Epithelial to Mesenchymal Transition: A Challenging Playground for Translational Research. Current Models and Focus on TWIST1 Relevance and Gastrointestinal Cancers
Abstract
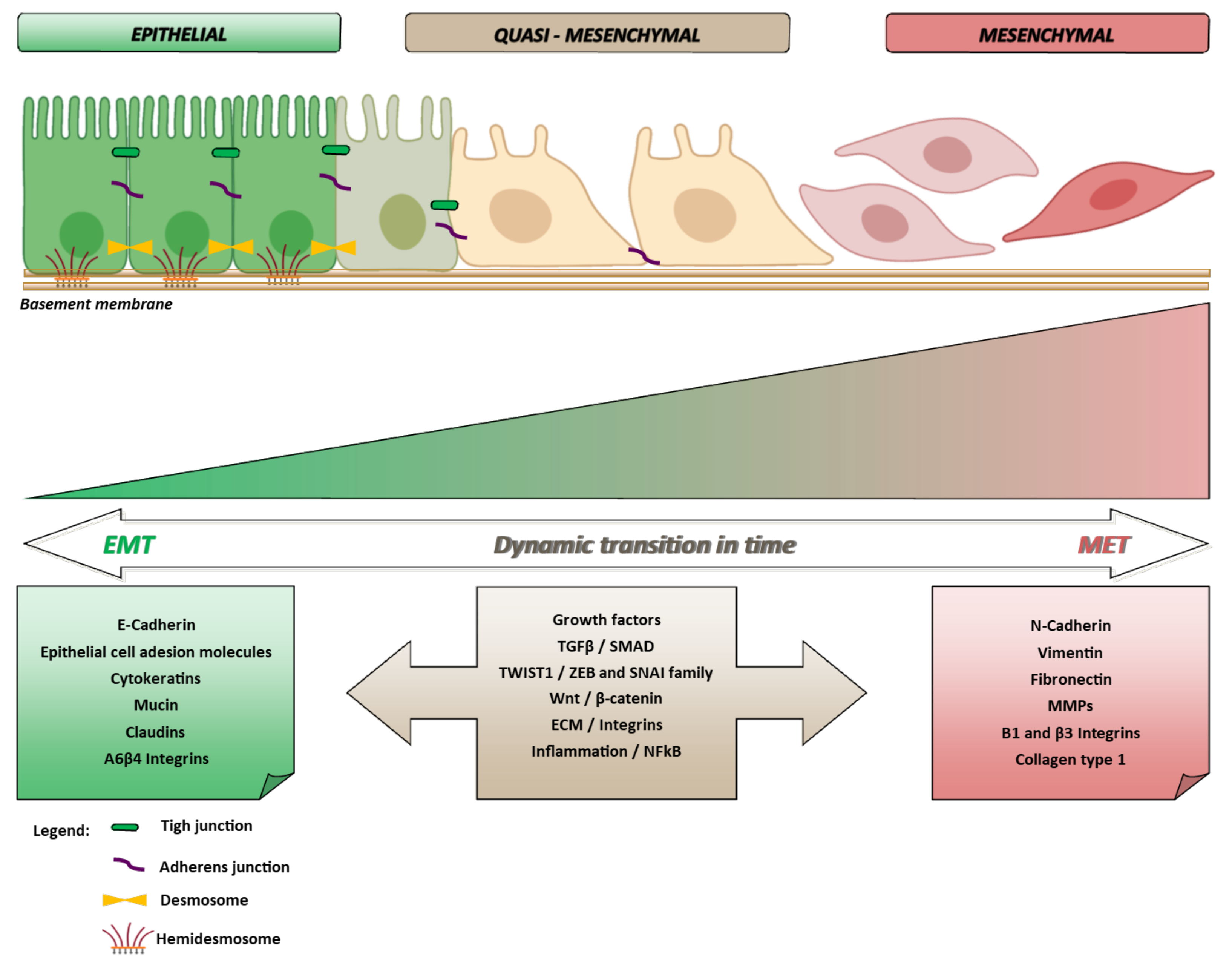
:1. Epithelial to Mesenchymal Transition and Cancer in Pills
1.1. EMT and Multistep Carcinogenesis
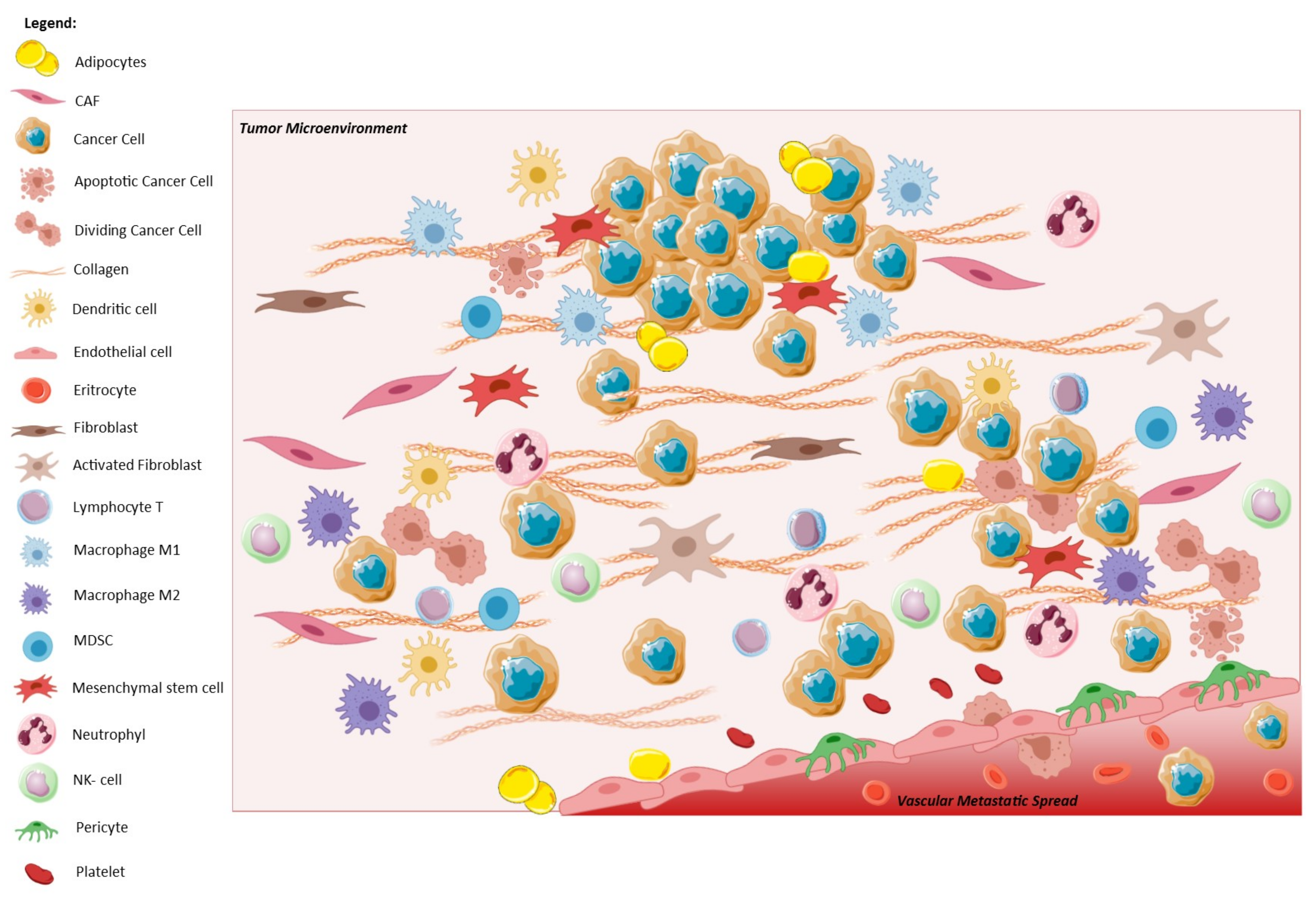
1.2. EMT and Tumor Microenvironment
2. The Molecular Biology of EMT
EMT and Crossing of Lineage-Specific Differentiation of Epithelial Cells in the Gastrointestinal Tract
3. EMT Involvement in Gastrointestinal Cancers: The Frame
4. EMT-Related Pathways
TWIST1, a Molecular Culprit
5. Models to Unravel the Role of EMT in Disease Progression
5.1. In Vitro Modeling of EMT
5.2. In Vivo Animal Models of EMT⇔MET, and Stemness Features
6. Tissue Expression of EMT-TFs and Their Potential as Biomarkers of Disease Progression in Translational Studies
6.1. Immunohistochemical Assessment
6.2. Expression Studies of mRNA Profiles
7. EMT, Stemness and CTC
8. EMT and Chemoresistance: An Open Issue
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.; Chen, W.-D.; Parmigiani, G.; Diehl, F.; Beerenwinkel, N.; Antal, T.; Traulsen, A.; Nowak, M.A.; Siegel, C.; Velculescu, V.; et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 4283–4288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Mani, S.A.; Donaher, J.L.; Ramaswamy, S.; Itzykson, R.A.; Come, C.; Savagner, P.; Gitelman, I.; Richardson, A.; Weinberg, R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004, 117, 927–939. [Google Scholar] [CrossRef] [Green Version]
- Chaffer, C.L.; San Juan, B.P.; Lim, E.; Weinberg, R.A. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016, 35, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Devecchi, A.; De Cecco, L.; Dugo, M.; Penso, D.; Dagrada, G.; Brich, S.; Stacchiotti, S.; Sensi, M.; Canevari, S.; Pilotti, S. The genomics of desmoplastic small round cell tumor reveals the deregulation of genes related to DNA damage response, epithelial-mesenchymal transition, and immune response. Cancer Commun. 2018, 38, 70. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, S.; Kumar, A.; Faheem, M.M.; Katoch, A.; Kumar, A.; Jamwal, V.L.; Nayak, D.; Golani, A.; Rasool, R.U.; Ahmad, S.M.; et al. Vimentin activation in early apoptotic cancer cells errands survival pathways during DNA damage inducer CPT treatment in colon carcinoma model. Cell Death Dis. 2019, 10, 467. [Google Scholar] [CrossRef] [Green Version]
- Pustovalova, M.; Alhaddad, L.; Blokhina, T.; Smetanina, N.; Chigasova, A.; Chuprov-Netochin, R.; Eremin, P.; Gilmutdinova, I.; Osipov, A.; Leonov, S. The CD44high Subpopulation of Multifraction Irradiation-Surviving NSCLC Cells Exhibits Partial EMT-Program Activation and DNA Damage Response Depending on Their p53 Status. Int. J. Mol. Sci. 2021, 22, 2369. [Google Scholar] [CrossRef]
- Grizzi, F.; Basso, G.; Borroni, E.M.; Cavalleri, T.; Bianchi, P.; Stifter, S.; Chiriva-Internati, M.; Malesci, A.; Laghi, L. Evolving notions on immune response in colorectal cancer and their implications for bi-omarker development. Inflamm. Res. 2018, 67, 375–389. [Google Scholar] [CrossRef]
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic instability in colorectal cancers. Nat. Cell Biol. 1997, 386, 623–627. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goossens, S.; Vandamme, N.; Van Vlierberghe, P.; Berx, G. EMT transcription factors in cancer development re-evaluated: Beyond EMT and MET. Biochim. Biophys. Acta Bioenerg. 2017, 1868, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.H.; Yang, J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013, 27, 2192–2206. [Google Scholar] [CrossRef] [Green Version]
- Mantovani, A.; Schioppa, T.; Porta, C.; Allavena, P.; Sica, A. Role of tumor-associated macrophages in tumor progression and invasion. Cancer Metastasis Rev. 2006, 25, 315–322. [Google Scholar] [CrossRef]
- Mantovani, A. The Yin-Yang of Tumor-Associated Neutrophils. Cancer Cell 2009, 16, 173–174. [Google Scholar] [CrossRef] [Green Version]
- Tabassum, D.P.; Polyak, K. Tumorigenesis: It takes a village. Nat. Rev. Cancer 2015, 15, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Cavalleri, T.; Bianchi, P.; Basso, G.; Celesti, G.; Grizzi, F.; Bossi, P.; Greco, L.; Pitrone, C.; Valtorta, E.; Mauri, G.; et al. Combined Low Densities of FoxP3+ and CD3+ Tumor-Infiltrating Lymphocytes Identify Stage II Colorectal Cancer at High Risk of Progression. Cancer Immunol. Res. 2019, 7, 751–758. [Google Scholar] [CrossRef] [Green Version]
- Pagès, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.-S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Malesci, A.; Bianchi, P.; Celesti, G.; Basso, G.; Marchesi, F.; Grizzi, F.; Di Caro, G.; Cavalleri, T.; Rimassa, L.; Palmqvist, R.; et al. Tumor-associated macrophages and response to 5-fluorouracil adjuvant therapy in stage III colorectal cancer. OncoImmunology 2017, 6, e1342918. [Google Scholar] [CrossRef]
- Di Caro, G.; Marchesi, F.; Laghi, L.; Grizzi, F. Immune cells: Plastic players along colorectal cancer progression. J. Cell. Mol. Med. 2013, 17, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Grizzi, F.; Bianchi, P.; Malesci, A.; Laghi, L. Prognostic value of innate and adaptive immunity in colorectal cancer. World J. Gastroenterol. 2013, 19, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Laghi, L.; Bianchi, P.; Miranda, E.; Balladore, E.; Pacetti, V.; Grizzi, F.; Allavena, P.; Torri, V.; Repici, A.; Santoro, A.; et al. CD3+ cells at the invasive margin of deeply invading (pT3–T4) colorectal cancer and risk of post-surgical metastasis: A longitudinal study. Lancet Oncol. 2009, 10, 877–884. [Google Scholar] [CrossRef]
