Curcumin Alleviates the Senescence of Canine Bone Marrow Mesenchymal Stem Cells during In Vitro Expansion by Activating the Autophagy Pathway
Abstract
1. Introduction
2. Results
2.1. Characteristics of cBMSCs
2.2. cBMSCs Progressively Display Senescent Features along Expansion In Vitro
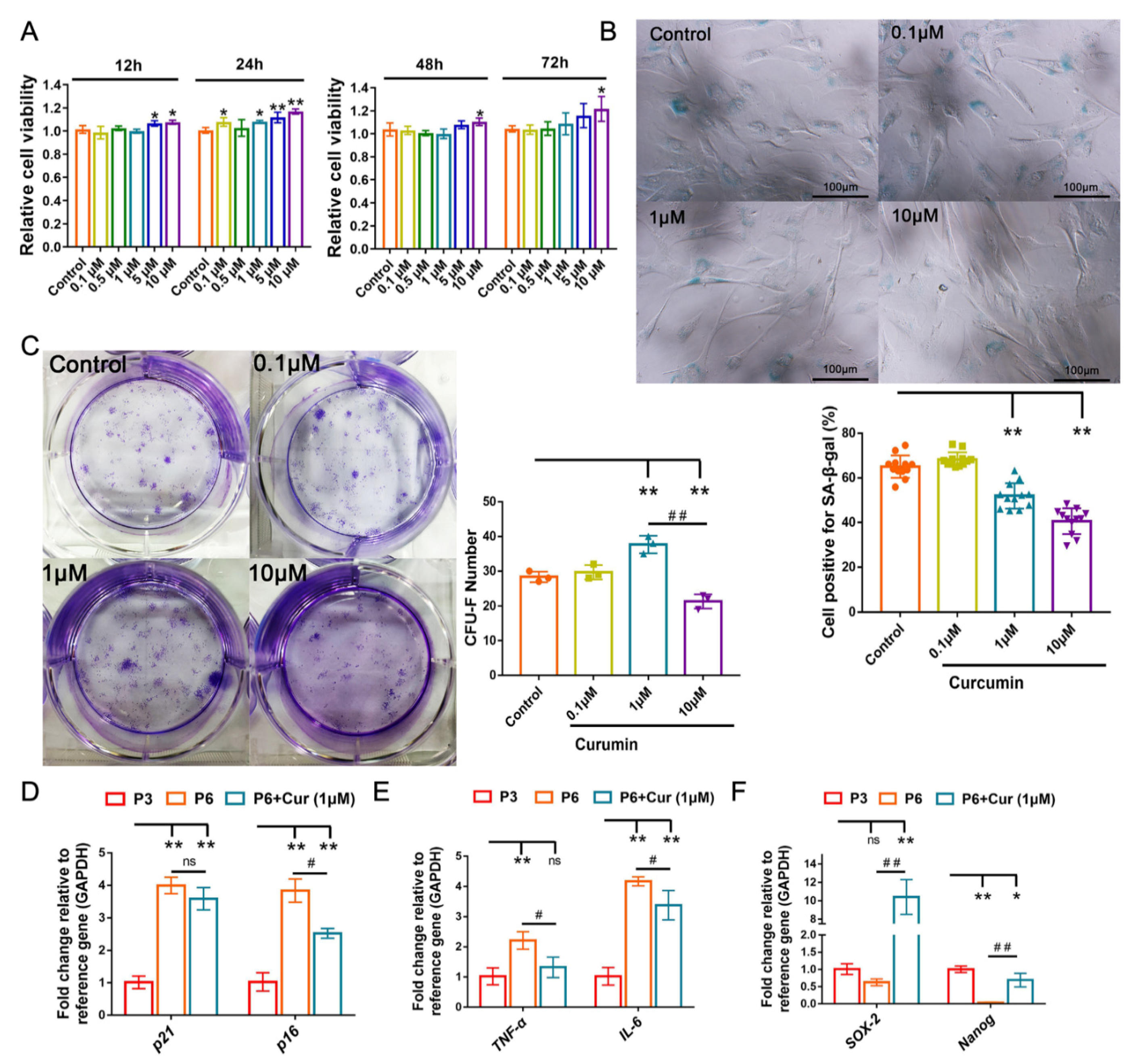
2.3. Cur Alleviates the Senescent State of cBMSCs
2.4. Cur Treatment Enhanced Autophagic Activity in cBMSCs
2.5. Autophagy Involves in Exerting the Protective Effect of Cur in cBMSC Senescence
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Preparation of Curcumin Solution
4.3. Cell Culture and Expansion
4.4. Cell Growth Curve
4.5. Detection of Immunophenotype of cBMSCs by Flow Cytometry
4.6. In Vitro Differentiation Assay
4.7. Effect of Cur on Cellular Viability
4.8. Colony Formation Assay
4.9. Beta-Galactosidase Staining Assay
4.10. Reverse Transcription Real-Time Quantitative PCR (RT-qPCR)
4.11. Tracking of Lysosomal Using LysoTracker
4.12. Immunofluorescence
4.13. Western Blotting Analysis
4.14. Transmission Electron Microscopy (TEM)
4.15. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3-MA | 3-Methyladenine |
| ATG | Autophagy-related gene |
| cBMSCs | canine bone marrow-derived mesenchymal stem cells |
| CCK-8 | cell counting kit-8 |
| CFU-F | Colony-forming unit-fibroblast |
| Cur | curcumin |
| IL | interleukin |
| LC3 | Microtubule-associatedprotein 1 light chain 3 |
| RAP | rapamycin |
| SA-β-gal | Senescence-associated beta-galactosidase |
| SASP | senescence-associated secretory phenotype |
| TEM | transmission electron microscopy |
| TNF | tumor necrosis Factor |
| ULK1 | unc51-like autophagy-activating kinase-1 |
References
- Zhan, X.S.; El-Ashram, S.; Luo, D.Z.; Luo, H.N.; Wang, B.Y.; Chen, S.F.; Bai, Y.S.; Chen, Z.S.; Liu, C.Y.; Ji, H.Q. A Comparative Study of Biological Characteristics and Transcriptome Profiles of Mesenchymal Stem Cells from Different Canine Tissues. Int. J. Mol. Sci. 2019, 20, 1485. [Google Scholar] [CrossRef]
- Liu, X.; Kumagai, G.; Wada, K.; Tanaka, T.; Asari, T.; Oishi, K.; Fujita, T.; Mizukami, H.; Furukawa, K.I.; Ishibashi, Y. High Osteogenic Potential of Adipose- and Muscle-derived Mesenchymal Stem Cells in Spinal-Ossification Model Mice. Spine 2017, 42, E1342–E1349. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Chen, X.; Dong, F.; Li, W.; Ren, X.; Zhang, Y.; Shi, Y. Concise review: Mesenchymal stem cells and translational medicine: Emerging issues. Stem Cells Transl. Med. 2012, 1, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, Q. Senescent Mesenchymal Stem Cells: Disease Mechanism and Treatment Strategy. Curr. Mol. Biol. Rep. 2020, 6, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Qi, M.; An, Y.; Zhang, L.; Yang, R.; Doro, D.H.; Liu, W.; Jin, Y. Autophagy controls mesenchymal stem cell properties and senescence during bone aging. Aging Cell 2018, 17, e12709. [Google Scholar] [CrossRef]
