabolism induced membrane instability plays important role in pericarp browning of refrigerated ‘Nanguo’ pears. Food Chem. 2020, 320. [Google Scholar] [CrossRef]
- Jiang, Y.M.; Joyce, D.C.; Jiang, W.B.; Lu, W.J. Effects of chilling temperatures on ethylene binding by banana fruit. Plant Growth Regul. 2004, 43, 109–115. [Google Scholar] [CrossRef]
- Zeng, W.F.; Pan, L.; Liu, H.; Niu, L.; Lu, Z.H.; Cui, G.C.; Wang, Z.Q. Characterization of 1-aminocyclopropane-1-carboxylic acid synthase (ACS) genes during nectarine fruit development and ripening. Tree Genet. Genomes 2015, 11. [Google Scholar] [CrossRef]
- Lurie, S.; Crisosto, C.H. Chilling injury in peach and nectarine. Postharvest Biol. Technol. 2005, 37, 195–208. [Google Scholar] [CrossRef]
- Fernandez-Trujillo, J.P.; Cano, A.; Artes, F. Physiological changes in peaches related to chilling injury and ripening. Postharvest Biol. Techy. 1998, 13, 109–119. [Google Scholar] [CrossRef]
- Zhou, H.W.; Dong, L.; Ben-Arie, R.; Lurie, S. The role of ethylene in the prevention of chilling injury in nectarines. J. Plant Physiol. 2001, 158, 55–61. [Google Scholar] [CrossRef]
- Bustamante, C.A.; Brotman, Y.; Monti, L.L.; Gabilondo, J.; Budde, C.O.; Lara, M.V.; Fernie, A.R.; Drincovich, M.F. Differential lipidome remodeling during postharvest of peach varieties with different susceptibility to chilling injury. Physiol. Plant 2018, 163, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.C.; Wang, K.; Wu, C.X.; Zhao, Y.; Yin, X.R.; Zhang, B.; Grierson, D.; Chen, K.S.; Xu, C.J. Effect of Ethylene on Cell Wall and Lipid Metabolism during Alleviation of Postharvest Chilling Injury in Peach. Cells 2019, 8, 1612. [Google Scholar] [CrossRef] [Green Version]
- Haji, T.; Yaegaki, H.; Yamaguchi, M. Softening of stony hard peach by ethylene and the induction of endogenous ethylene by 1-aminocyclopropane-1-carboxylic acid (ACC). J. Jpn. Soc. Hortic. Sci. 2003, 72, 212–217. [Google Scholar] [CrossRef] [Green Version]
- Haji, T.; Yaegaki, H.; Yamaguchi, M. Varietal differences in the relationship between maturation characteristics, storage life and ethylene production in peach fruit. J. Jpn. Soc. Hortic. Sci. 2004, 73, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Pan, L.; Zeng, W.F.; Niu, L.; Lu, Z.H.; Liu, H.; Cui, G.C.; Zhu, Y.Q.; Chu, J.F.; Li, W.P.; Fang, W.C.; et al. PpYUC11, a strong candidate gene for the stony hard phenotype in peach (Prunus persica L. Batsch), participates in IAA biosynthesis during fruit ripening. J. Exp. Bot. 2015, 66, 7031–7044. [Google Scholar] [CrossRef] [Green Version]
- Tatsuki, M.; Haji, T.; Yamaguchi, M. The involvement of 1-aminocyclopropane-1-carboxylic acid synthase isogene, Pp-ACS1, in peach fruit softening. J. Exp. Bot. 2006, 57, 1281–1289. [Google Scholar] [CrossRef]
- Yin, X.R.; Allan, A.C.; Chen, K.S.; Ferguson, I.B. Kiwifruit EIL and ERF Genes Involved in Regulating Fruit Ripening. Plant Physiol. 2010, 153, 1280–1292. [Google Scholar] [CrossRef] [Green Version]
- Nambara, E.; Marion-Poll, A. ABA action and interactions in seeds. Trends Plant Sci. 2003, 8, 213–217. [Google Scholar] [CrossRef]