- Van Pelt, G.W.; Sandberg, T.P.; Morreau, H.; Gelderblom, H.; van Krieken, J.H.J.M.; Tollenaar, R.A.E.M.; Mesker, W.E. The tumour-stroma ratio in colon cancer: The biological role and its prognostic impact. Histopathology 2018, 73, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Vennin, C.; Mélénec, P.; Rouet, R.; Nobis, M.; Cazet, A.S.; Murphy, K.J.; Herrmann, D.; Reed, D.A.; Lucas, M.C.; Warren, S.C.; et al. CAF hierarchy driven by pancreatic cancer cell p53-status creates a pro-metastatic and chemoresistant environment via perlecan. Nat. Commun. 2019, 10, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tauriello, D.V.F.; Batlle, E. Targeting the Microenvironment in Advanced Colorectal Cancer. Trends Cancer 2016, 2, 495–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bridelance, J.; Drebert, Z.; De Wever, O.; Bracke, M.; Beck, I.M. When Neighbors Talk: Colon Cancer Cell Invasion and Tumor Microenvironment Myofibroblasts. Curr. Drug Targets 2017, 18, 964–982. [Google Scholar] [CrossRef]
- Ueno, H.; Kanemitsu, Y.; Sekine, S.; Ishiguro, M.; Ito, E.; Hashiguchi, Y.; Kondo, F.; Shimazaki, H.; Mochizuki, S.; Kajiwara, Y.; et al. Desmoplastic Pattern at the Tumor Front Defines Poor-prognosis Subtypes of Colorectal Cancer. Am. J. Surg. Pathol. 2017, 41, 1506–1512. [Google Scholar] [CrossRef] [PubMed]
- Celesti, G.; Di Caro, G.; Bianchi, P.; Grizzi, F.; Basso, G.; Marchesi, F.; Doni, A.; Marra, G.; Roncalli, M.; Mantovani, A.; et al. Presence of Twist1-positive neoplastic cells in the stroma of chromosome-unstable colo-rectal tumors. Gastroenterology 2013, 145, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating Breast Tumor Cells Exhibit Dynamic Changes in Epithelial and Mesenchymal Composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Liu, S.; Liu, Z.; Huang, J.; Pu, X.; Li, J.; Yang, D.; Deng, H.; Yang, N.; Xu, J. Classification of Circulating Tumor Cells by Epithelial-Mesenchymal Transition Markers. PLoS ONE 2015, 10, e0123976. [Google Scholar] [CrossRef] [PubMed]
- Chea, H.K.; Wright, C.V.; Swalla, B.J. Nodal signaling and the evolution of deuterostome gastrulation. Dev. Dyn. 2005, 234, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Zeisberg, M.; Neilson, E.G. Biomarkers for epithelial-mesenchymal transitions. J. Clin. Investig. 2009, 119, 1429–1437. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.-L.; Luo, M.; Huang, C.; Chen, H.-N.; Zhou, Z.-G. Epigenetic Regulation of Epithelial to Mesenchymal Transition in the Cancer Metastatic Cas-cade: Implications for Cancer Therapy. Front. Oncol. 2021, 11, 657546. [Google Scholar] [CrossRef]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.-J.; Nieto, M.A. Epithelial-Mesenchymal Transitions in Development and Disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef]
- Polyak, K.; Weinberg, R.A. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat. Rev. Cancer 2009, 9, 265–273. [Google Scholar] [CrossRef]
- Kukcinaviciute, E.; Jonusiene, V.; Sasnauskiene, A.; Dabkeviciene, D.; Eidenaite, E.; Laurinavicius, A. Significance of Notch and Wnt signaling for chemoresistance of colorectal cancer cells HCT. J. Cell. Biochem. 2018, 119, 5913–5920. [Google Scholar] [CrossRef]
- Heppert, J.K.; Davison, J.M.; Kelly, C.; Mercado, G.P.; Lickwar, C.R.; Rawls, J.F. Transcriptional programmes underlying cellular identity and microbial responsiveness in the intestinal epithelium. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 7–23. [Google Scholar] [CrossRef]
- Kaemmerer, E.; Jeon, M.K.; Berndt, A.; Liedtke, C.; Gassler, N. Targeting Wnt Signaling via Notch in Intestinal Carcinogenesis. Cancers 2019, 11, 555. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Q.; Wang, J.; Chen, C.; Zhang, Y.; Feng, D.; Zhao, R.; Chen, T. ASCL2 expression contributes to gastric tumor migration and invasion by downregulating miR223 and inducing EMT. Mol. Med. Rep. 2018, 18, 3751–3759. [Google Scholar] [CrossRef]
- Kim, W.K.; Kwon, Y.; Jang, M.; Park, M.; Kim, J.; Cho, S.; Jang, D.G.; Lee, W.-B.; Jung, S.H.; Choi, H.J.; et al. β-catenin activation down-regulates cell-cell junction-related genes and induces epithelial-to-mesenchymal transition in colorectal cancers. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopan, R.; Ilagan, M.X.G. The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef] [Green Version]
- Demitrack, E.S.; Samuelson, L.C. Notch regulation of gastrointestinal stem cells. J. Physiol. 2016, 594, 4791–4803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyagi, A.; Sharma, A.K.; Damodaran, C. A Review on Notch Signaling and Colorectal Cancer. Cells 2020, 9, 1549. [Google Scholar] [CrossRef]
- Zhu, W.; Cai, M.-Y.; Tong, Z.-T.; Dong, S.-S.; Mai, S.-J.; Liao, Y.; Bian, X.-W.; Lin, M.C.; Kung, H.-F.; Zeng, Y.-X.; et al. Overexpression of EIF5A2 promotes colorectal carcinoma cell aggressiveness by upregulating MTA1 through C-myc to induce epithelial–mesenchymaltransition. Gut 2011, 61, 562–575. [Google Scholar] [CrossRef]
- Spaderna, S.; Schmalhofer, O.; Hlubek, F.; Berx, G.; Eger, A.; Merkel, S.; Jung, A.; Kirchner, T.; Brabletz, T. A Transient, EMT-Linked Loss of Basement Membranes Indicates Metastasis and Poor Survival in Colorectal Cancer. Gastroenterology 2006, 131, 830–840. [Google Scholar] [CrossRef]