- Nivet, E. Modifiers of Neural Stem Cells and Aging: Pulling the Trigger of a Neurogenic Decline. Curr. Stem Cell Rep. 2016, 2, 273–281. [Google Scholar] [CrossRef][Green Version]
- Sepúlveda, J.C.; Tomé, M.; Fernández, M.E.; Delgado, M.; Campisi, J.; Bernad, A.; González, M.A. Cell senescence abrogates the therapeutic potential of human mesenchymal stem cells in the lethal endotoxemia model. Stem Cells 2014, 32, 1865–1877. [Google Scholar] [CrossRef]
- Liu, M.; Lei, H.; Dong, P.; Fu, X.; Yang, Z.; Yang, Y.; Ma, J.; Liu, X.; Cao, Y.; Xiao, R. Adipose-Derived Mesenchymal Stem Cells from the Elderly Exhibit Decreased Migration and Differentiation Abilities with Senescent Properties. Cell Transplant. 2017, 26, 1505–1519. [Google Scholar] [CrossRef]
- Ridzuan, N.; Al Abbar, A.; Yip, W.K.; Maqbool, M.; Ramasamy, R. Characterization and Expression of Senescence Marker in Prolonged Passages of Rat Bone Marrow-Derived Mesenchymal Stem Cells. Stem Cells Int. 2016, 2016, 8487264. [Google Scholar] [CrossRef]
- Cosme-Blanco, W.; Shen, M.F.; Lazar, A.J.; Pathak, S.; Lozano, G.; Multani, A.S.; Chang, S. Telomere dysfunction suppresses spontaneous tumorigenesis in vivo by initiating p53-dependent cellular senescence. EMBO Rep. 2007, 8, 497–503. [Google Scholar] [CrossRef]
- Fumagalli, M.; Rossiello, F.; Clerici, M.; Barozzi, S.; Cittaro, D.; Kaplunov, J.M.; Bucci, G.; Dobreva, M.; Matti, V.; Beausejour, C.M.; et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat. Cell Biol. 2012, 14, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Chen, X.; Liu, T.; Gong, Y.; Chen, S.; Pan, G.; Cui, W.; Luo, Z.P.; Pei, M.; Yang, H.; et al. Melatonin reverses H2O2 -induced premature senescence in mesenchymal stem cells via the SIRT1-dependent pathway. J. Pineal Res. 2015, 59, 190–205. [Google Scholar] [CrossRef] [PubMed]
- Palmero, I.; Serrano, M. Induction of senescence by oncogenic Ras. Methods Enzymol. 2001, 333, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ding, Y.; Liu, Z.; Liang, X. Senescence in Mesenchymal Stem Cells: Functional Alterations, Molecular Mechanisms, and Rejuvenation Strategies. Front. Cell Dev. Biol. 2020, 8, 258. [Google Scholar] [CrossRef] [PubMed]