- Qin, X.Q.; Zeevaart, J.A.D. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 2002, 128, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.L.; Wang, J.N.; Shan, W.; Fan, J.G.; Kuang, J.F.; Wu, K.Q.; Li, X.P.; He, F.Y.; Chen, J.Y.; Lu, W.J. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 2013, 36, 30–51. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Duan, G.H.; Li, C.Q.; Liu, L.; Han, G.Y.; Zhang, Y.L.; Wang, C.M. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Welti, R.; Li, W.Q.; Li, M.Y.; Sang, Y.M.; Biesiada, H.; Zhou, H.E.; Rajashekar, C.B.; Williams, T.D.; Wang, X.M. Profiling membrane lipids in plant stress responses—Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 2002, 277, 31994–32002. [Google Scholar] [CrossRef] [Green Version]
- Chintalapati, S.; Kiran, M.D.; Shivaji, S. Role of membrane lipid fatty acids in cold adaptation. Cell Mol. Biol. 2004, 50, 631–642. [Google Scholar] [PubMed]
- Zhang, C.F.; Tian, S.P. Crucial contribution of membrane lipids’ unsaturation to acquisition of chilling-tolerance in peach fruit stored at 0 degrees C. Food Chem. 2009, 115, 405–411. [Google Scholar] [CrossRef]
- Ernst, R.; Ejsing, C.S.; Antonny, B. Homeoviscous Adaptation and the Regulation of Membrane Lipids. J. Mol. Biol. 2016, 428, 4776–4791. [Google Scholar] [CrossRef] [Green Version]
- Liang, H.; Yao, N.; Song, L.T.; Luo, S.; Lu, H.; Greenberg, L.T. Ceramides modulate programmed cell death in plants. Genes Dev. 2003, 17, 2636–2641. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.H.; Bielawski, J.; Mu, J.Y.; Dong, H.L.; Teng, C.; Zhang, J.; Yang, X.H.; Tomishige, N.; Hanada, K.; Hannun, Y.A.; et al. Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Genes Dev. 2007, 17, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Markham, J.E.; Cahoon, E.B. Sphingolipid delta 8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J. 2012, 69, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Rajashekar, C.B.; Zhou, H.E.; Zhang, Y.W.; Li, W.Q.; Wang, X.M. Suppression of phospholipase D alpha 1 induces freezing tolerance in Arabidopsis: Response of coldresponsive genes and osmolyte accumulation. J. Plant Physiol. 2006, 163, 916–926. [Google Scholar] [CrossRef]
- Zhang, L.N.; Zhang, L.C.; Xia, C.; Zhao, G.Y.; Liu, J.; Jia, J.Z.; Kong, X.Y. A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Plant Physiol. 2015, 153, 538–554. [Google Scholar] [CrossRef] [PubMed]
- Shan, W.; Kuang, J.F.; Lu, W.J.; Chen, J.Y. Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant Cell Environ. 2014, 37, 2116–2127. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.L.; Ba, L.J.; Shan, W.; Kuang, J.F.; Lu, W.J.; Chen, J.Y. Involvement of WRKY Transcription Factors in Abscisic-Acid-Induced Cold Tolerance of Banana Fruit. J. Agr. Food Chem. 2017, 65, 3627–3635. [Google Scholar] [CrossRef]
- Bolt, S.; Zuther, E.; Zintl, S.; Hincha, D.K.; Schmulling, T. ERF105 is a transcription factor gene of Arabidopsis thaliana required for freezing tolerance and cold acclimation. Plant Cell Environ. 2017, 40, 108–120. [Google Scholar] [CrossRef]
- Liu, Y.D.; Zhang, L.; Meng, S.D.; Liu, Y.F.; Zhao, X.M.; Pang, C.P.; Zhang, H.D.; Xu, T.; He, Y.; Qi, M.F.; et al. Expression of galactinol synthase from Ammopiptanthus nanus in tomato improves tolerance to cold stress. J. Exp. Bot. 2020, 71, 435–449. [Google Scholar] [CrossRef]
- Artlip, T.; McDermaid, A.; Ma, Q.; Wisniewski, M. Differential gene expression in non-transgenic and transgenic “M.26” apple overexpressing a peach CBF gene during the transition from eco-dormancy to bud break. Hort. Res. 2019, 6. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.R.; Han, X.; Yang, M.L.; Zhang, M.H.; Pan, J.J.; Yu, D.Q. The Transcription Factor INDUCER of CBF EXPRESSION1 Interacts with ABSCISIC ACID INSENSITIVE5 and DELLA Proteins to Fine-Tune Abscisic Acid Signaling during Seed Germination in Arabidopsis. Plant Cell 2019, 31, 1520–1538. [Google Scholar] [CrossRef]
