Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities
Abstract
:1. Introduction
2. Antimicrobial Mechanism of AMPs
2.1. Mechanism of Cell Wall Targeting
2.2. Mechanism of Membrane Targeting
2.2.1. Transmembrane Pore Model
2.2.2. Non-Membrane Pore Model
| Action Model | Mode of Action | Represents AMPs | |
|---|---|---|---|
| Transmembrane pore model | Barrel-stave model | Holes | Alamethicin, pardaxin, and protegrins [56,57,58] |
| Toroidal-pore model | Holes | Lacticin Q and melittin [59,60] | |
| Nonmembrane pore model | Carpet model/Detergent-like mode | Splitting | Cecropin P1 and aurein 1.2 [61,62] |
| Agglutination model | Devour | Thanatin [55] | |
2.3. Intracellular Targeting Mechanism of Action
2.3.1. Mechanism of Translocation
Energy-Independent Direct Permeation of the Plasma Membrane
Energy-Dependent Endocytosis
2.3.2. Intracellular Mechanism of Action
2.3.3. Development and Significance of Intracellular Targeted AMPs
2.4. Dual or Multiple Mechanisms of Action
3. Other Mechanisms
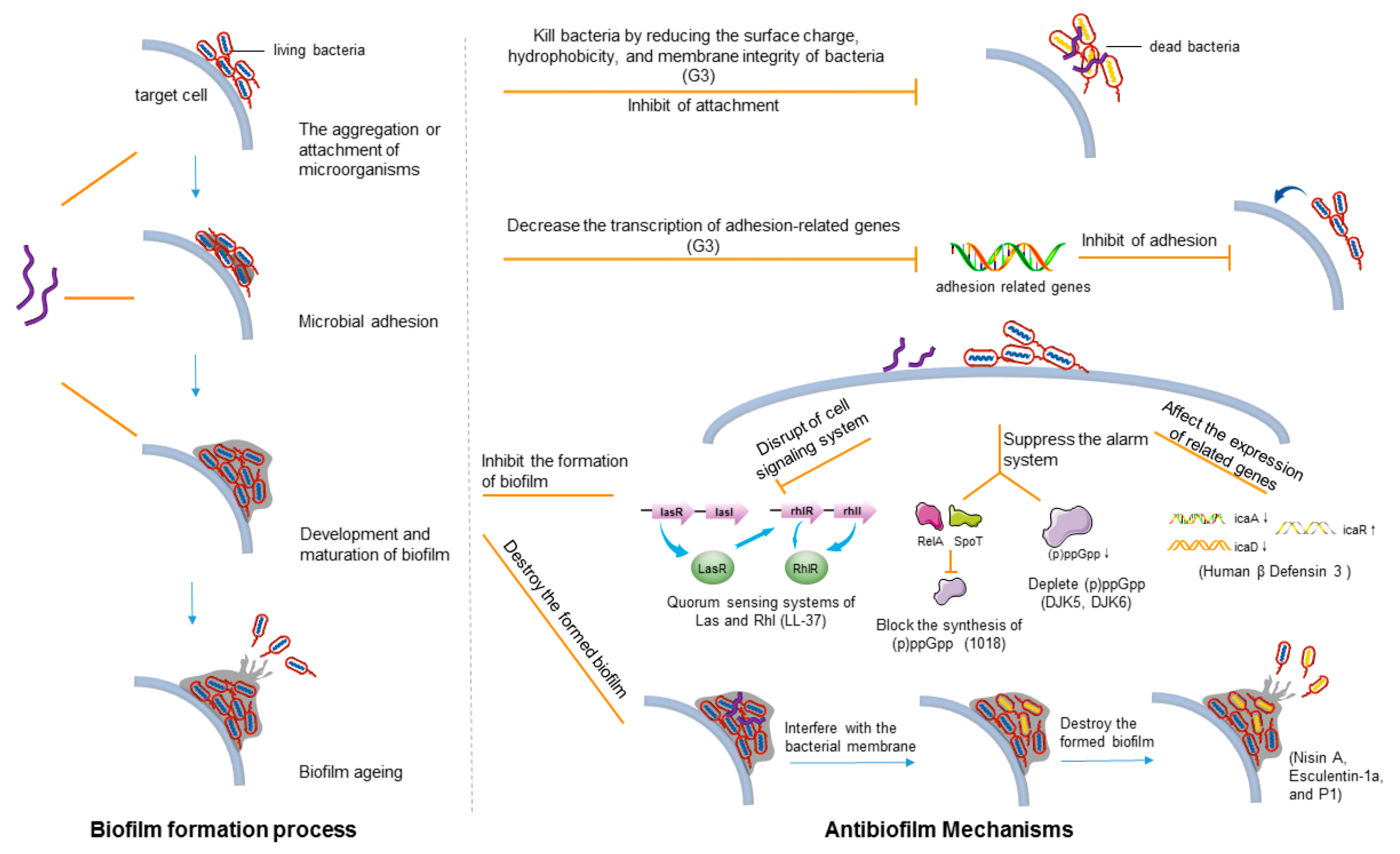
3.1. Antibiofilm Mechanism
3.1.1. Biofilm Formation Process
3.1.2. Main Mechanism of AMPs against Biofilms
| AMPs | Microorganisms | Mechanism of Action | References |
|---|---|---|---|
| LL-37 | Pseudomonas aeruginosa | Inhibit bacterial adhesion; disruption of cell signaling system | [126] |
| DJK5 and DJK6 | Pseudomonas aeruginosa | Suppress the alarm system | [131,132] |
| 1081 | A series of G+ and G− (Pseudomonas aeruginosa, Escherichia coli, etc.) | Suppress the alarm system; eradication of mature biofilms | [130] |
| Human β-defensin 3 | Staphylococcus epidermidis | Downregulate the expression of binding protein transport genes responsible for biofilm formation | [133,134] |
| 1037 | Pseudomonas aeruginosa | Downregulate the expression of binding protein transport genes responsible for biofilm formation | [135] |
| Nisin A | MRSA | Interfere with the bacterial membrane potential in the biofilm | [125] |
| Esculentin (1–21) | Pseudomonas aeruginosa | Interfere with the bacterial membrane potential in the biofilm | [137] |
| G3 | Streptococcus mutans | Inhibit bacterial adhesion; degrade EPSs | [141] |
| P1 | Streptococcus mutans | Degrade EPSs | [138] |
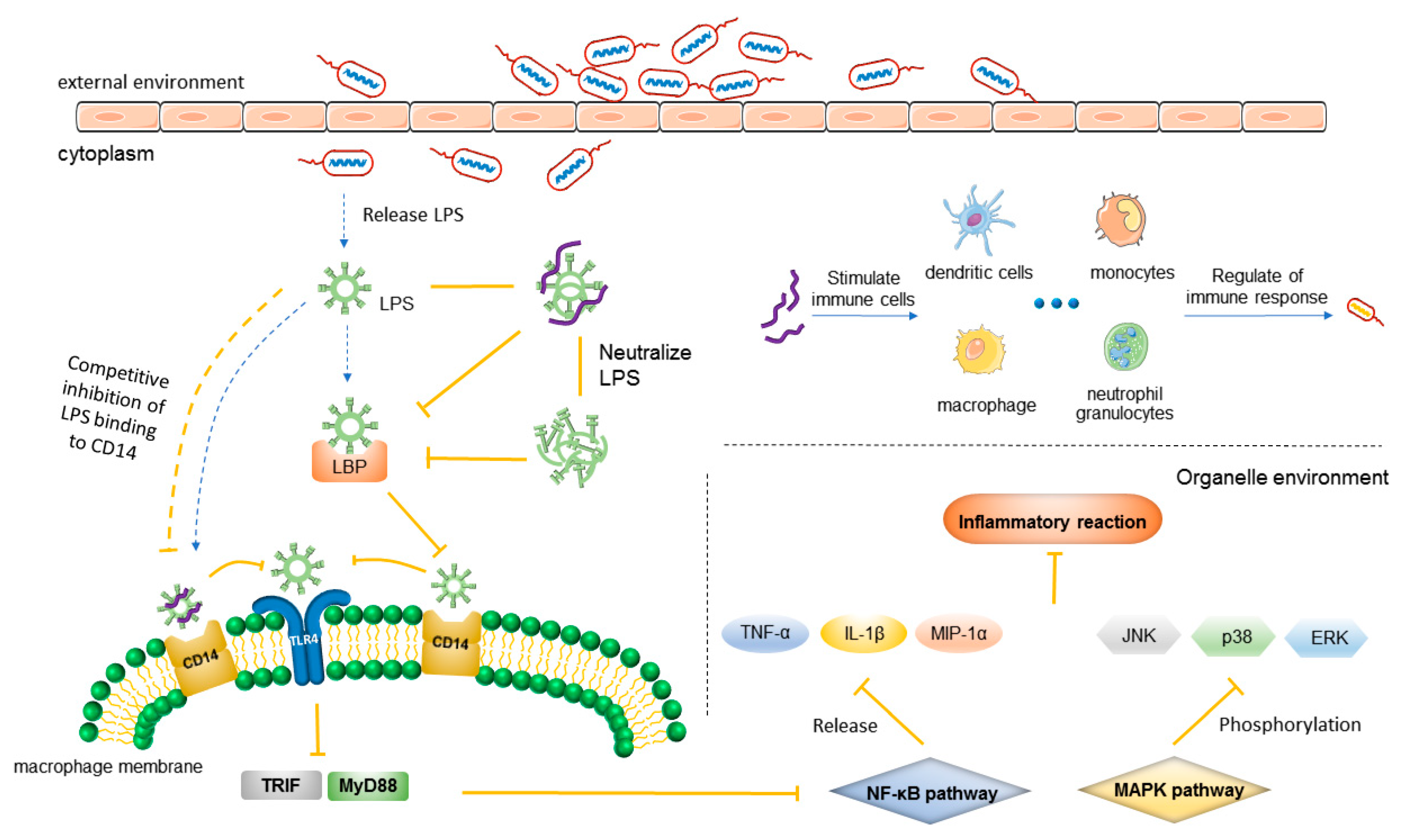
3.2. Anti-Inflammatory Mechanism
3.2.1. Mechanism of Inflammation
3.2.2. Anti-Inflammatory Mechanism of AMPs
| AMP | Mechanism of Action | References |
|---|---|---|
| LL-37 | Binds to LPS receptors (CD14 and TLR4) expressed on cells and inhibits TNF-α; neutralizes LPS; suppresses the macrophage pyroptosis that induces the release of pro-inflammatory cytokines; releases neutrophil extracellular traps; stimulates neutrophils to release antimicrobial microvesicles | [178,179,180] |
| CAP18 | Binds to LPS, inhibits the interaction between LPS and LPS-binding protein, and attaches to CD14 molecule, thus inhibiting the expression of LPS-binding CD14 (+) cells to reduce the production of TNF-α by these cells | [165] |
| dCATH 12-4 and dCATH 12-5 | Bind with LPS oligomers leading to the dissociation of LPS aggregates, which prevents LPS from binding to LBP or alternatively to macrophage CD14 receptor | [163] |
| PA-13 | Neutralize LPS; inhibit LPS-mediated TLR activation | [181] |
| SET-M33 and SET-M33D | Neutralize LPS; reduce the release of TNF-α, IL6, COX-2, and other inflammatory factors | [182,183] |
| γ-AA | Inhibits LPS-activated TLR4 signal transduction | [184] |
| OIR3 | Inhibits pro-inflammatory factors TNF-α, IL-1β, and IL-6 release | [185] |
| LB-PG, CA-PG | Inhibit the expression of pro-inflammatory cytokines and chemokines induced by LPS, such as TNF-α, iNOS, MIP-1α, and monocytes | [186] |
| GW-A2 | Inhibits No, iNOS and TNF-α, and IL-6 in LPS-activated macrophages; reduces NF- κB activation increase; inhibits LPS- and ATP-induced NLRP3 inflammasome activation; neutralizes LPS and ATP | [157] |
| WALK11.3 | Inhibits the expression of inflammatory mediators, including No, IL-1β, IL-6, INF- β, and TNF-α; specifically inhibits TLR4 endocytosis | [187] |
| Ps-K18 | Inhibits TLR4-mediated NF- κB pathway, activating innate defense | [154] |
| Papiliocin | Inhibits expression of the NF- κB pathway | [188] |
| CLP-19 | Inhibits LPS–LBP binding and subsequent MAPK signaling | [189] |
| CecropinA | Inhibits ERK, JNK, and p38 phosphorylation in the MAPK pathway | [190] |
| Human beta-defensin (hBD)-3 and hBD-4 | Mediate phosphorylation of ERK-1/2 and p38; activate mast cells, degranulate mast cells, and increase vascular permeability, thereby regulating active defense and enhancing anti-inflammatory effects | [176] |
| IDR-1 | Activates FPR1 chemotactic neutrophils to participate in immune regulation | [175] |
4. Concluding Remarks and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Jia, F.; Wang, J.; Zhang, L.; Zhou, J.; He, Y.; Lu, Y.; Liu, K.; Yan, W.; Wang, K. Multiple action mechanism and in vivo antimicrobial efficacy of antimicrobial peptide Jelleine-I. J. Pept. Sci. 2021, 27, e3294. [Google Scholar] [CrossRef]
- Shi, Y.; Li, C.; Wang, M.; Chen, Z.; Luo, Y.; Xia, X.S.; Song, Y.; Sun, Y.; Zhang, A.M. Cathelicidin-DM is an Antimicrobial Peptide from Duttaphrynus melanostictus and Has Wound-Healing Therapeutic Potential. ACS Omega 2020, 5, 9301–9310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014, 12, 35–48. [Google Scholar] [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Savini, F.; Loffredo, M.R.; Troiano, C.; Bobone, S.; Malanovic, N.; Eichmann, T.O.; Caprio, L.; Canale, V.C.; Park, Y.; Mangoni, M.L.; et al. Binding of an antimicrobial peptide to bacterial cells: Interaction with different species, strains and cellular components. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183291. [Google Scholar] [CrossRef]