- Abruzzo, P.M.; Canaider, S.; Pizzuti, V.; Pampanella, L.; Casadei, R.; Facchin, F.; Ventura, C. Herb-Derived Products: Natural Tools to Delay and Counteract Stem Cell Senescence. Stem Cells Int. 2020, 2020, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Cao, X.; Hu, X.; Li, S.; Wang, J. The anti-apoptotic, antioxidant and anti-inflammatory effects of curcumin on acrylamide-induced neurotoxicity in rats. BMC Pharmacol. Toxicol. 2020, 21, 62. [Google Scholar] [CrossRef] [PubMed]
- Pouliquen, D.L.; Boissard, A.; Henry, C.; Blandin, S.; Coqueret, O.; Guette, C. Lymphoid Organ Proteomes Identify Therapeutic Efficacy Biomarkers Following the Intracavitary Administration of Curcumin in a Highly Invasive Rat Model of Peritoneal Mesothelioma. Int. J. Mol. Sci. 2021, 22, 8566. [Google Scholar] [CrossRef]
- Srirod, S.; Tewtrakul, S. Anti-inflammatory and wound healing effects of cream containing Curcuma mangga extract. J. Ethnopharmacol. 2019, 238, 111828. [Google Scholar] [CrossRef]
- Bielak-Zmijewska, A.; Grabowska, W.; Ciolko, A.; Bojko, A.; Mosieniak, G.; Bijoch, L.; Sikora, E. The Role of Curcumin in the Modulation of Ageing. Int. J. Mol. Sci. 2019, 20, 1239. [Google Scholar] [CrossRef]
- Zia, A.; Farkhondeh, T.; Pourbagher-Shahri, A.M.; Samarghandian, S. The role of curcumin in aging and senescence: Molecular mechanisms. Biomed. Pharmacother. 2021, 134, 111119. [Google Scholar] [CrossRef]
- Santos-Parker, J.R.; Lubieniecki, K.L.; Rossman, M.J.; Van Ark, H.J.; Bassett, C.J.; Strahler, T.R.; Chonchol, M.B.; Justice, J.N.; Seals, D.R. Curcumin supplementation and motor-cognitive function in healthy middle-aged and older adults. Nutr. Healthy Aging 2018, 4, 323–333. [Google Scholar] [CrossRef]
- Takano, K.; Tatebe, J.; Washizawa, N.; Morita, T. Curcumin Inhibits Age-Related Vascular Changes in Aged Mice Fed a High-Fat Diet. Nutrients 2018, 10, 1476. [Google Scholar] [CrossRef]
- Azami, S.H.; Nazarian, H.; Abdollahifar, M.A.; Eini, F.; Farsani, M.A.; Novin, M.G. The antioxidant curcumin postpones ovarian aging in young and middle-aged mice. Reprod. Fertil. Dev. 2020, 32, 292–303. [Google Scholar] [CrossRef]
- Buhrmann, C.; Honarvar, A.; Setayeshmehr, M.; Karbasi, S.; Shakibaei, M.; Valiani, A. Herbal Remedies as Potential in Cartilage Tissue Engineering: An Overview of New Therapeutic Approaches and Strategies. Molecules 2020, 25, 3075. [Google Scholar] [CrossRef]
- Moore, T.L.; Bowley, B.G.E.; Shultz, P.L.; Calderazzo, S.M.; Shobin, E.J.; Uprety, A.R.; Rosene, D.L.; Moss, M.B. Oral curcumin supplementation improves fine motor function in the middle-aged rhesus monkey. Somatosens. Mot. Res. 2018, 35, 1–10. [Google Scholar] [CrossRef]
- Receno, C.N.; Liang, C.; Korol, D.L.; Atalay, M.; Heffernan, K.S.; Brutsaert, T.D.; DeRuisseau, K.C. Effects of Prolonged Dietary Curcumin Exposure on Skeletal Muscle Biochemical and Functional Responses of Aged Male Rats. Int. J. Mol. Sci. 2019, 20, 1178. [Google Scholar] [CrossRef]