- Orth, M.; Metzger, P.; Gerum, S.; Mayerle, J.; Schneider, G.; Belka, C.; Schnurr, M.; Lauber, K. Pancreatic ductal adenocarcinoma: Biological hallmarks, current status, and future perspectives of combined modality treatment approaches. Radiat. Oncol. 2019, 14, 1–20. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Becht, E.; De Reyniès, A.; Giraldo, N.; Pilati, C.; Buttard, B.; Lacroix, L.; Selves, J.; Sautes-Fridman, C.; Laurent-Puig, P.; Fridman, W.H. Immune and Stromal Classification of Colorectal Cancer Is Associated with Molecular Subtypes and Relevant for Precision Immunotherapy. Clin. Cancer Res. 2016, 22, 4057–4066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ieda, T.; Tazawa, H.; Okabayashi, H.; Yano, S.; Shigeyasu, K.; Kuroda, S.; Ohara, T.; Noma, K.; Kishimoto, H.; Nishizaki, M.; et al. Visualization of epithelial-mesenchymal transition in an inflammatory microenvironment–colorectal cancer network. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Nishimura, H.; Kameoka, S.; Saito, N. Clinical significance of fibronectin expression in colorectal cancer. Mol. Med. Rep. 2008, 1, 77–81. [Google Scholar] [CrossRef]
- Yan, X.; Yan, L.; Liu, S.; Shan, Z.; Tian, Y.; Jin, Z. N-cadherin, a novel prognostic biomarker, drives malignant progression of colorectal cancer. Mol. Med. Rep. 2015, 12, 2999–3006. [Google Scholar] [CrossRef] [Green Version]
- Xiang, L.; Xie, G.; Ou, J.; Wei, X.; Pan, F.; Liang, H. The Extra Domain A of Fibronectin Increases VEGF-C Expression in Colorectal Carcinoma Involving the PI3K/AKT Signaling Pathway. PLoS ONE 2012, 7, e35378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, W.; Yang, L.; Li, T.; Zhang, Y. Cadherin Signaling in Cancer: Its Functions and Role as a Therapeutic Target. Front. Oncol. 2019, 9, 989. [Google Scholar] [CrossRef]
- Pickup, M.; Novitskiy, S.; Moses, H.L. The roles of TGFbeta in the tumour microenvironment. Nat. Rev. Cancer 2013, 13, 788–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pino, M.S.; Kikuchi, H.; Zeng, M.; Herraiz, M.M.-T.; Sperduti, I.; Berger, D.; Park, D.D.-Y.D.; Iafrate, A.J.; Zukerberg, L.R.; Chung, D.C. Epithelial to mesenchymal transition is impaired in colon cancer cells with microsatellite instability. Gastroenterology 2010, 138, 1406–1417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helm, O.; Held-Feindt, J.; Grage-Griebenow, E.; Reiling, N.; Ungefroren, H.; Vogel, I.; Krüger, U.; Becker, T.; Ebsen, M.; Röcken, C.; et al. Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int. J. Cancer 2014, 135, 843–861. [Google Scholar] [CrossRef]
- Li, N.; Babaei-Jadidi, R.; Lorenzi, F.; Spencer-Dene, B.; Clarke, P.; Domingo, E.; Tulchinsky, E.; Vries, R.G.J.; Kerr, D.; Pan, Y.; et al. An FBXW7-ZEB2 axis links EMT and tumour microenvironment to promote colorectal cancer stem cells and chemoresistance. Oncogenesis 2019, 8, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Shibue, T.; Weinberg, T.S.R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Dong, B.; Li, S.; Zhu, S.; Yi, M.; Luo, S.; Wu, K. MiRNA-mediated EMT and CSCs in cancer chemoresistance. Exp. Hematol. Oncol. 2021, 10, 1–12. [Google Scholar] [CrossRef]
- Wang, F.; Rong, L.; Zhang, Z.; Li, M.; Ma, L.; Ma, Y.; Xie, X.; Tian, X.; Yang, Y. LncRNA H19-Derived miR-675-3p Promotes Epithelial-Mesenchymal Transition and Stemness in Human Pancreatic Cancer Cells by targeting the STAT3 Pathway. J. Cancer 2020, 11, 4771–4782. [Google Scholar] [CrossRef]
- Shen, W.; Cui, L.; Chen, W. DKK4 is important in Snail1-induced chemoresistance to fluorouracil in colorectal cancer. Transl. Cancer Res. 2017, 6, 304–311. [Google Scholar] [CrossRef]
- Cho, Y.-H.; Ro, E.J.; Yoon, J.-S.; Mizutani, T.; Kang, D.-W.; Park, J.-C.; Kim, T.I.; Clevers, H.; Choi, K.-Y. 5-FU promotes stemness of colorectal cancer via p53-mediated WNT/β-catenin pathway activation. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Horn, L.A.; Fousek, K.; Palena, C. Tumor Plasticity and Resistance to Immunotherapy. Trends Cancer 2020, 6, 432–441. [Google Scholar] [CrossRef]
- Zhong, Y.; Ma, T.; Qiao, T.; Hu, H.; Li, Z.; Luo, K.; Wang, Y.; Tang, Q.; Wang, G.; Huang, R.; et al. Role of Phenotypes of Circulating Tumor Cells in the Diagnosis and Treatment of Colorectal Cancer. Cancer Manag. Res. 2021, 13, 7077–7085. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Feng, M.; Bai, L.; Liao, W.; Zhou, K.; Zhang, M.; Wu, Q.; Wen, F.; Lei, W.; Zhang, P.; et al. Comprehensive analysis of EMT-related genes and lncRNAs in the prognosis, immunity, and drug treatment of colorectal cancer. J. Transl. Med. 2021, 19, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Shi, M.; Li, K.; Ma, Y.; Jiang, L.; Chen, H.; Peng, C. Development and validation of a cancer stem cell-related signature for prognostic prediction in pancreatic ductal adenocarcinoma. J. Transl. Med. 2020, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shaul, Y.D.; Freinkman, E.; Comb, W.C.; Cantor, J.R.; Tam, W.L.; Thiru, P.; Kim, D.; Kanarek, N.; Pacold, M.E.; Chen, W.W.; et al. Dihydropyrimidine Accumulation Is Required for the Epithelial-Mesenchymal Transition. Cell 2014, 158, 1094–1109. [Google Scholar] [CrossRef] [Green Version]
- Viswanathan, V.S.; Ryan, M.J.; Dhruv, H.D.; Gill, S.; Eichhoff, O.M.; Seashore-Ludlow, B.; Kaffenberger, S.D.; Eaton, J.K.; Shimada, K.; Aguirre, A.J.; et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 2017, 547, 453–457. [Google Scholar] [CrossRef]