- Bassetti, M.; Merelli, M.; Temperoni, C.; Astilean, A. New antibiotics for bad bugs: Where are we? Ann. Clin. Microbiol. Antimicrob. 2013, 12, 22. [Google Scholar] [CrossRef] [Green Version]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Kostyanev, T.; Bonten, M.J.; O’Brien, S.; Steel, H.; Ross, S.; François, B.; Tacconelli, E.; Winterhalter, M.; Stavenger, R.A.; Karlén, A.; et al. The Innovative Medicines Initiative’s New Drugs for Bad Bugs programme: European public-private partnerships for the development of new strategies to tackle antibiotic resistance. J. Antimicrob. Chemother. 2016, 71, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Karam, G.; Chastre, J.; Wilcox, M.H.; Vincent, J.L. Antibiotic strategies in the era of multidrug resistance. Crit. Care 2016, 20, 136. [Google Scholar] [CrossRef] [Green Version]
- Bragginton, E.C.; Piddock, L.J. UK and European Union public and charitable funding from 2008 to 2013 for bacteriology and antibiotic research in the UK: An observational study. Lancet Infect. Dis. 2014, 14, 857–868. [Google Scholar] [CrossRef]
- Carlet, J.; Collignon, P.; Goldmann, D.; Goossens, H.; Gyssens, I.C.; Harbarth, S.; Jarlier, V.; Levy, S.B.; N’Doye, B.; Pittet, D.; et al. Society’s failure to protect a precious resource: Antibiotics. Lancet 2011, 378, 369–371. [Google Scholar] [CrossRef]
- van Hoek, M.L. Antimicrobial peptides in reptiles. Pharmaceuticals 2014, 7, 723–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawrot, R.; Barylski, J.; Nowicki, G.; Broniarczyk, J.; Buchwald, W.; Goździcka-Józefiak, A. Plant antimicrobial peptides. Folia Microbiol. 2014, 59, 181–196. [Google Scholar] [CrossRef] [Green Version]
- Lehrer, R.I.; Ganz, T. Antimicrobial peptides in mammalian and insect host defence. Curr. Opin. Immunol. 1999, 11, 23–27. [Google Scholar] [CrossRef]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Park, S.; Park, S.H.; Ahn, H.C.; Kim, S.; Kim, S.S.; Lee, B.J.; Lee, B.J. Structural study of novel antimicrobial peptides, nigrocins, isolated from Rana nigromaculata. FEBS Lett. 2001, 507, 95–100. [Google Scholar] [CrossRef] [Green Version]
- Boman, H.G.; Hultmark, D. Cell-free immunity in insects. Annu. Rev. Microbiol. 1987, 41, 103–126. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Samperio, P. Recent advances in the field of antimicrobial peptides in inflammatory diseases. Adv. Biomed. Res. 2013, 2, 50. [Google Scholar] [CrossRef]
- Diamond, G.; Beckloff, N.; Weinberg, A.; Kisich, K.O. The roles of antimicrobial peptides in innate host defense. Curr. Pharm. Des. 2009, 15, 2377–2392. [Google Scholar] [CrossRef] [Green Version]
- Reddy, K.V.; Yedery, R.D.; Aranha, C. Antimicrobial peptides: Premises and promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [Green Version]
- Chung, P.Y.; Khanum, R. Antimicrobial peptides as potential anti-biofilm agents against multidrug-resistant bacteria. J. Microbiol. Immunol. Infect. 2017, 50, 405–410. [Google Scholar] [CrossRef]
- Mygind, P.H.; Fischer, R.L.; Schnorr, K.M.; Hansen, M.T.; Sönksen, C.P.; Ludvigsen, S.; Raventós, D.; Buskov, S.; Christensen, B.; De Maria, L.; et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature 2005, 437, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Liu, Q.; Wang, D.; Li, J. Characterization and antimicrobial mechanism of CF-14, a new antimicrobial peptide from the epidermal mucus of catfish. Fish Shellfish. Immunol. 2019, 92, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.B.; Seo, J. Antimicrobial peptides under clinical investigation. Pept. Sci. 2019, 111, e24122. [Google Scholar] [CrossRef]
- Radek, K.; Gallo, R. Antimicrobial peptides: Natural effectors of the innate immune system. Semin. Immunopathol. 2007, 29, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- Wang, J.; Dou, X.; Song, J.; Lyu, Y.; Zhu, X.; Xu, L.; Li, W.; Shan, A. Antimicrobial peptides: Promising alternatives in the post feeding antibiotic era. Med. Res. Rev. 2019, 39, 831–859. [Google Scholar] [CrossRef]
- Haney, E.F.; Straus, S.K.; Hancock, R.E.W. Reassessing the Host Defense Peptide Landscape. Front. Chem. 2019, 7, 43. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, T.A.E.; Hammami, R. Recent insights into structure-function relationships of antimicrobial peptides. J. Food Biochem. 2019, 43, e12546. [Google Scholar] [CrossRef] [Green Version]
- Raheem, N.; Straus, S.K. Mechanisms of Action for Antimicrobial Peptides With Antibacterial and Antibiofilm Functions. Front. Microbiol. 2019, 10, 2866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, C.F.; Fang, C.M.; Sekaran, S.D. Intracellular Targeting Mechanisms by Antimicrobial Peptides. Antimicrob. Agents Chemother 2017, 61, e02340-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [Green Version]
- Ulmschneider, J.P. Charged Antimicrobial Peptides Can Translocate across Membranes without Forming Channel-like Pores. Biophys. J. 2017, 113, 73–81. [Google Scholar] [CrossRef] [Green Version]
- Scocchi, M.; Tossi, A.; Gennaro, R. Proline-rich antimicrobial peptides: Converging to a non-lytic mechanism of action. Cell Mol. Life Sci. 2011, 68, 2317–2330. [Google Scholar] [CrossRef]
- Jean-François, F.; Castano, S.; Desbat, B.; Odaert, B.; Roux, M.; Metz-Boutigue, M.H.; Dufourc, E.J. Aggregation of cateslytin beta-sheets on negatively charged lipids promotes rigid membrane domains. A new mode of action for antimicrobial peptides? Biochemistry 2008, 47, 6394–6402. [Google Scholar] [CrossRef] [PubMed]
- Graf, M.; Wilson, D.N. Intracellular Antimicrobial Peptides Targeting the Protein Synthesis Machinery. Adv. Exp. Med. Biol. 2019, 1117, 73–89. [Google Scholar] [CrossRef]
- Park, J.; Kang, H.K.; Choi, M.C.; Chae, J.D.; Son, B.K.; Chong, Y.P.; Seo, C.H.; Park, Y. Antibacterial activity and mechanism of action of analogues derived from the antimicrobial peptide mBjAMP1 isolated from Branchiostoma japonicum. J. Antimicrob. Chemother. 2018, 73, 2054–2063. [Google Scholar] [CrossRef]
- Zong, L.; Teng, D.; Wang, X.; Mao, R.; Yang, N.; Hao, Y.; Wang, J. Mechanism of action of a novel recombinant peptide, MP1102, against Clostridium perfringens type C. Appl. Microbiol. Biotechnol. 2016, 100, 5045–5057. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Moravej, H.; Moravej, Z.; Yazdanparast, M.; Heiat, M.; Mirhosseini, A.; Moosazadeh Moghaddam, M.; Mirnejad, R. Antimicrobial Peptides: Features, Action, and Their Resistance Mechanisms in Bacteria. Microb. Drug Resist. 2018, 24, 747–767. [Google Scholar] [CrossRef]