- Buhrmann, C.; Brockmueller, A.; Mueller, A.L.; Shayan, P.; Shakibaei, M. Curcumin Attenuates Environment-Derived Osteoarthritis by Sox9/NF-kB Signaling Axis. Int. J. Mol. Sci. 2021, 22, 7645. [Google Scholar] [CrossRef]
- Buhrmann, C.; Mobasheri, A.; Matis, U.; Shakibaei, M. Curcumin mediated suppression of nuclear factor-kappaB promotes chondrogenic differentiation of mesenchymal stem cells in a high-density co-culture microenvironment. Arthritis Res. Ther. 2010, 12, R127. [Google Scholar] [CrossRef]
- Forouzanfar, F.; Read, M.I.; Barreto, G.E.; Sahebkar, A. Neuroprotective effects of curcumin through autophagy modulation. IUBMB Life 2020, 72, 652–664. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Chen, C.; Zhang, X.; Li, X.; Chen, Z.; Yang, C.; Liang, X.; Zhu, G.; Xu, Z. Neuroprotective Effect of Curcumin against Cerebral Ischemia-Reperfusion via Mediating Autophagy and Inflammation. J. Mol. Neurosci. 2018, 64, 129–139. [Google Scholar] [CrossRef]
- Wang, Y.L.; Liu, X.S.; Wang, S.S.; Xue, P.; Zeng, Z.L.; Yang, X.P.; Zhang, S.M.; Zheng, W.; Hua, L.; Li, J.F.; et al. Curcumin-Activated Mesenchymal Stem Cells Derived from Human Umbilical Cord and Their Effects on MPTP-Mouse Model of Parkinson’s Disease: A New Biological Therapy for Parkinson’s Disease. Stem Cells Int. 2020, 2020, 4636397. [Google Scholar] [CrossRef] [PubMed]
- Wanjiang, W.; Xin, C.; Yaxing, C.; Jie, W.; Hongyan, Z.; Fei, N.; Chengmin, L.; Chengjian, F.; Jichao, Y.; Jiangkai, L. Curcumin Improves Human Umbilical Cord-Derived Mesenchymal Stem Cell Survival via ERK1/2 Signaling and Promotes Motor Outcomes After Spinal Cord Injury. Cell Mol. Neurobiol. 2020, 15, 658. [Google Scholar] [CrossRef]
- Gorabi, A.M.; Kiaie, N.; Hajighasemi, S.; Jamialahmadi, T.; Majeed, M.; Sahebkar, A. The Effect of Curcumin on the Differentiation of Mesenchymal Stem Cells into Mesodermal Lineage. Molecules 2019, 24, 4029. [Google Scholar] [CrossRef] [PubMed]
- Khezri, K.; Maleki Dizaj, S.; Rahbar Saadat, Y.; Sharifi, S.; Shahi, S.; Ahmadian, E.; Eftekhari, A.; Dalir Abdolahinia, E.; Lotfipour, F. Osteogenic Differentiation of Mesenchymal Stem Cells via Curcumin-Containing Nanoscaffolds. Stem Cells Int. 2021, 2021, 1520052. [Google Scholar] [CrossRef]
- Ayadilord, M.; Nasseri, S.; Emadian Razavi, F.; Saharkhiz, M.; Rostami, Z.; Naseri, M. Immunomodulatory effects of phytosomal curcumin on related-micro RNAs, CD200 expression and inflammatory pathways in dental pulp stem cells. Cell Biochem. Funct. 2021, 39, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Pirmoradi, S.; Fathi, E.; Farahzadi, R.; Pilehvar-Soltanahmadi, Y.; Zarghami, N. Curcumin Affects Adipose Tissue-Derived Mesenchymal Stem Cell Aging Through TERT Gene Expression. Drug Res. 2018, 68, 213–221. [Google Scholar] [CrossRef]