- Mathow, D.; Chessa, F.; Rabionet, M.; Kaden, S.; Jennemann, R.; Sandhoff, R.; Gröne, H.; Feuerborn, A. Zeb1 affects epithelial cell adhesion by diverting glycosphingolipid metabolism. EMBO Rep. 2015, 16, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Ramesh, V.; Brabletz, T.; Ceppi, P. Targeting EMT in Cancer with Repurposed Metabolic Inhibitors. Trends Cancer 2020, 6, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Hong, J.; Du, W.; Lin, Y.-W.; Ren, L.-L.; Wang, Y.-C.; Su, W.-Y.; Wang, J.-L.; Cui, Y.; Wang, Z.-H.; et al. Roles of STAT3 and ZEB1 Proteins in E-cadherin Down-regulation and Human Colorectal Cancer Epithelial-Mesenchymal Transition. J. Biol. Chem. 2012, 287, 5819–5832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Tillo, E.; de Barrios, O.; Siles, L.; Cuatrecasas, M.; Castells, A.; Postigo, A. -catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc. Natl. Acad. Sci. USA 2011, 108, 19204–19209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahlgren, C.; Gustafsson, M.V.; Jin, S.; Poellinger, L.; Lendahl, U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl. Acad. Sci. USA 2008, 105, 6392–6397. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Deng, J.; Rychahou, P.; Qiu, S.; Evers, B.M.; Zhou, B.P. Stabilization of Snail by NF-κB Is Required for Inflammation-Induced Cell Migration and Invasion. Cancer Cell 2009, 15, 416–428. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Xu, E.; Liu, H.; Wan, L.; Lai, M. Epithelial–mesenchymal transition in colorectal cancer metastasis: A system review. Pathol. Res. Pract. 2015, 211, 557–569. [Google Scholar] [CrossRef]
- Niessen, K.; Fu, Y.; Chang, L.; Hoodless, P.A.; McFadden, D.; Karsan, A. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J. Cell Biol. 2008, 182, 315–325. [Google Scholar] [CrossRef] [Green Version]
- Xie, M.; Zhang, L.; He, C.-S.; Xu, F.; Liu, J.-L.; Hu, Z.-H.; Zhao, L.-P.; Tian, Y. Activation of Notch-1 enhances epithelial-mesenchymal transition in gefitinib-acquired resistant lung cancer cells. J. Cell. Biochem. 2012, 113, 1501–1513. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Calin, G.; Croce, C.M. MicroRNA Signatures in Human Cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Hur, K.; Toiyama, Y.; Takahashi, M.; Balaguer, F.; Nagasaka, T.; Koike, J.; Hemmi, H.; Koi, M.; Boland, C.R.; Goel, A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut 2012, 62, 1315–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008, 9, 582–589. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Omura, N.; Hong, S.-M.; Vincent, A.; Walter, K.; Griffith, M.; Borges, M.; Goggins, M. Pancreatic Cancers Epigenetically Silence SIP1 and Hypomethylate and Overexpress miR-200a/200b in Association with Elevated Circulating miR-200a and miR-200b Levels. Cancer Res. 2010, 70, 5226–5237. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Xu, Y.; Ding, L.; Yao, H.; Yu, H.; Zhou, T.; Si, J. Down-regulation of miR-141 in gastric cancer and its involvement in cell growth. J. Gastroenterol. 2009, 44, 556–561. [Google Scholar] [CrossRef]
- López, A.D.; Diaz-Martin, J.; Moreno-Bueno, G.; Cuevas, E.P.; Santos, V.; Olmeda, D.; Portillo, F.; Palacios, J.; Cano, A. Zeb1 and Snail1 engage miR-200f transcriptional and epigenetic regulation during EMT. Int. J. Cancer 2014, 136, E62–E73. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Martín, J.; Díaz-López, A.; Moreno-Bueno, G.; Castilla, M.Á.; Rosa-Rosa, J.M.; Cano, A.; Palacios, J. A core microRNA signature associated with inducers of the epithelial-to-mesenchymal transition. J. Pathol. 2014, 232, 319–329. [Google Scholar] [CrossRef]
- Bourguignon, L.Y.; Wong, G.; Earle, C.; Krueger, K.; Spevak, C.C. Hyaluronan-CD44 Interaction Promotes c-Src-mediated Twist Signaling, MicroRNA-10b Expression, and RhoA/RhoC Up-regulation, Leading to Rho-kinase-associated Cytoskeleton Activation and Breast Tumor Cell Invasion. J. Biol. Chem. 2010, 285, 36721–36735. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhang, Y.; Zhang, H.; Liu, X.; Gong, T.; Li, M.; Sun, L.; Ji, G.; Shi, Y.; Han, Z.; et al. miRNA-223 Promotes Gastric Cancer Invasion and Metastasis by Targeting Tumor Suppressor EPB41L3. Mol. Cancer Res. 2011, 9, 824–833. [Google Scholar] [CrossRef] [Green Version]
- Dehai, C.; Bo, P.; Qiang, T.; Lihua, S.; Fang, L.; Shi, J.; Jingyan, C.; Yan, Y.; Guangbin, W.; Zhenjun, Y. Enhanced invasion of lung adenocarcinoma cells after co-culture with THP-1-derived macrophages via the induction of EMT by IL-6. Immunol. Lett. 2014, 160, 1–10. [Google Scholar] [CrossRef]
- Singh, R.; Shankar, B.S.; Sainis, K.B. TGF-β1–ROS–ATM–CREB signaling axis in macrophage mediated migration of human breast cancer MCF7 cells. Cell. Signal. 2014, 26, 1604–1615. [Google Scholar] [CrossRef]
- Ohta, M.; Kitadai, Y.; Tanaka, S.; Yoshihara, M.; Yasui, W.; Mukaida, N.; Haruma, K.; Chayama, K. Monocyte chemoattractant protein-1 expression correlates with macrophage infiltration and tumor vascularity in human gastric carcinomas. Int. J. Oncol. 2003, 22, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Ishigami, S.; Natsugoe, S.; Tokuda, K.; Nakajo, A.; Okumura, H.; Matsumoto, M.; Miyazono, F.; Hokita, S.; Aikou, T. Tumor-associated macrophage (TAM) infiltration in gastric cancer. Anticancer. Res. 2003, 23, 4079–4083. [Google Scholar]
- Väyrynen, S.A.; Zhang, J.; Yuan, C.; Väyrynen, J.P.; Costa, A.D.; Williams, H.; Morales-Oyarvide, V.; Lau, M.C.; Rubinson, D.A.; Dunne, R.F.; et al. Composition, Spatial Characteristics, and Prognostic Significance of Myeloid Cell Infiltration in Pancreatic Cancer. Clin. Cancer Res. 2021, 27, 1069–1081. [Google Scholar] [CrossRef]