- Navarre, W.W.; Schneewind, O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 1999, 63, 174–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, A.L. Bacterial wall as target for attack: Past, present, and future research. Clin. Microbiol. Rev. 2003, 16, 673–687. [Google Scholar] [CrossRef] [Green Version]
- Omardien, S.; Brul, S.; Zaat, S.A. Antimicrobial Activity of Cationic Antimicrobial Peptides against Gram-Positives: Current Progress Made in Understanding the Mode of Action and the Response of Bacteria. Front. Cell Dev. Biol. 2016, 4, 111. [Google Scholar] [CrossRef] [PubMed]
- Münch, D.; Sahl, H.G. Structural variations of the cell wall precursor lipid II in Gram-positive bacteria—Impact on binding and efficacy of antimicrobial peptides. Biochim. Biophys. Acta 2015, 1848, 3062–3071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, T.; Kruse, T.; Wimmer, R.; Wiedemann, I.; Sass, V.; Pag, U.; Jansen, A.; Nielsen, A.K.; Mygind, P.H.; Raventós, D.S.; et al. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 2010, 328, 1168–1172. [Google Scholar] [CrossRef] [Green Version]
- Wenzel, M.; Chiriac, A.I.; Otto, A.; Zweytick, D.; May, C.; Schumacher, C.; Gust, R.; Albada, H.B.; Penkova, M.; Krämer, U.; et al. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc. Natl. Acad. Sci. USA 2014, 111, E1409–E1418. [Google Scholar] [CrossRef] [Green Version]
- Ho, S.; Pothoulakis, C.; Koon, H.W. Antimicrobial peptides and colitis. Curr. Pharm. Des. 2013, 19, 40–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epand, R.M.; Walker, C.; Epand, R.F.; Magarvey, N.A. Molecular mechanisms of membrane targeting antibiotics. Biochim. Biophys. Acta 2016, 1858, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Ciumac, D.; Gong, H.; Hu, X.; Lu, J.R. Membrane targeting cationic antimicrobial peptides. J. Colloid Interface Sci. 2019, 537, 163–185. [Google Scholar] [CrossRef]
- Sinha, S.; Zheng, L.; Mu, Y.; Ng, W.J.; Bhattacharjya, S. Structure and Interactions of A Host Defense Antimicrobial Peptide Thanatin in Lipopolysaccharide Micelles Reveal Mechanism of Bacterial Cell Agglutination. Sci. Rep. 2017, 7, 17795. [Google Scholar] [CrossRef] [Green Version]
- Hallock, K.J.; Lee, D.K.; Omnaas, J.; Mosberg, H.I.; Ramamoorthy, A. Membrane composition determines pardaxin’s mechanism of lipid bilayer disruption. Biophys. J. 2002, 83, 1004–1013. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Harroun, T.A.; Weiss, T.M.; Ding, L.; Huang, H.W. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 2001, 81, 1475–1485. [Google Scholar] [CrossRef] [Green Version]
- Henderson, J.M.; Waring, A.J.; Separovic, F.; Lee, K.Y.C. Antimicrobial Peptides Share a Common Interaction Driven by Membrane Line Tension Reduction. Biophys. J. 2016, 111, 2176–2189. [Google Scholar] [CrossRef] [Green Version]
- Yoneyama, F.; Imura, Y.; Ohno, K.; Zendo, T.; Nakayama, J.; Matsuzaki, K.; Sonomoto, K. Peptide-lipid huge toroidal pore, a new antimicrobial mechanism mediated by a lactococcal bacteriocin, lacticin Q. Antimicrob. Agents Chemother. 2009, 53, 3211–3217. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, D.; Leontiadou, H.; Mark, A.E.; Marrink, S.J. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim. Biophys. Acta 2008, 1778, 2308–2317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, D.I.; Le Brun, A.P.; Whitwell, T.C.; Sani, M.A.; James, M.; Separovic, F. The antimicrobial peptide aurein 1.2 disrupts model membranes via the carpet mechanism. Phys. Chem. Chem. Phys. 2012, 14, 15739–15751. [Google Scholar] [CrossRef] [PubMed]
- Gazit, E.; Boman, A.; Boman, H.G.; Shai, Y. Interaction of the mammalian antibacterial peptide cecropin P1 with phospholipid vesicles. Biochemistry 1995, 34, 11479–11488. [Google Scholar] [CrossRef]
- Neundorf, I. Antimicrobial and Cell-Penetrating Peptides: How to Understand Two Distinct Functions Despite Similar Physicochemical Properties. Adv. Exp. Med. Biol. 2019, 1117, 93–109. [Google Scholar] [CrossRef]
- Henriques, S.T.; Melo, M.N.; Castanho, M.A. Cell-penetrating peptides and antimicrobial peptides: How different are they? Biochem. J. 2006, 399, 1–7. [Google Scholar] [CrossRef]
- Budagavi, D.P.; Chugh, A. Antibacterial properties of Latarcin 1 derived cell-penetrating peptides. Eur. J. Pharm. Sci. 2018, 115, 43–49. [Google Scholar] [CrossRef]
- Buccini, D.F.; Cardoso, M.H.; Franco, O.L. Antimicrobial Peptides and Cell-Penetrating Peptides for Treating Intracellular Bacterial Infections. Front. Cell Infect. Microbiol. 2020, 10, 612931. [Google Scholar] [CrossRef]
- Bechara, C.; Sagan, S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 2013, 587, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Madani, F.; Lindberg, S.; Langel, U.; Futaki, S.; Gräslund, A. Mechanisms of cellular uptake of cell-penetrating peptides. J. Biophys. 2011, 2011, 414729. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.W.; Chen, F.Y.; Lee, M.T. Molecular mechanism of Peptide-induced pores in membranes. Phys. Rev. Lett. 2004, 92, 198304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K. Magainins as paradigm for the mode of action of pore forming polypeptides. Biochim. Biophys. Acta 1998, 1376, 391–400. [Google Scholar] [CrossRef]
- Nicolas, P. Multifunctional host defense peptides: Intracellular-targeting antimicrobial peptides. FEBS J. 2009, 276, 6483–6496. [Google Scholar] [CrossRef]
- Jean-François, F.; Elezgaray, J.; Berson, P.; Vacher, P.; Dufourc, E.J. Pore formation induced by an antimicrobial peptide: Electrostatic effects. Biophys. J. 2008, 95, 5748–5756. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, S.; Chikushi, A.; Tougu, S.; Imura, Y.; Nishida, M.; Yano, Y.; Matsuzaki, K. Membrane translocation mechanism of the antimicrobial peptide buforin 2. Biochemistry 2004, 43, 15610–15616. [Google Scholar] [CrossRef]
- Hasan, M.; Moghal, M.M.R.; Saha, S.K.; Yamazaki, M. The role of membrane tension in the action of antimicrobial peptides and cell-penetrating peptides in biomembranes. Biophys. Rev. 2019, 11, 431–448. [Google Scholar] [CrossRef]