- Mato-Basalo, R.; Morente-Lopez, M.; Arntz, O.J.; van de Loo, F.A.J.; Fafian-Labora, J.; Arufe, M.C. Therapeutic Potential for Regulation of the Nuclear Factor Kappa-B Transcription Factor p65 to Prevent Cellular Senescence and Activation of Pro-Inflammatory in Mesenchymal Stem Cells. Int. J. Mol. Sci. 2021, 22, 3367. [Google Scholar] [CrossRef]
- Yan, P.; Sun, X.; Chen, X.; Chen, Y.; Wang, X.; Su, D.; Zhou, H.; Gao, L.; Lu, L.; Wang, J.; et al. The Polyphenolic Compound Curcumin Conjugation with an Alkyne Moiety in the Process of Autophagy. Am. J. Chin. Med. 2018, 46, 673–687. [Google Scholar] [CrossRef]
- Shakeri, A.; Cicero, A.F.G.; Panahi, Y.; Mohajeri, M.; Sahebkar, A. Curcumin: A naturally occurring autophagy modulator. J. Cell. Physiol. 2019, 234, 5643–5654. [Google Scholar] [CrossRef]
- Han, J.; Pan, X.Y.; Xu, Y.; Xiao, Y.; An, Y.; Tie, L.; Pan, Y.; Li, X.J. Curcumin induces autophagy to protect vascular endothelial cell survival from oxidative stress damage. Autophagy 2012, 8, 812–825. [Google Scholar] [CrossRef]
- Huang, Z.; Ye, B.; Dai, Z.; Wu, X.; Lu, Z.; Shan, P.; Huang, W. Curcumin inhibits autophagy and apoptosis in hypoxia/reoxygenation-induced myocytes. Mol. Med. Rep. 2015, 11, 4678–4684. [Google Scholar] [CrossRef]
- Geissler, S.; Textor, M.; Kuhnisch, J.; Konnig, D.; Klein, O.; Ode, A.; Pfitzner, T.; Adjaye, J.; Kasper, G.; Duda, G.N. Functional comparison of chronological and in vitro aging: Differential role of the cytoskeleton and mitochondria in mesenchymal stromal cells. PLoS ONE 2012, 7, e52700. [Google Scholar] [CrossRef]
- Zhang, D.; Chen, Y.; Xu, X.; Xiang, H.; Shi, Y.; Gao, Y.; Wang, X.; Jiang, X.; Li, N.; Pan, J. Autophagy inhibits the mesenchymal stem cell aging induced by D-galactose through ROS/JNK/p38 signalling. Clin. Exp. Pharmacol. Physiol. 2020, 47, 466–477. [Google Scholar] [CrossRef]
- Zheng, Y.; Hu, C.J.; Zhuo, R.H.; Lei, Y.S.; Han, N.N.; He, L. Inhibition of autophagy alleviates the senescent state of rat mesenchymal stem cells during long-term culture. Mol. Med. Rep. 2014, 10, 3003–3008. [Google Scholar] [CrossRef]
- Zheng, Y.; Lei, Y.; Hu, C.; Hu, C. p53 regulates autophagic activity in senescent rat mesenchymal stromal cells. Exp. Gerontol. 2016, 75, 64–71. [Google Scholar] [CrossRef]