- Zhao, Z.; Rahman, M.A.; Chen, Z.G.; Shin, D.M. Multiple biological functions of Twist1 in various cancers. Oncotarget 2017, 8, 20380–20393. [Google Scholar] [CrossRef] [Green Version]
- Amin, A.R.; Karpowicz, P.A.; Carey, T.; Arbiser, J.; Nahta, R.; Chen, Z.G.; Dong, J.-T.; Kucuk, O.; Khan, G.N.; Huang, G.; et al. Evasion of anti-growth signaling: A key step in tumorigenesis and potential target for treatment and prophylaxis by natural compounds. Semin. Cancer Biol. 2015, 35, S55–S77. [Google Scholar] [CrossRef] [PubMed]
- Kwok, W.K.; Ling, M.-T.; Yuen, H.F.; Wong, Y.-C.; Wang, X. Role of p14ARF in TWIST-mediated senescence in prostate epithelial cells. Carcinogenesis 2007, 28, 2467–2475. [Google Scholar] [CrossRef] [Green Version]
- Vichalkovski, A.; Gresko, E.; Hess, D.; Restuccia, D.; Hemmings, B.A. PKB/AKT phosphorylation of the transcription factor Twist-1 at Ser42 inhibits p53 activity in response to DNA damage. Oncogene 2010, 29, 3554–3565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maestro, R.; Dei Tos, A.P.; Hamamori, Y.; Krasnokutsky, S.; Sartorelli, V.; Kedes, L.; Doglioni, C.; Beach, D.H.; Hannon, G.J. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999, 13, 2207–2217. [Google Scholar] [CrossRef]
- Valsesia-Wittmann, S.; Magdeleine, M.; Dupasquier, S.; Garin, E.; Jallas, A.-C.; Combaret, V.; Krause, A.; Leissner, P.; Puisieux, A. Oncogenic cooperation between H-Twist and N-Myc overrides failsafe programs in cancer cells. Cancer Cell 2004, 6, 625–630. [Google Scholar] [CrossRef] [Green Version]
- Piccinin, S.; Tonin, E.; Sessa, S.; Demontis, S.; Rossi, S.; Pecciarini, L.; Zanatta, L.; Pivetta, F.; Grizzo, A.; Sonego, M.; et al. A “Twist box” Code of p53 Inactivation: Twist box: p53 Interaction Promotes p53 Degradation. Cancer Cell 2012, 22, 404–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stasinopoulos, I.A.; Mironchik, Y.; Raman, A.; Wildes, F.; Winnard, P.; Raman, V. HOXA5-Twist Interaction Alters p53 Homeostasis in Breast Cancer Cells. J. Biol. Chem. 2005, 280, 2294–2299. [Google Scholar] [CrossRef] [Green Version]
- Pinho, A.V.; Rooman, I.; Real, F.X. p53-dependent regulation of growth, epithelial-mesenchymal transition and stemness in normal pancreatic epithelial cells. Cell Cycle 2011, 10, 1312–1321. [Google Scholar] [CrossRef] [PubMed]
- Ansieau, S.; Bastid, J.; Doreau, A.; Morel, A.-P.; Bouchet, B.P.; Thomas, C.; Fauvet, F.; Puisieux, I.; Doglioni, C.; Piccinin, S.; et al. Induction of EMT by Twist Proteins as a Collateral Effect of Tumor-Promoting Inactivation of Premature Senescence. Cancer Cell 2008, 14, 79–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, B.; Lapouge, G.; Rorive, S.; Drogat, B.; Desaedelaere, K.; Delafaille, S.; Dubois, C.; Salmon, I.; Willekens, K.; Marine, J.-C.; et al. Different Levels of Twist1 Regulate Skin Tumor Initiation, Stemness, and Progression. Cell Stem Cell 2015, 16, 67–79. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.-Q.; Ma, C.; Wang, Q.; Song, Y.; Lv, T. The role of TWIST1 in epithelial-mesenchymal transition and cancers. Tumor Biol. 2016, 37, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Chanrion, M.; Kuperstein, I.; Barrière, C.; El Marjou, F.; Cohen, D.; Vignjevic, D.M.; Stimmer, L.; Paul-Gilloteaux, P.; Bieche, I.; Tavares, S.D.R.; et al. Concomitant Notch activation and p53 deletion trigger epithelial-to-mesenchymal transition and metastasis in mouse gut. Nat. Commun. 2014, 5, 5005. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.-J.; Zhang, W.; Xu, X.-M.; Zhang, F.; Tao, W.-P.; Ye, J.-J.; Ge, W. Twist mediates an aggressive phenotype in human colorectal cancer cells. Int. J. Oncol. 2016, 48, 1117–1124. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-R.; Liang, L.; Zhao, J.M.; Zhang, Y.; Zhang, M.; Zhong, W.-L.; Zhang, Q.; Wei, J.-J.; Li, M.; Yuan, J.; et al. Twist1 confers multidrug resistance in colon cancer through upregulation of ATP-binding cassette transporters. Oncotarget 2017, 8, 52901–52912. [Google Scholar] [CrossRef] [Green Version]
- Gomez, I.; Pena, C.; Herrera, M.; Muñoz, C.; Larriba, M.J.; Garcia, V.; Dominguez, G.; Silva, J.; Rodriguez, R.; De Herreros, A.G.; et al. TWIST1 Is Expressed in Colorectal Carcinomas and Predicts Patient Survival. PLoS ONE 2011, 6, e18023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fattahi, F.; Zanjani, L.S.; Vafaei, S.; Shams, Z.H.; Kiani, J.; Naseri, M.; Gheytanchi, E.; Madjd, Z. Expressions of TWIST1 and CD105 markers in colorectal cancer patients and their association with metastatic potential and prognosis. Diagn. Pathol. 2021, 16, 1–15. [Google Scholar] [CrossRef]
- Locke, W.; Guanzon, D.; Ma, C.; Liew, Y.J.; Duesing, K.; Fung, K.; Ross, J.P. DNA Methylation Cancer Biomarkers: Translation to the Clinic. Front. Genet. 2019, 10, 1150. [Google Scholar] [CrossRef] [PubMed]
- Leygo, C.; Williams, M.; Jin, H.C.; Chan, M.; Chu, W.K.; Grusch, M.; Cheng, Y.Y. DNA Methylation as a Noninvasive Epigenetic Biomarker for the Detection of Cancer. Dis. Markers 2017, 2017, 1–13. [Google Scholar] [CrossRef]
- Okugawa, Y.; Grady, W.M.; Goel, A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015, 149, 1204–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, T.; Suehiro, Y.; Ueno, K.; Mitomori, S.; Kaneko, S.; Nishioka, M.; Okayama, N.; Sakai, K.; Higaki, S.; Hazama, S.; et al. TWIST1hypermethylation is observed frequently in colorectal tumors and its overexpression is associated with unfavorable outcomes in patients with colorectal cancer. Genes Chromosom. Cancer 2010, 49, 452–462. [Google Scholar] [CrossRef]