- Elmore, D.E. Insights into buforin II membrane translocation from molecular dynamics simulations. Peptides 2012, 38, 357–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardoso, M.H.; Meneguetti, B.T.; Costa, B.O.; Buccini, D.F.; Oshiro, K.G.N.; Preza, S.L.E.; Carvalho, C.M.E.; Migliolo, L.; Franco, O.L. Non-Lytic Antibacterial Peptides That Translocate Through Bacterial Membranes to Act on Intracellular Targets. Int. J. Mol. Sci. 2019, 20, 4877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Järver, P.; Mäger, I.; Langel, Ü. In vivo biodistribution and efficacy of peptide mediated delivery. Trends Pharmacol. Sci. 2010, 31, 528–535. [Google Scholar] [CrossRef]
- Epand, R.M.; Epand, R.F. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim. Biophys. Acta 2009, 1788, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Baxter, A.A.; Lay, F.T.; Poon, I.K.H.; Kvansakul, M.; Hulett, M.D. Tumor cell membrane-targeting cationic antimicrobial peptides: Novel insights into mechanisms of action and therapeutic prospects. Cell Mol. Life Sci. 2017, 74, 3809–3825. [Google Scholar] [CrossRef]
- Magzoub, M.; Pramanik, A.; Gräslund, A. Modeling the endosomal escape of cell-penetrating peptides: Transmembrane pH gradient driven translocation across phospholipid bilayers. Biochemistry 2005, 44, 14890–14897. [Google Scholar] [CrossRef] [PubMed]
- Terrone, D.; Sang, S.L.; Roudaia, L.; Silvius, J.R. Penetratin and related cell-penetrating cationic peptides can translocate across lipid bilayers in the presence of a transbilayer potential. Biochemistry 2003, 42, 13787–13799. [Google Scholar] [CrossRef] [PubMed]
- Runti, G.; Lopez Ruiz Mdel, C.; Stoilova, T.; Hussain, R.; Jennions, M.; Choudhury, H.G.; Benincasa, M.; Gennaro, R.; Beis, K.; Scocchi, M. Functional characterization of SbmA, a bacterial inner membrane transporter required for importing the antimicrobial peptide Bac7(1–35). J. Bacteriol. 2013, 195, 5343–5351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armas, F.; Di Stasi, A.; Mardirossian, M.; Romani, A.A.; Benincasa, M.; Scocchi, M. Effects of Lipidation on a Proline-Rich Antibacterial Peptide. Int. J. Mol. Sci. 2021, 22, 7959. [Google Scholar] [CrossRef]
- Hansen, A.M.; Bonke, G.; Larsen, C.J.; Yavari, N.; Nielsen, P.E.; Franzyk, H. Antibacterial Peptide Nucleic Acid-Antimicrobial Peptide (PNA-AMP) Conjugates: Antisense Targeting of Fatty Acid Biosynthesis. Bioconjug. Chem. 2016, 27, 863–867. [Google Scholar] [CrossRef]
- Guida, F.; Benincasa, M.; Zahariev, S.; Scocchi, M.; Berti, F.; Gennaro, R.; Tossi, A. Effect of size and N-terminal residue characteristics on bacterial cell penetration and antibacterial activity of the proline-rich peptide Bac7. J. Med. Chem. 2015, 58, 1195–1204. [Google Scholar] [CrossRef]
- Li, W.F.; Ma, G.X.; Zhou, X.X. Apidaecin-type peptides: Biodiversity, structure-function relationships and mode of action. Peptides 2006, 27, 2350–2359. [Google Scholar] [CrossRef]
- Lele, D.S.; Kaur, G.; Thiruvikraman, M.; Kaur, K.J. Comparing naturally occurring glycosylated forms of proline rich antibacterial peptide, Drosocin. Glycoconj J. 2017, 34, 613–624. [Google Scholar] [CrossRef]
- Alves, I.D.; Goasdoué, N.; Correia, I.; Aubry, S.; Galanth, C.; Sagan, S.; Lavielle, S.; Chassaing, G. Membrane interaction and perturbation mechanisms induced by two cationic cell penetrating peptides with distinct charge distribution. Biochim. Biophys. Acta 2008, 1780, 948–959. [Google Scholar] [CrossRef]
- Van den Berg, A.; Dowdy, S.F. Protein transduction domain delivery of therapeutic macromolecules. Curr. Opin. Biotechnol. 2011, 22, 888–893. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Ulvatne, H.; Samuelsen, Ø.; Haukland, H.H.; Krämer, M.; Vorland, L.H. Lactoferricin B inhibits bacterial macromolecular synthesis in Escherichia coli and Bacillus subtilis. FEMS Microbiol. Lett. 2004, 237, 377–384. [Google Scholar] [CrossRef] [Green Version]
- Ramamourthy, G.; Park, J.; Seo, C.H.; Vogel, H.J.; Park, Y. Antifungal and Antibiofilm Activities and the Mechanism of Action of Repeating Lysine-Tryptophan Peptides against Candida albicans. Microorganisms 2020, 8, 758. [Google Scholar] [CrossRef]
- Braffman, N.R.; Piscotta, F.J.; Hauver, J.; Campbell, E.A.; Link, A.J.; Darst, S.A. Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin. Proc. Natl. Acad. Sci. USA 2019, 116, 1273–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, S.W.; Wang, G.H.; Yue, B.; Zhou, S.; Zhang, M. TO17: A teleost antimicrobial peptide that induces degradation of bacterial nucleic acids and inhibits bacterial infection in red drum, Sciaenops ocellatus. Fish Shellfish Immunol. 2018, 72, 639–645. [Google Scholar] [CrossRef]
- Rahnamaeian, M.; Cytryńska, M.; Zdybicka-Barabas, A.; Dobslaff, K.; Wiesner, J.; Twyman, R.M.; Zuchner, T.; Sadd, B.M.; Regoes, R.R.; Schmid-Hempel, P.; et al. Insect antimicrobial peptides show potentiating functional interactions against Gram-negative bacteria. Proc. Biol. Sci. 2015, 282, 20150293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chileveru, H.R.; Lim, S.A.; Chairatana, P.; Wommack, A.J.; Chiang, I.L.; Nolan, E.M. Visualizing attack of Escherichia coli by the antimicrobial peptide human defensin 5. Biochemistry 2015, 54, 1767–1777. [Google Scholar] [CrossRef] [Green Version]
- Maria-Neto, S.; Cândido Ede, S.; Rodrigues, D.R.; de Sousa, D.A.; da Silva, E.M.; de Moraes, L.M.; Otero-Gonzalez Ade, J.; Magalhães, B.S.; Dias, S.C.; Franco, O.L. Deciphering the magainin resistance process of Escherichia coli strains in light of the cytosolic proteome. Antimicrob. Agents Chemother. 2012, 56, 1714–1724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, S.; Takeshima, K.; Park, C.B.; Kim, S.C.; Matsuzaki, K. Interactions of the novel antimicrobial peptide buforin 2 with lipid bilayers: Proline as a translocation promoting factor. Biochemistry 2000, 39, 8648–8654. [Google Scholar] [CrossRef]
- Subbalakshmi, C.; Sitaram, N. Mechanism of antimicrobial action of indolicidin. FEMS Microbiol. Lett. 1998, 160, 91–96. [Google Scholar] [CrossRef]