- Turinetto, V.; Vitale, E.; Giachino, C. Senescence in Human Mesenchymal Stem Cells: Functional Changes and Implications in Stem Cell-Based Therapy. Int. J. Mol. Sci. 2016, 17, 1164. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Cuervo, A.M.; Ravikumar, B.; Sarkar, S.; Korolchuk, V.; Kaushik, S.; Klionsky, D.J. In search of an “autophagomometer”. Autophagy 2009, 5, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.; Ezaki, J.; Murata, S.; et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Z.; Hong, C.G.; Hu, W.B.; Chen, M.L.; Duan, R.; Li, H.M.; Yue, T.; Cao, J.; Wang, Z.X.; Chen, C.Y.; et al. Autophagy receptor OPTN (optineurin) regulates mesenchymal stem cell fate and bone-fat balance during aging by clearing FABP3. Autophagy 2020, 15, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Malaise, O.; Tachikart, Y.; Constantinides, M.; Mumme, M.; Ferreira-Lopez, R.; Noack, S.; Krettek, C.; Noel, D.; Wang, J.; Jorgensen, C.; et al. Mesenchymal stem cell senescence alleviates their intrinsic and seno-suppressive paracrine properties contributing to osteoarthritis development. Aging 2019, 11, 9128–9146. [Google Scholar] [CrossRef]
- Kapetanos, K.; Asimakopoulos, D.; Christodoulou, N.; Vogt, A.; Khan, W. Chronological Age Affects MSC Senescence In Vitro—A Systematic Review. Int. J. Mol. Sci. 2021, 22, 7945. [Google Scholar] [CrossRef]
- Oja, S.; Komulainen, P.; Penttila, A.; Nystedt, J.; Korhonen, M. Automated image analysis detects aging in clinical-grade mesenchymal stromal cell cultures. Stem Cell. Res. Ther. 2018, 9, 6. [Google Scholar] [CrossRef]
- Qu, Y.N.; Zhang, L.; Wang, T.; Zhang, H.Y.; Yang, Z.J.; Yuan, F.F.; Wang, Y.; Li, S.W.; Jiang, X.X.; Xie, X.H. Vitamin C Treatment Rescues Prelamin A-Induced Premature Senescence of Subchondral Bone Mesenchymal Stem Cells. Stem Cells Int. 2020, 2020, 3150716. [Google Scholar] [CrossRef]
- Yun, S.P.; Han, Y.S.; Lee, J.H.; Kim, S.M.; Lee, S.H. Melatonin Rescues Mesenchymal Stem Cells from Senescence Induced by the Uremic Toxin p-Cresol via Inhibiting mTOR-Dependent Autophagy. Biomol. Ther. 2018, 26, 389–398. [Google Scholar] [CrossRef]
- Constanze, B.; Popper, B.; Aggarwal, B.B.; Shakibaei, M. Evidence that TNF-beta suppresses osteoblast differentiation of mesenchymal stem cells and resveratrol reverses it through modulation of NF-kappaB, Sirt1 and Runx2. Cell Tissue Res. 2020, 381, 83–98. [Google Scholar] [CrossRef]
- Wang, N.; Wang, F.; Gao, Y.; Yin, P.; Pan, C.; Liu, W.; Zhou, Z.; Wang, J. Curcumin protects human adipose-derived mesenchymal stem cells against oxidative stress-induced inhibition of osteogenesis. J. Pharmacol. Sci. 2016, 132, 192–200. [Google Scholar] [CrossRef]
- Ghufran, H.; Mehmood, A.; Azam, M.; Butt, H.; Ramzan, A.; Yousaf, M.A.; Ejaz, A.; Tarar, M.N.; Riazuddin, S. Curcumin preconditioned human adipose derived stem cells co-transplanted with platelet rich plasma improve wound healing in diabetic rats. Life Sci. 2020, 257, 118091. [Google Scholar] [CrossRef]