- Lin, P.-C.; Lin, J.-K.; Lin, C.-H.; Lin, H.-H.; Yang, S.-H.; Jiang, J.-K.; Chen, W.-S.; Chou, C.-C.; Tsai, S.-F.; Chang, S.-C. Clinical Relevance of Plasma DNA Methylation in Colorectal Cancer Patients Identified by Using a Genome-Wide High-Resolution Array. Ann. Surg. Oncol. 2014, 22, 1419–1427. [Google Scholar] [CrossRef] [PubMed]
- Galván, J.A.; Helbling, M.; Koelzer, V.; Tschan, M.P.; Berger, M.D.; Hädrich, M.; Schnüriger, B.; Karamitopoulou, E.; Dawson, H.; Inderbitzin, D.; et al. TWIST1 and TWIST2 promoter methylation and protein expression in tumor stroma influence the epithelial-mesenchymal transition-like tumor budding phenotype in colorectal cancer. Oncotarget 2014, 6, 874–885. [Google Scholar] [CrossRef] [Green Version]
- Faraji, F.; Eissenberg, J.C. Seed and Soil: A Conceptual Framework of Metastasis for Clinicians. Mo. Med. 2013, 110, 302–308. [Google Scholar]
- Chen, S.; Chen, X.; Li, W.; Shan, T.; Lin, W.R.; Ma, J.; Cui, X.; Yang, W.; Cao, G.; Li, Y.; et al. Conversion of epithelial-to-mesenchymal transition to mesenchymal-to-epithelial transition is mediated by oxygen concentration in pancreatic cancer cells. Oncol. Lett. 2018, 15, 7144–7152. [Google Scholar] [CrossRef]
- Cayrefourcq, L.; Mazard, T.; Joosse, S.; Solassol, J.; Ramos, J.; Assenat, E.; Schumacher, U.; Costes, V.; Maudelonde, T.; Pantel, K.; et al. Establishment and Characterization of a Cell Line from Human Circulating Colon Cancer Cells. Cancer Res. 2015, 75, 892–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Q.; Chen, J.; Di, Z.; Yuan, W.; Zhou, Z.; Liu, Z.; Han, S.; Liu, Y.; Ying, G.; Shu, X.; et al. TM4SF1 promotes EMT and cancer stemness via the Wnt/β-catenin/SOX2 pathway in colorectal cancer. J. Exp. Clin. Cancer Res. 2020, 39, 1–17. [Google Scholar] [CrossRef]
- Koike, Y.; Yozaki, M.; Utani, A.; Murota, H. Fibroblast growth factor 2 accelerates the epithelial–mesenchymal transition in keratinocytes during wound healing process. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Yu, Y.; Xiao, C.-H.; Tan, L.-D.; Wang, Q.-S.; Li, X.-Q.; Feng, Y.-M. Cancer-associated fibroblasts induce epithelial–mesenchymal transition of breast cancer cells through paracrine TGF-β signalling. Br. J. Cancer 2014, 110, 724–732. [Google Scholar] [CrossRef] [Green Version]
- Choe, C.; Shin, Y.-S.; Kim, S.-H.; Jeon, M.-J.; Choi, S.-J.; Lee, J.; Kim, J. Tumor-stromal interactions with direct cell contacts enhance motility of non-small cell lung cancer cells through the hedgehog signaling pathway. Anticancer Res. 2013, 33, 3715–3723. [Google Scholar] [PubMed]
- Chitcholtan, K.; Asselin, E.; Parent, S.; Sykes, P.; Evans, J.J. Differences in growth properties of endometrial cancer in three dimensional (3D) culture and 2D cell monolayer. Exp. Cell Res. 2013, 319, 75–87. [Google Scholar] [CrossRef]
- Santos, E.; Hernandez, R.M.; Pedraz, J.L.; Orive, G. Novel advances in the design of three-dimensional bio-scaffolds to control cell fate: Translation from 2D to 3D. Trends Biotechnol. 2012, 30, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Amann, A.; Zwierzina, M.; Gamerith, G.; Bitsche, M.; Huber, J.M.; Vogel, G.-F.; Blumer, M.; Koeck, S.; Pechriggl, E.J.; Kelm, J.M.; et al. Development of an Innovative 3D Cell Culture System to Study Tumour—Stroma Interactions in Non-Small Cell Lung Cancer Cells. PLoS ONE 2014, 9, e92511. [Google Scholar] [CrossRef] [Green Version]
- Lochter, A.; Galosy, S.; Muschler, J.; Freedman, N.; Werb, Z.; Bissell, M.J. Matrix Metalloproteinase Stromelysin-1 Triggers a Cascade of Molecular Alterations That Leads to Stable Epithelial-to-Mesenchymal Conversion and a Premalignant Phenotype in Mammary Epithelial Cells. J. Cell Biol. 1997, 139, 1861–1872. [Google Scholar] [CrossRef]
- Pinho, S.; Oliveira, P.; Cabral, J.; Carvalho, S.; Huntsman, D.; Gärtner, F.; Seruca, R.; Reis, C.A.; Oliveira, C. Loss and Recovery of Mgat3 and GnT-III Mediated E-cadherin N-glycosylation Is a Mechanism Involved in Epithelial-Mesenchymal-Epithelial Transitions. PLoS ONE 2012, 7, e33191. [Google Scholar] [CrossRef] [Green Version]
- Olivera, P. Through the looking glass: The reversion of EMT. Eur. J. Hum. Genet. 2015, 3. [Google Scholar]
- Crnic, I.; Christofori, G. Novel technologies and recent advances in metastasis research. Int. J. Dev. Biol. 2004, 48, 573–581. [Google Scholar] [CrossRef]
- Eger, A.; Mikulits, W. Models of epithelial–mesenchymal transition. Drug Discov. Today Dis. Model. 2005, 2, 57–63. [Google Scholar] [CrossRef]
- Jacks, T.; Weinberg, R.A. Taking the Study of Cancer Cell Survival to a New Dimension. Cell 2002, 111, 923–925. [Google Scholar] [CrossRef] [Green Version]
- Date, S.; Sato, T. Mini-Gut Organoids: Reconstitution of the Stem Cell Niche. Annu. Rev. Cell Dev. Biol. 2015, 31, 269–289. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Clevers, H. SnapShot: Growing Organoids from Stem Cells. Cell 2015, 161, 1700. [Google Scholar] [CrossRef] [Green Version]
- Fujii, M.; Clevers, H.; Sato, T. Modeling Human Digestive Diseases with CRISPR-Cas9–Modified Organoids. Gastroenterology 2019, 156, 562–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Kim, R.; Gunasekara, D.B.; Reed, M.I.; DiSalvo, M.; Nguyen, D.L.; Bultman, S.J.; Sims, C.E.; Magness, S.T.; Allbritton, N.L. Formation of Human Colonic Crypt Array by Application of Chemical Gradients Across a Shaped Epithelial Monolayer. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 113–130. [Google Scholar] [CrossRef] [Green Version]