- Sousa, D.A.; Porto, W.F.; Silva, M.Z.; da Silva, T.R.; Franco, O.L. Influence of Cysteine and Tryptophan Substitution on DNA-Binding Activity on Maize α-Hairpinin Antimicrobial Peptide. Molecules 2016, 21, 1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.K.; Park, S.C.; Hahm, K.S.; Park, Y. Antimicrobial HPA3NT3 peptide analogs: Placement of aromatic rings and positive charges are key determinants for cell selectivity and mechanism of action. Biochim. Biophys. Acta 2013, 1828, 443–454. [Google Scholar] [CrossRef] [Green Version]
- Ko, S.J.; Kang, N.H.; Kim, M.K.; Park, J.; Park, E.; Park, G.H.; Kang, T.W.; Na, D.E.; Park, J.B.; Yi, Y.E.; et al. Antibacterial and anti-biofilm activity, and mechanism of action of pleurocidin against drug resistant Staphylococcus aureus. Microb. Pathog. 2019, 127, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Teng, D.; Mao, R.; Yang, N.; Hao, Y.; Wang, J. Combined Systems Approaches Reveal a Multistage Mode of Action of a Marine Antimicrobial Peptide against Pathogenic Escherichia coli and Its Protective Effect against Bacterial Peritonitis and Endotoxemia. Antimicrob. Agents Chemother. 2017, 61, e01056-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, W.; Li, C.; Li, M.; Zong, X.; Han, D.; Chen, Y. Antimicrobial peptide melittin against Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen in rice. Appl. Microbiol. Biotechnol. 2016, 100, 5059–5067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Wang, K.; Dang, W.; Chen, R.; Xie, J.; Zhang, B.; Song, J.; Wang, R. Two hits are better than one: Membrane-active and DNA binding-related double-action mechanism of NK-18, a novel antimicrobial peptide derived from mammalian NK-lysin. Antimicrob. Agents Chemother. 2013, 57, 220–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makobongo, M.O.; Gancz, H.; Carpenter, B.M.; McDaniel, D.P.; Merrell, D.S. The oligo-acyl lysyl antimicrobial peptide C12K-2β12 exhibits a dual mechanism of action and demonstrates strong in vivo efficacy against Helicobacter pylori. Antimicrob. Agents Chemother. 2012, 56, 378–390. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Mohapatra, S.; Weisshaar, J.C. Rigidification of the Escherichia coli cytoplasm by the human antimicrobial peptide LL-37 revealed by superresolution fluorescence microscopy. Proc. Natl. Acad. Sci. USA 2019, 116, 1017–1026. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Tan, C. Dead bacterial absorption of antimicrobial peptides underlies collective tolerance. J. R. Soc. Interface 2019, 16, 20180701. [Google Scholar] [CrossRef] [Green Version]
- Snoussi, M.; Talledo, J.P.; Del Rosario, N.A.; Mohammadi, S.; Ha, B.Y.; Košmrlj, A.; Taheri-Araghi, S. Heterogeneous absorption of antimicrobial peptide LL37 in Escherichia coli cells enhances population survivability. eLife 2018, 7, e38174. [Google Scholar] [CrossRef]
- Yan, J.; Liang, X.; Liu, C.; Cheng, Y.; Zhou, L.; Wang, K.; Zhao, L. Influence of Proline Substitution on the Bioactivity of Mammalian-Derived Antimicrobial Peptide NK-2. Probiotics Antimicrob. Proteins 2018, 10, 118–127. [Google Scholar] [CrossRef]
- Lin, L.; Nonejuie, P.; Munguia, J.; Hollands, A.; Olson, J.; Dam, Q.; Kumaraswamy, M.; Rivera, H., Jr.; Corriden, R.; Rohde, M.; et al. Azithromycin Synergizes with Cationic Antimicrobial Peptides to Exert Bactericidal and Therapeutic Activity Against Highly Multidrug-Resistant Gram-Negative Bacterial Pathogens. EBioMedicine 2015, 2, 690–698. [Google Scholar] [CrossRef] [Green Version]
- Zerweck, J.; Strandberg, E.; Kukharenko, O.; Reichert, J.; Bürck, J.; Wadhwani, P.; Ulrich, A.S. Molecular mechanism of synergy between the antimicrobial peptides PGLa and magainin 2. Sci. Rep. 2017, 7, 13153. [Google Scholar] [CrossRef]
- Marxer, M.; Vollenweider, V.; Schmid-Hempel, P. Insect antimicrobial peptides act synergistically to inhibit a trypanosome parasite. Philos. Trans R. Soc. Lond. B. Biol. Sci. 2016, 371, 20150302. [Google Scholar] [CrossRef]
- Song, Y.; Ji, S.; Liu, W.; Yu, X.; Meng, Q.; Lai, R. Different expression profiles of bioactive peptides in Pelophylax nigromaculatus from distinct regions. Biosci. Biotechnol. Biochem. 2013, 77, 1075–1079. [Google Scholar] [CrossRef] [PubMed]
- Mangoni, M.L.; Miele, R.; Renda, T.G.; Barra, D.; Simmaco, M. The synthesis of antimicrobial peptides in the skin of Rana esculenta is stimulated by microorganisms. FASEB J. 2001, 15, 1431–1432. [Google Scholar] [CrossRef] [PubMed]
- Lai, P.K.; Tresnak, D.T.; Hackel, B.J. Identification and elucidation of proline-rich antimicrobial peptides with enhanced potency and delivery. Biotechnol. Bioeng. 2019, 116, 2439–2450. [Google Scholar] [CrossRef]
- Staubitz, P.; Peschel, A.; Nieuwenhuizen, W.F.; Otto, M.; Götz, F.; Jung, G.; Jack, R.W. Structure-function relationships in the tryptophan-rich, antimicrobial peptide indolicidin. J. Pept. Sci. 2001, 7, 552–564. [Google Scholar] [CrossRef]
- Di Somma, A.; Recupido, F.; Cirillo, A.; Romano, A.; Romanelli, A.; Caserta, S.; Guido, S.; Duilio, A. Antibiofilm Properties of Temporin-L on Pseudomonas fluorescens in Static and In-Flow Conditions. Int. J. Mol. Sci. 2020, 21, 8526. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [Green Version]
- Seviour, T.; Derlon, N.; Dueholm, M.S.; Flemming, H.C.; Girbal-Neuhauser, E.; Horn, H.; Kjelleberg, S.; van Loosdrecht, M.C.M.; Lotti, T.; Malpei, M.F.; et al. Extracellular polymeric substances of biofilms: Suffering from an identity crisis. Water Res. 2019, 151, 1–7. [Google Scholar] [CrossRef]
- More, T.T.; Yadav, J.S.; Yan, S.; Tyagi, R.D.; Surampalli, R.Y. Extracellular polymeric substances of bacteria and their potential environmental applications. J. Environ. Manag. 2014, 144, 1–25. [Google Scholar] [CrossRef]
- Di Somma, A.; Moretta, A.; Canè, C.; Cirillo, A.; Duilio, A. Antimicrobial and Antibiofilm Peptides. Biomolecules 2020, 10, 652. [Google Scholar] [CrossRef] [Green Version]
- Okuda, K.; Zendo, T.; Sugimoto, S.; Iwase, T.; Tajima, A.; Yamada, S.; Sonomoto, K.; Mizunoe, Y. Effects of bacteriocins on methicillin-resistant Staphylococcus aureus biofilm. Antimicrob. Agents Chemother. 2013, 57, 5572–5579. [Google Scholar] [CrossRef] [Green Version]
- Overhage, J.; Campisano, A.; Bains, M.; Torfs, E.C.; Rehm, B.H.; Hancock, R.E. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 2008, 76, 4176–4182. [Google Scholar] [CrossRef] [Green Version]