- Taguchi, R.; Ishikawa, M. Precise and global identification of phospholipid molecular species by an Orbitrap mass spectrometer and automated search engine Lipid Search. J. Chromatogr. 2010, 1217, 4229–4239. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Siddiqua, M.; Braybrook, S.; Nassuth, A. Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant Cell Environ. 2006, 29, 1410–1421. [Google Scholar] [CrossRef] [Green Version]
- Steponkus, P.L.; Uemura, M.; Balsamo, R.A.; Arvinte, T.; Lynch, D.V. Transformation of the cryobehavior of rye protoplasts by modifification of the plasma membrane lipid composition. Proc. Natl. Acad. Sci. USA 1988, 85, 9026–9030. [Google Scholar] [CrossRef] [Green Version]
- Degenkolbe, T.; Giavalisco, P.; Zuther, E.; Seiwert, B.; Hincha, D.K.; Willmitzer, L. Differential remodeling of the lipidome during cold acclimation in natural accessions of Arabidopsis thaliana. Plant J. 2012, 72, 972–982. [Google Scholar] [CrossRef]
- Gao, H. Research on Chilling Injury and Mechanism of Chilling Injury Physiology of Nectarines. Ph.D. Thesis, Northwest A&F University, Xianyang, China, 2007. [Google Scholar]
- García-Pastor, M.E.; Falagán, N.; Giné-Bordonaba, J.; Dorota, A.W.; Leon, A.T.; Carmen, M. Cultivar and tissue-specific changes of abscisic acid, its catabolites and individual sugars during postharvest handling of flat peaches (Prunus persica cv. platycarpa). Postharvest Biol. Tec. 2021, 181, 111688. [Google Scholar] [CrossRef]
- Shiroguchi, K.; Jia, T.Z.; Sims, P.A.; Xie, X.S. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc. Natl. Acad. Sci. USA 2012, 109, 1347–1352. [Google Scholar] [CrossRef] [Green Version]
- Smith, T.; Heger, A.; Sudbery, I. UMI-tools: Modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017, 27, 491–499. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Mao, X.Z.; Cai, T.; Olyarchuk, J.G.; Wei, L.P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.C.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Deng, L.; Meng, J.; Niu, L.; Pan, L.; Lu, Z.; Cui, G.; Wang, Z.; Zeng, W. Transcriptomic and Metabolic Analyses Reveal the Mechanism of Ethylene Production in Stony Hard Peach Fruit during Cold Storage. Int. J. Mol. Sci. 2021, 22, 11308. https://doi.org/10.3390/ijms222111308
Wang Y, Deng L, Meng J, Niu L, Pan L, Lu Z, Cui G, Wang Z, Zeng W. Transcriptomic and Metabolic Analyses Reveal the Mechanism of Ethylene Production in Stony Hard Peach Fruit during Cold Storage. International Journal of Molecular Sciences. 2021; 22(21):11308. https://doi.org/10.3390/ijms222111308
Chicago/Turabian StyleWang, Yan, Li Deng, Junren Meng, Liang Niu, Lei Pan, Zhenhua Lu, Guochao Cui, Zhiqiang Wang, and Wenfang Zeng. 2021. "Transcriptomic and Metabolic Analyses Reveal the Mechanism of Ethylene Production in Stony Hard Peach Fruit during Cold Storage" International Journal of Molecular Sciences 22, no. 21: 11308. https://doi.org/10.3390/ijms222111308
APA StyleWang, Y., Deng, L., Meng, J., Niu, L., Pan, L., Lu, Z., Cui, G., Wang, Z., & Zeng, W. (2021). Transcriptomic and Metabolic Analyses Reveal the Mechanism of Ethylene Production in Stony Hard Peach Fruit during Cold Storage. International Journal of Molecular Sciences, 22(21), 11308. https://doi.org/10.3390/ijms222111308