- Seino, T.; Kawasaki, S.; Shimokawa, M.; Tamagawa, H.; Toshimitsu, K.; Fujii, M.; Ohta, Y.; Matano, M.; Nanki, K.; Kawasaki, K.; et al. Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell 2018, 22, 454–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Öhlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A.S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T.E.; Hearn, S.A.; Lee, E.J.; et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017, 214, 579–596. [Google Scholar] [CrossRef] [PubMed]
- Kabiri, Z.; Greicius, G.; Madan, B.; Biechele, S.; Zhong, Z.; Zaribafzadeh, H.; Edison; Aliyev, J.; Wu, Y.; Bunte, R.; et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development 2014, 141, 2206–2215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nozaki, K.; Mochizuki, W.; Matsumoto, Y.; Matsumoto, T.; Fukuda, M.; Mizutani, T.; Watanabe, M.; Nakamura, T. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J. Gastroenterol. 2016, 51, 206–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; Van De Haar, J.; Fanchi, L.F.; Slagter, M.; Van Der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ootani, A.; Li, X.; Sangiorgi, E.; Ho, Q.T.; Ueno, H.; Toda, S.; Sugihara, H.; Fujimoto, K.; Weissman, I.L.; Capecchi, M.R.; et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 2009, 15, 701–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Yamamoto, Y.; Wilson, L.H.; Zhang, T.; Howitt, B.; Farrow, M.A.; Kern, F.; Ning, G.; Hong, Y.; Khor, C.C.; et al. Cloning and variation of ground state intestinal stem cells. Nat. Cell Biol. 2015, 522, 173–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gagliano, N.; Celesti, G.; Tacchini, L.; Pluchino, S.; Sforza, C.; Rasile, M.; Valerio, V.; Laghi, L.; Conte, V.; Procacci, P. Epithelial-to-mesenchymal transition in pancreatic ductal adenocarcinoma: Characterization in a 3D-cell culture model. World J. Gastroenterol. 2016, 22, 4466–4483. [Google Scholar] [CrossRef]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H.K. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.D.; Luitel, K.; Kim, M.; Zhang, K.; Longmore, G.D.; Tran, D.D. Transient SNAIL1 Expression Is Necessary for Metastatic Competence in Breast Cancer. Cancer Res. 2014, 74, 6330–6340. [Google Scholar] [CrossRef] [Green Version]
- Tsai, J.H.; Donaher, J.L.; Murphy, D.A.; Chau, S.; Yang, J. Spatiotemporal Regulation of Epithelial-Mesenchymal Transition Is Essential for Squamous Cell Carcinoma Metastasis. Cancer Cell 2012, 22, 725–736. [Google Scholar] [CrossRef] [Green Version]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.C.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Carstens, J.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.-C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nat. Cell Biol. 2015, 527, 525–530. [Google Scholar] [CrossRef] [Green Version]
- Krebs, A.M.; Mitschke, J.; Losada, M.L.; Schmalhofer, O.; Boerries, M.; Busch, H.; Böttcher, M.; Mougiakakos, D.; Reichardt, W.; Bronsert, P.; et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017, 19, 518–529. [Google Scholar] [CrossRef] [Green Version]
- Taube, J.; Herschkowitz, J.I.; Komurov, K.; Zhou, A.; Gupta, S.; Yang, J.; Hartwell, K.; Onder, T.; Gupta, P.B.; Evans, K.W.; et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. USA 2010, 107, 15449–15454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morel, A.-P.; Hinkal, G.W.; Thomas, C.; Fauvet, F.; Courtois-Cox, S.; Wierinckx, A.; Devouassoux-Shisheboran, M.; Treilleux, I.; Tissier, A.; Gras, B.; et al. EMT Inducers Catalyze Malignant Transformation of Mammary Epithelial Cells and Drive Tumorigenesis towards Claudin-Low Tumors in Transgenic Mice. PLoS Genet. 2012, 8, e1002723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhim, A.D.; Aiello, N.M.; Mirek, E.T.; Stanger, B.Z. Abstract IA5: EMT and dissemination precede pancreatic tumor formation. Tumor Biol. 2012, 148, 349–361. [Google Scholar] [CrossRef]
- Mueller, S.; Engleitner, T.; Maresch, R.; Zukowska, M.; Lange, S.; Kaltenbacher, T.; Konukiewitz, B.; Öllinger, R.; Zwiebel, M.; Strong, A.; et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nat. Cell Biol. 2018, 554, 62–68. [Google Scholar] [CrossRef]
- Schild, T.; Low, V.; Blenis, J.; Gomes, A.P. Unique Metabolic Adaptations Dictate Distal Organ-Specific Metastatic Colonization. Cancer Cell 2018, 33, 347–354. [Google Scholar] [CrossRef] [Green Version]
- Lehuédé, C.; Dupuy, F.; Rabinovitch, R.; Jones, R.G.; Siegel, P.M. Metabolic Plasticity as a Determinant of Tumor Growth and Metastasis. Cancer Res. 2016, 76, 5201–5208. [Google Scholar] [CrossRef] [Green Version]
- Nimmakayala, R.K.; Leon, F.; Rachagani, S.; Rauth, S.; Nallasamy, P.; Marimuthu, S.; Shailendra, G.K.; Chhonker, Y.S.; Chugh, S.; Chirravuri, R.; et al. Metabolic programming of distinct cancer stem cells promotes metastasis of pancreatic ductal adenocarcinoma. Oncogene 2021, 40, 215–231. [Google Scholar] [CrossRef]
- Reichert, M.; Bakir, B.; Moreira, L.; Pitarresi, J.R.; Feldmann, K.; Simon, L.; Suzuki, K.; Maddipati, R.; Rhim, A.D.; Schlitter, A.M.; et al. Regulation of Epithelial Plasticity Determines Metastatic Organotropism in Pancreatic Cancer. Dev. Cell 2018, 45, 696–711. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Fang, E.; Rong, Y.; Han, H.; Gong, Q.; Xiao, Y.; Li, H.; Mei, P.; Li, H.; Zhu, Z.; et al. Hypoxia-induced lncRNA CASC9 enhances glycolysis and the epithelial-mesenchymal transi-tion of pancreatic cancer by a positive feedback loop with AKT/HIF-1α signaling. Am. J. Cancer Res. 2021, 11, 123–137. [Google Scholar]