- Wolz, C.; Geiger, T.; Goerke, C. The synthesis and function of the alarmone (p)ppGpp in firmicutes. Int. J. Med. Microbiol. 2010, 300, 142–147. [Google Scholar] [CrossRef]
- Potrykus, K.; Cashel, M. (p)ppGpp: Still magical? Annu. Rev. Microbiol. 2008, 62, 35–51. [Google Scholar] [CrossRef] [Green Version]
- Crosse, A.M.; Greenway, D.L.; England, R.R. Accumulation of ppGpp and ppGp in Staphylococcus aureus 8325-4 following nutrient starvation. Lett. Appl. Microbiol. 2000, 31, 332–337. [Google Scholar] [CrossRef] [Green Version]
- de la Fuente-Núñez, C.; Reffuveille, F.; Haney, E.F.; Straus, S.K.; Hancock, R.E. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 2014, 10, e1004152. [Google Scholar] [CrossRef] [Green Version]
- De la Fuente-Núñez, C.; Reffuveille, F.; Mansour, S.C.; Reckseidler-Zenteno, S.L.; Hernández, D.; Brackman, G.; Coenye, T.; Hancock, R.E. D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 2015, 22, 196–205. [Google Scholar] [CrossRef]
- Pletzer, D.; Wolfmeier, H.; Bains, M.; Hancock, R.E.W. Synthetic Peptides to Target Stringent Response-Controlled Virulence in a Pseudomonas aeruginosa Murine Cutaneous Infection Model. Front. Microbiol. 2017, 8, 1867. [Google Scholar] [CrossRef] [Green Version]
- Sutton, J.M.; Pritts, T.A. Human beta-defensin 3: A novel inhibitor of Staphylococcus-produced biofilm production. Commentary on “Human β-defensin 3 inhibits antibiotic-resistant Staphylococcus biofilm formation”. J. Surg. Res. 2014, 186, 99–100. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Tan, H.; Cheng, T.; Shen, H.; Shao, J.; Guo, Y.; Shi, S.; Zhang, X. Human β-defensin 3 inhibits antibiotic-resistant Staphylococcus biofilm formation. J. Surg. Res. 2013, 183, 204–213. [Google Scholar] [CrossRef]
- De la Fuente-Núñez, C.; Korolik, V.; Bains, M.; Nguyen, U.; Breidenstein, E.B.; Horsman, S.; Lewenza, S.; Burrows, L.; Hancock, R.E. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 2012, 56, 2696–2704. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.Y.; Lin, L.; Tan, L.S.; Yu, H.Y.; Cheng, J.W.; Pan, Y.P. Molecular pathways underlying inhibitory effect of antimicrobial peptide Nal-P-113 on bacteria biofilms formation of Porphyromonas gingivalis W83 by DNA microarray. BMC Microbiol. 2017, 17, 37. [Google Scholar] [CrossRef] [Green Version]
- Luca, V.; Stringaro, A.; Colone, M.; Pini, A.; Mangoni, M.L. Esculentin(1-21), an amphibian skin membrane-active peptide with potent activity on both planktonic and biofilm cells of the bacterial pathogen Pseudomonas aeruginosa. Cell Mol. Life Sci. 2013, 70, 2773–2786. [Google Scholar] [CrossRef]
- Ansari, J.M.; Abraham, N.M.; Massaro, J.; Murphy, K.; Smith-Carpenter, J.; Fikrig, E. Anti-Biofilm Activity of a Self-Aggregating Peptide against Streptococcus mutans. Front. Microbiol. 2017, 8, 488. [Google Scholar] [CrossRef]
- Xie, J.; Li, Y.; Guo, X.; Rao, J.; Yan, T.; Mou, L.; Wu, X.; Xie, X.; Yang, W.; Zhang, B. CPF-C1 analog with effective antimicrobial and antibiofilm activities against Staphylococcus aureus including MRSA. Biochimie 2020, 176, 1–11. [Google Scholar] [CrossRef]
- Wang, H.; He, H.; Chen, X.; Zhou, M.; Wei, M.; Xi, X.; Ma, C.; Du, Q.; Chen, T.; Shaw, C.; et al. A Novel Antimicrobial Peptide (Kassinatuerin-3) Isolated from the Skin Secretion of the African Frog, Kassina senegalensis. Biology 2020, 9, 148. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, C.; Chen, J.; Zhou, S.; Zhao, Y.; Xu, M.; Xu, H. Dual Mode of Anti-Biofilm Action of G3 against Streptococcus mutans. ACS Appl. Mater. Interfaces 2020, 12, 27866–27875. [Google Scholar] [CrossRef]
- Weiss, U. Inflammation. Nature 2008, 454, 427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha e Silva, M. A brief survey of the history of inflammation. Agents Actions 1978, 8, 45–49. [Google Scholar] [CrossRef]
- Medzhitov, R. Inflammation 2010: New adventures of an old flame. Cell 2010, 140, 771–776. [Google Scholar] [CrossRef] [Green Version]
- Caroff, M.; Karibian, D.; Cavaillon, J.M.; Haeffner-Cavaillon, N. Structural and functional analyses of bacterial lipopolysaccharides. Microbes Infect. 2002, 4, 915–926. [Google Scholar] [CrossRef]
- Venkataranganayaka Abhilasha, K.; Kedihithlu Marathe, G. Bacterial lipoproteins in sepsis. Immunobiology 2021, 226, 152128. [Google Scholar] [CrossRef]
- Oliveira, J.; Reygaert, W.C. Gram Negative Bacteria; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2021. [Google Scholar]
- Kong, W.; Kang, K.; Gao, Y.; Liu, H.; Meng, X.; Yang, S.; Yu, K.; Zhao, M. Dexmedetomidine alleviates LPS-induced septic cardiomyopathy via the cholinergic anti-inflammatory pathway in mice. Am. J. Transl. Res. 2017, 9, 5040–5047. [Google Scholar] [PubMed]
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008, 454, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.K.; Kim, S.J.; Rah, S.H.; Kang, J.I.; Jung, H.E.; Lee, D.; Lee, H.K.; Lee, J.O.; Park, B.S.; Yoon, T.Y.; et al. Reconstruction of LPS Transfer Cascade Reveals Structural Determinants within LBP, CD14, and TLR4-MD2 for Efficient LPS Recognition and Transfer. Immunity 2017, 46, 38–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciesielska, A.; Matyjek, M.; Kwiatkowska, K. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell Mol. Life Sci. 2021, 78, 1233–1261. [Google Scholar] [CrossRef] [PubMed]
- Płóciennikowska, A.; Hromada-Judycka, A.; Borzęcka, K.; Kwiatkowska, K. Co-operation of TLR4 and raft proteins in LPS-induced pro-inflammatory signaling. Cell Mol. Life Sci. 2015, 72, 557–581. [Google Scholar] [CrossRef] [Green Version]
- Haziot, A.; Tsuberi, B.Z.; Goyert, S.M. Neutrophil CD14: Biochemical properties and role in the secretion of tumor necrosis factor-alpha in response to lipopolysaccharide. J. Immunol. 1993, 150, 5556–5565. [Google Scholar]
- Jang, M.; Kim, J.; Choi, Y.; Bang, J.; Kim, Y. Antiseptic Effect of Ps-K18: Mechanism of Its Antibacterial and Anti-Inflammatory Activities. Int. J. Mol. Sci. 2019, 20, 4895. [Google Scholar] [CrossRef] [Green Version]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Yang, L.; Weiss, T.M.; Waring, A.J.; Lehrer, R.I.; Huang, H.W. Interaction of antimicrobial peptides with lipopolysaccharides. Biochemistry 2003, 42, 12251–12259. [Google Scholar] [CrossRef] [Green Version]