- Yang, Q.; Leong, S.A.; Chan, K.P.; Yuan, X.L.; Ng, T.K. Complex effect of continuous curcumin exposure on human bone marrow-derived mesenchymal stem cell regenerative properties through matrix metalloproteinase regulation. Basic Clin. Pharmacol. Toxicol. 2021, 128, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Zhong, L.; Zhou, Z.; Gu, C.; Huang, X.; Shen, L.; Cao, S.; Ren, Z.; Zuo, Z.; Deng, J.; et al. Autophagy: A promising therapeutic target for improving mesenchymal stem cell biological functions. Mol. Cell Biochem. 2021, 476, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Han, X.; Qu, G.; Su, L.; Zhao, B.; Miao, J. A pH probe inhibits senescence in mesenchymal stem cells. Stem Cell. Res. Ther. 2018, 9, 343. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Jeong, J.K.; Park, S.Y. AMPK Activation Mediated by Hinokitiol Inhibits Adipogenic Differentiation of Mesenchymal Stem Cells through Autophagy Flux. Int. J. Endocrinol. 2018, 2018, 2014192. [Google Scholar] [CrossRef] [PubMed]
- Stacchiotti, A.; Corsetti, G. Natural Compounds and Autophagy: Allies Against Neurodegeneration. Front. Cell Dev. Biol. 2020, 8, 555409. [Google Scholar] [CrossRef] [PubMed]
- Rubinsztein, D.C.; Marino, G.; Kroemer, G. Autophagy and aging. Cell 2011, 146, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Molaei, S.; Roudkenar, M.H.; Amiri, F.; Harati, M.D.; Bahadori, M.; Jaleh, F.; Jalili, M.A.; Mohammadi Roushandeh, A. Down-regulation of the autophagy gene, ATG7, protects bone marrow-derived mesenchymal stem cells from stressful conditions. Blood Res. 2015, 50, 80–86. [Google Scholar] [CrossRef]
- Kim, D.W.; Choi, C.H.; Park, J.P.; Lee, S.J. Nanospheres Loaded with Curcumin Improve the Bioactivity of Umbilical Cord Blood-Mesenchymal Stem Cells via c-Src Activation During the Skin Wound Healing Process. Cells 2020, 9, 1467. [Google Scholar] [CrossRef]
- Golchin, A.; Hosseinzadeh, S.; Jouybar, A.; Staji, M.; Soleimani, M.; Ardeshirylajimi, A.; Khojasteh, A. Wound healing improvement by curcumin-loaded electrospun nanofibers and BFP-MSCs as a bioactive dressing. Polym. Adv. Technol. 2020, 31, 1519–1531. [Google Scholar] [CrossRef]
- Zhou, Z.H.; Gu, C.W.; Li, J.; Huang, X.Y.; Deng, J.Q.; Shen, L.H.; Cao, S.Z.; Deng, J.L.; Zuo, Z.C.; Wang, Y.; et al. 17 beta-estradiol affects proliferation and apoptosis of canine bone marrow mesenchymal stem cells in vitro. Pol. J. Vet. Sci. 2020, 23, 235–245. [Google Scholar] [CrossRef]