- Li, L.H.; Ju, T.C.; Hsieh, C.Y.; Dong, W.C.; Chen, W.T.; Hua, K.F.; Chen, W.J. A synthetic cationic antimicrobial peptide inhibits inflammatory response and the NLRP3 inflammasome by neutralizing LPS and ATP. PLoS ONE 2017, 12, e0182057. [Google Scholar] [CrossRef] [Green Version]
- Gutsmann, T.; Razquin-Olazaran, I.; Kowalski, I.; Kaconis, Y.; Howe, J.; Bartels, R.; Hornef, M.; Schurholz, T.; Rossle, M.; Sanchez-Gomez, S.; et al. New antiseptic peptides to protect against endotoxin-mediated shock. Antimicrob. Agents Chemother. 2010, 54, 3817–3824. [Google Scholar] [CrossRef] [Green Version]
- Kaconis, Y.; Kowalski, I.; Howe, J.; Brauser, A.; Richter, W.; Razquin-Olazaran, I.; Inigo-Pestana, M.; Garidel, P.; Rossle, M.; Martinez de Tejada, G.; et al. Biophysical mechanisms of endotoxin neutralization by cationic amphiphilic peptides. Biophys. J. 2011, 100, 2652–2661. [Google Scholar] [CrossRef] [Green Version]
- Heinbockel, L.; Sanchez-Gomez, S.; Martinez de Tejada, G.; Domming, S.; Brandenburg, J.; Kaconis, Y.; Hornef, M.; Dupont, A.; Marwitz, S.; Goldmann, T.; et al. Preclinical investigations reveal the broad-spectrum neutralizing activity of peptide Pep19-2.5 on bacterial pathogenicity factors. Antimicrob. Agents Chemother. 2013, 57, 1480–1487. [Google Scholar] [CrossRef] [Green Version]
- Correa, W.; Heinbockel, L.; Behrends, J.; Kaconis, Y.; Barcena-Varela, S.; Gutsmann, T.; Mauss, K.; Schurholz, T.; Schromm, A.B.; Martinez de Tejada, G.; et al. Antibacterial action of synthetic antilipopolysaccharide peptides (SALP) involves neutralization of both membrane-bound and free toxins. FEBS J. 2019, 286, 1576–1593. [Google Scholar] [CrossRef]
- Rosenfeld, Y.; Papo, N.; Shai, Y. Endotoxin (lipopolysaccharide) neutralization by innate immunity host-defense peptides. Peptide properties and plausible modes of action. J. Biol. Chem. 2006, 281, 1636–1643. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.D.; Shin, S.Y. Antimicrobial and anti-inflammatory activities of short dodecapeptides derived from duck cathelicidin: Plausible mechanism of bactericidal action and endotoxin neutralization. Eur. J. Med. Chem. 2020, 204, 112580. [Google Scholar] [CrossRef]
- Uppu, D.S.; Haldar, J. Lipopolysaccharide Neutralization by Cationic-Amphiphilic Polymers through Pseudoaggregate Formation. Biomacromolecules 2016, 17, 862–873. [Google Scholar] [CrossRef]
- Nagaoka, I.; Hirota, S.; Niyonsaba, F.; Hirata, M.; Adachi, Y.; Tamura, H.; Tanaka, S.; Heumann, D. Augmentation of the lipopolysaccharide-neutralizing activities of human cathelicidin CAP18/LL-37-derived antimicrobial peptides by replacement with hydrophobic and cationic amino acid residues. Clin. Diagn. Lab. Immunol. 2002, 9, 972–982. [Google Scholar] [CrossRef] [Green Version]
- Martinez de Tejada, G.; Heinbockel, L.; Ferrer-Espada, R.; Heine, H.; Alexander, C.; Barcena-Varela, S.; Goldmann, T.; Correa, W.; Wiesmuller, K.H.; Gisch, N.; et al. Lipoproteins/peptides are sepsis-inducing toxins from bacteria that can be neutralized by synthetic anti-endotoxin peptides. Sci. Rep. 2015, 5, 14292. [Google Scholar] [CrossRef] [Green Version]
- Heinbockel, L.; Weindl, G.; Correa, W.; Brandenburg, J.; Reiling, N.; Wiesmuller, K.H.; Schurholz, T.; Gutsmann, T.; Martinez de Tejada, G.; Mauss, K.; et al. Anti-Infective and Anti-Inflammatory Mode of Action of Peptide 19-2.5. Int. J. Mol. Sci. 2021, 22, 1465. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. TLR signaling. Semin. Immunol. 2007, 19, 24–32. [Google Scholar] [CrossRef]
- Barton, G.M.; Medzhitov, R. Toll-like receptor signaling pathways. Science 2003, 300, 1524–1525. [Google Scholar] [CrossRef]
- Lin, S.C.; Lo, Y.C.; Wu, H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 2010, 465, 885–890. [Google Scholar] [CrossRef] [Green Version]
- Motshwene, P.G.; Moncrieffe, M.C.; Grossmann, J.G.; Kao, C.; Ayaluru, M.; Sandercock, A.M.; Robinson, C.V.; Latz, E.; Gay, N.J. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J. Biol. Chem. 2009, 284, 25404–25411. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.Y.; Richardson, B.C. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005, 6, 322–327. [Google Scholar] [CrossRef]
- Liu, L.; Guo, H.; Song, A.; Huang, J.; Zhang, Y.; Jin, S.; Li, S.; Zhang, L.; Yang, C.; Yang, P. Progranulin inhibits LPS-induced macrophage M1 polarization via NF-кB and MAPK pathways. BMC Immunol. 2020, 21, 32. [Google Scholar] [CrossRef]
- Lai, J.L.; Liu, Y.H.; Liu, C.; Qi, M.P.; Liu, R.N.; Zhu, X.F.; Zhou, Q.G.; Chen, Y.Y.; Guo, A.Z.; Hu, C.M. Indirubin Inhibits LPS-Induced Inflammation via TLR4 Abrogation Mediated by the NF-kB and MAPK Signaling Pathways. Inflammation 2017, 40, 1–12. [Google Scholar] [CrossRef]
- Lee, H.Y.; Bae, Y.S. The anti-infective peptide, innate defense-regulator peptide, stimulates neutrophil chemotaxis via a formyl peptide receptor. Biochem. Biophys. Res. Commun. 2008, 369, 573–578. [Google Scholar] [CrossRef]
- Chen, X.; Niyonsaba, F.; Ushio, H.; Hara, M.; Yokoi, H.; Matsumoto, K.; Saito, H.; Nagaoka, I.; Ikeda, S.; Okumura, K.; et al. Antimicrobial peptides human beta-defensin (hBD)-3 and hBD-4 activate mast cells and increase skin vascular permeability. Eur. J. Immunol. 2007, 37, 434–444. [Google Scholar] [CrossRef]
- Kim, Y.I.; Park, S.W.; Kang, I.J.; Shin, M.K.; Lee, M.H. Activin suppresses LPS-induced Toll-like receptor, cytokine and inducible nitric oxide synthase expression in normal human melanocytes by inhibiting NF-κB and MAPK pathway activation. Int. J. Mol. Med. 2015, 36, 1165–1172. [Google Scholar] [CrossRef]
- Suzuki, K.; Murakami, T.; Kuwahara-Arai, K.; Tamura, H.; Hiramatsu, K.; Nagaoka, I. Human anti-microbial cathelicidin peptide LL-37 suppresses the LPS-induced apoptosis of endothelial cells. Int. Immunol. 2011, 23, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Mookherjee, N.; Brown, K.L.; Bowdish, D.M.; Doria, S.; Falsafi, R.; Hokamp, K.; Roche, F.M.; Mu, R.; Doho, G.H.; Pistolic, J.; et al. Modulation of the TLR-mediated inflammatory response by the endogenous human host defense peptide LL-37. J. Immunol. 2006, 176, 2455–2464. [Google Scholar] [CrossRef] [Green Version]