- Hou, J.; Han, Z.P.; Jing, Y.Y.; Yang, X.; Zhang, S.S.; Sun, K.; Hao, C.; Meng, Y.; Yu, F.H.; Liu, X.Q.; et al. Autophagy prevents irradiation injury and maintains stemness through decreasing ROS generation in mesenchymal stem cells. Cell Death Dis. 2013, 4, e844. [Google Scholar] [CrossRef]






| Surface Antigens | CD31 | CD34 | CD45 | CD90 | CD105 | ITGB1 |
|---|---|---|---|---|---|---|
| Positive cell rate (%) | 6.69 ± 1.62 | 3.09 ± 0.77 | 1.50 ± 0.09 | 98.77 ± 0.80 | 98.17 ± 0.52 | 99.63 ± 0.36 |
| Primers | Forward Primer Sequence (5′-3′) | Reverse Primer Sequence (5′-3′) | Product Size (bp) |
|---|---|---|---|
| GAPDH | TCCCGCCAACATCAAA | TCACGCCCATCACAAAC | 163 |
| SOX-2 | AACCCCAAGATGCACAACTC | CGGGGCCGGTATTTATAATC | 171 |
| Nanog | CCTGCATCCTTGCCAATGTC | TCCGGGCTGTCCTGAGTAAG | 98 |
| p16 | CGGAGCCCGATTCAGGTCAT | CACCAGCGTGTCCAGGAAGC | 150 |
| p21 | CATCCCTCATGGCAGCAAG | AGGCAGGGAGACCTTGGACA | 208 |
| IL-6 | TGATGGCTACTGCTTTCCCTACC | CCAGTGCCTCTTTGCTGTCTTC | 195 |
| INF-α | GCCTCTTCTCCTTCCTCCTC | GCTACTGGCTTGTCACTTGG | 169 |
| LC3 | AGAGCAGCATCCTACCAA | CCATCTTCATCCTTCTCACT | 249 |
| ATG7 | ACGCCAATATCTCCTACTCCAA | CTGCTCTAGTTGCTCCACATC | 230 |
| ATG12 | ATGGCTGAGGAGTCGGAGT | TGGTTCGGGTTCGCTCTAC | 241 |
| ULK1 | TGGAGCAAGAGCACACGGAGA | GGATCTGGTCAATGGCGGTCTG | 258 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, J.; Ouyang, P.; Li, W.; Zhong, L.; Gu, C.; Shen, L.; Cao, S.; Yin, L.; Ren, Z.; Zuo, Z.; et al. Curcumin Alleviates the Senescence of Canine Bone Marrow Mesenchymal Stem Cells during In Vitro Expansion by Activating the Autophagy Pathway. Int. J. Mol. Sci. 2021, 22, 11356. https://doi.org/10.3390/ijms222111356
Deng J, Ouyang P, Li W, Zhong L, Gu C, Shen L, Cao S, Yin L, Ren Z, Zuo Z, et al. Curcumin Alleviates the Senescence of Canine Bone Marrow Mesenchymal Stem Cells during In Vitro Expansion by Activating the Autophagy Pathway. International Journal of Molecular Sciences. 2021; 22(21):11356. https://doi.org/10.3390/ijms222111356
Chicago/Turabian StyleDeng, Jiaqiang, Ping Ouyang, Weiyao Li, Lijun Zhong, Congwei Gu, Liuhong Shen, Suizhong Cao, Lizi Yin, Zhihua Ren, Zhicai Zuo, and et al. 2021. "Curcumin Alleviates the Senescence of Canine Bone Marrow Mesenchymal Stem Cells during In Vitro Expansion by Activating the Autophagy Pathway" International Journal of Molecular Sciences 22, no. 21: 11356. https://doi.org/10.3390/ijms222111356
APA StyleDeng, J., Ouyang, P., Li, W., Zhong, L., Gu, C., Shen, L., Cao, S., Yin, L., Ren, Z., Zuo, Z., Deng, J., Yan, Q., & Yu, S. (2021). Curcumin Alleviates the Senescence of Canine Bone Marrow Mesenchymal Stem Cells during In Vitro Expansion by Activating the Autophagy Pathway. International Journal of Molecular Sciences, 22(21), 11356. https://doi.org/10.3390/ijms222111356