- Nagaoka, I.; Tamura, H.; Reich, J. Therapeutic Potential of Cathelicidin Peptide LL-37, an Antimicrobial Agent, in a Murine Sepsis Model. Int. J. Mol. Sci. 2020, 21, 5973. [Google Scholar] [CrossRef]
- Klubthawee, N.; Adisakwattana, P.; Hanpithakpong, W.; Somsri, S.; Aunpad, R. A novel, rationally designed, hybrid antimicrobial peptide, inspired by cathelicidin and aurein, exhibits membrane-active mechanisms against Pseudomonas aeruginosa. Sci. Rep. 2020, 10, 9117. [Google Scholar] [CrossRef]
- Brunetti, J.; Carnicelli, V.; Ponzi, A.; Di Giulio, A.; Lizzi, A.R.; Cristiano, L.; Cresti, L.; Cappello, G.; Pollini, S.; Mosconi, L.; et al. Antibacterial and Anti-Inflammatory Activity of an Antimicrobial Peptide Synthesized with D Amino Acids. Antibiotics 2020, 9, 840. [Google Scholar] [CrossRef]
- Brunetti, J.; Roscia, G.; Lampronti, I.; Gambari, R.; Quercini, L.; Falciani, C.; Bracci, L.; Pini, A. Immunomodulatory and Anti-inflammatory Activity in Vitro and in Vivo of a Novel Antimicrobial Candidate. J. Biol. Chem. 2016, 291, 25742–25748. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Smith, C.; Wu, H.; Padhee, S.; Manoj, N.; Cardiello, J.; Qiao, Q.; Cao, C.; Yin, H.; Cai, J. Lipidated cyclic γ-AApeptides display both antimicrobial and anti-inflammatory activity. ACS Chem. Biol. 2014, 9, 211–217. [Google Scholar] [CrossRef] [Green Version]
- Dong, N.; Wang, C.; Li, X.; Guo, Y.; Li, X. Simplified Head-to-Tail Cyclic Polypeptides as Biomaterial-Associated Antimicrobials with Endotoxin Neutralizing and Anti-Inflammatory Capabilities. Int. J. Mol. Sci. 2019, 20, 5904. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xia, X.; Xu, L.; Wang, Y. Design of hybrid β-hairpin peptides with enhanced cell specificity and potent anti-inflammatory activity. Biomaterials 2013, 34, 237–250. [Google Scholar] [CrossRef]
- Shim, D.W.; Heo, K.H.; Kim, Y.K.; Sim, E.J.; Kang, T.B.; Choi, J.W.; Sim, D.W.; Cheong, S.H.; Lee, S.H.; Bang, J.K.; et al. Anti-Inflammatory Action of an Antimicrobial Model Peptide That Suppresses the TRIF-Dependent Signaling Pathway via Inhibition of Toll-Like Receptor 4 Endocytosis in Lipopolysaccharide-Stimulated Macrophages. PLoS ONE 2015, 10, e0126871. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.K.; Lee, E.; Shin, S.; Jeong, K.W.; Lee, J.Y.; Bae, S.Y.; Kim, S.H.; Lee, J.; Kim, S.R.; Lee, D.G.; et al. Structure and function of papiliocin with antimicrobial and anti-inflammatory activities isolated from the swallowtail butterfly, Papilio xuthus. J. Biol. Chem. 2011, 286, 41296–41311. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Ni, B.; Ren, J.D.; Chen, J.H.; Tian, Z.Q.; Tang, M.; Li, D.; Xia, P. Cyclic Limulus anti-lipopolysaccharide (LPS) factor-derived peptide CLP-19 antagonizes LPS function by blocking binding to LPS binding protein. Biol. Pharm. Bull. 2011, 34, 1678–1683. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.; Shin, A.; Kim, Y. Anti-inflammatory activities of cecropin A and its mechanism of action. Arch. Insect. Biochem. Physiol. 2015, 88, 31–44. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, J.; Tong, Z.; Jia, Y.; Yang, B.; Wang, Z. The revitalization of antimicrobial peptides in the resistance era. Pharmacol. Res. 2021, 163, 105276. [Google Scholar] [CrossRef]
- Zou, P.; Chen, W.T.; Sun, T.; Gao, Y.; Li, L.L.; Wang, H. Recent advances: Peptides and self-assembled peptide-nanosystems for antimicrobial therapy and diagnosis. Biomater. Sci. 2020, 8, 4975–4996. [Google Scholar] [CrossRef] [PubMed]

| Specific Mechanism of Action | AMPs | Action Site | References |
|---|---|---|---|
| Induce degradation of genomic DNA and total RNA | TO17 | Nucleic acid | [96] |
| Bind with nucleic acids and finally inhibit the synthesis of DNA, RNA, and proteins | Buforin-2 and indolicidin | Nucleic acid | [100,101] |
| Bind with nucleic acids | A series of derived peptides, such as HPA3NT3-A2, MBP-1, IARR-Anal10, and KW4 | Nucleic acid | [40,94,102,103] |
| Bind to RNA polymerase and inhibit the activity of RNA polymerase | Microcin J25 and capistruin | Nucleic acid synthetases | [95] |
| Act on the termination process of translation. Inhibit protein synthesis by capturing the release factor on the 70S ribosome after hydrolysis of the new polypeptide chain | Apidaecin 1b and Api137 | Ribosome | [39] |
| Transfer of aa-tRNA from EF-Tu to ribosome; a site blocked to inhibit protein synthesis | Bac7, Onc112, pyrrhocoricin, and metalnikowin | Ribosome | [39] |
| Inhibit the protein synthesis of 70S ribosome and interact with DnaK to inhibit the necessary ATPase activity or protein folding activity | Bac7 | Molecular chaperone DnaK | [77] |
| Inhibit DnaK activity | Abaecin | Molecular chaperone DnaK | [97] |
| Affect cell cycle, inhibit DNA synthesis, and prevent cell division | Indolicidin | Nucleic acid; cell division | [101] |
| Affect cell cycle and inhibit cell division | HD5ox | Cell division | [98] |
| Destruct organelles and inhibit mitochondrial respiration to destroy mitochondria | His-rich AMPs | Mitochondria | [42] |
| Inhibit the activity of energy metabolism proteins to affect energy metabolism | Magainin 1 | Energy metabolism protein | [99] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Y.; Song, Y. Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities. Int. J. Mol. Sci. 2021, 22, 11401. https://doi.org/10.3390/ijms222111401
Luo Y, Song Y. Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities. International Journal of Molecular Sciences. 2021; 22(21):11401. https://doi.org/10.3390/ijms222111401
Chicago/Turabian StyleLuo, Ying, and Yuzhu Song. 2021. "Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities" International Journal of Molecular Sciences 22, no. 21: 11401. https://doi.org/10.3390/ijms222111401
APA StyleLuo, Y., & Song, Y. (2021). Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities. International Journal of Molecular Sciences, 22(21), 11401. https://doi.org/10.3390/ijms222111401