- Liu, J.; Shen, J.; Hu, J.; Dou, X.; Zhang, G. The role of epithelial-mesenchymal transition in invasion and metastasis of breast cancers. OA Cancer 2013, 1. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef] [Green Version]
- Mironchik, Y.; Winnard, P.T., Jr.; Vesuna, F.; Kato, Y.; Wildes, F.; Pathak, A.P.; Kominsky, S.; Artemov, D.; Bhujwalla, Z.; Van Diest, P.; et al. Twist Overexpression Induces In vivo Angiogenesis and Correlates with Chromosomal Instability in Breast Cancer. Cancer Res. 2005, 65, 10801–10809. [Google Scholar] [CrossRef] [Green Version]
- Gaiani, F.; Marchesi, F.; Negri, F.; Greco, L.; Malesci, A.; De’Angelis, G.; Laghi, L. Heterogeneity of Colorectal Cancer Progression: Molecular Gas and Brakes. Int. J. Mol. Sci. 2021, 22, 5246. [Google Scholar] [CrossRef] [PubMed]
- Laghi, L.; Negri, F.; Gaiani, F.; Cavalleri, T.; Grizzi, F.; Angelis, G.L.D.; Malesci, A. Prognostic and Predictive Cross-Roads of Microsatellite Instability and Immune Response to Colon Cancer. Int. J. Mol. Sci. 2020, 21, 9680. [Google Scholar] [CrossRef]
- Malesci, A.; Laghi, L.; Bianchi, P.; Delconte, G.; Randolph, A.; Torri, V.; Carnaghi, C.; Doci, R.; Rosati, R.; Montorsi, M.; et al. Reduced Likelihood of Metastases in Patients with Microsatellite-Unstable Colorectal Cancer. Clin. Cancer Res. 2007, 13, 3831–3839. [Google Scholar] [CrossRef] [Green Version]
- Laghi, L.; Malesci, A. Microsatellite Instability and Therapeutic Consequences in Colorectal Cancer. Dig. Dis. 2012, 30, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Foroutan, M.; Cursons, J.; Hediyeh-Zadeh, S.; Thompson, E.W.; Davis, M.J. A Transcriptional Program for Detecting TGFβ-Induced EMT in Cancer. Mol. Cancer Res. 2017, 15, 619–631. [Google Scholar] [CrossRef] [Green Version]
- Malesci, A.; Basso, G.; Bianchi, P.; Fini, L.; Grizzi, F.; Celesti, G.; Di Caro, G.; Delconte, G.; Dattola, F.; Repici, A.; et al. Molecular heterogeneity and prognostic implications of synchronous advanced colorectal neoplasia. Br. J. Cancer 2014, 110, 1228–1235. [Google Scholar] [CrossRef] [Green Version]
- Cereda, M.; Gambardella, G.; Benedetti, L.; Iannelli, F.; Patel, D.; Basso, G.; Guerra, R.F.; Mourikis, T.P.; Puccio, I.; Sinha, S.; et al. Patients with genetically heterogeneous synchronous colorectal cancer carry rare damaging germline mutations in immune-related genes. Nat. Commun. 2016, 7, 12072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Fang, H.; Cheng, Y.; Li, L.; Sun, X.; Fu, T.; Huang, P.; Zhang, A.; Feng, Z.; Li, C.; et al. The molecular landscape of synchronous colorectal cancer reveals genetic heterogeneity. Carcinogenesis 2018, 39, 708–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ten Hoorn, S.; Trinh, A.; de Jong, J.; Koens, L.; Vermeulen, L. Classification of Colorectal Cancer in Molecular Subtypes by Immunohistochemistry. Methods Mol. Biol. 2018, 1765, 179–191. [Google Scholar] [PubMed]
- Bellovin, D.I.; Simpson, K.J.; Danilov, T.; Maynard, E.; Rimm, D.L.; Oettgen, P.; Mercurio, A.M. Reciprocal regulation of RhoA and RhoC characterizes the EMT and identifies RhoC as a prognostic marker of colon carcinoma. Oncogene 2006, 25, 6959–6967. [Google Scholar] [CrossRef] [Green Version]
- Bates, R.C.; Bellovin, D.I.; Brown, C.; Maynard, E.; Wu, B.; Kawakatsu, H.; Sheppard, D.; Oettgen, P.; Mercurio, A.M. Transcriptional activation of integrin beta6 during the epithelial-mesenchymal transition defines a novel prognostic indicator of aggressive colon carcinoma. J. Clin. Investig. 2005, 115, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, M.; Kouvaras, E.; Papamichali, R.; Samara, M.; Chiotoglou, I.; Koukoulis, G. Smad4 and epithelial–mesenchymal transition proteins in colorectal carcinoma: An immunohistochemical study. J. Mol. Histol. 2018, 49, 235–244. [Google Scholar] [CrossRef]
- Alexander, N.R.; Tran, N.L.; Rekapally, H.; Summers, C.E.; Glackin, C.; Heimark, R.L. N-cadherin gene expression in prostate carcinoma is modulated by integ-rin-dependent nuclear translocation of Twist1. Cancer Res. 2006, 66, 3365–3369. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Gramolini, A.O. Characterization of sequences in human TWIST required for nuclear localization. BMC Cell Biol. 2009, 10, 47. [Google Scholar] [CrossRef] [Green Version]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Nam, E.-H.; Lee, Y.; Moon, B.; Lee, J.W.; Kim, S. Twist1 and AP-1 cooperatively upregulate integrin α5 expression to induce invasion and the epithelial–mesenchymal transition. Carcinogenesis 2015, 36, 327–337. [Google Scholar] [CrossRef]
- Wei, S.; Fattet, L.; Tsai, J.H.; Guo, Y.; Pai, V.H.; Majeski, H.E.; Chen, A.C.; Sah, R.L.; Taylor, S.S.; Engler, A.; et al. Matrix stiffness drives epithelial–mesenchymal transition and tumour metastasis through a TWIST1–G3BP2 mechanotransduction pathway. Nat. Cell Biol. 2015, 17, 678–688. [Google Scholar] [CrossRef]
- Ren, J.; Crowley, S.D. Twist1: A Double-Edged Sword in Kidney Diseases. Kidney Dis. 2020, 6, 247–257. [Google Scholar] [CrossRef]
- Rasti, A.; Madjd, Z.; Abolhasani, M.; Mehrazma, M.; Janani, L.; Zanjani, L.S.; Asgari, M. Cytoplasmic expression of Twist1, an EMT-related transcription factor, is associated with higher grades renal cell carcinomas and worse progression-free survival in clear cell renal cell carcinoma. Clin. Exp. Med. 2018, 18, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Ueno, H.; Shinto, E.; Kajiwara, Y.; Fukazawa, S.; Shimazaki, H.; Yamamoto, J.K.; Hase, K. Prognostic impact of histological categorisation of epithelial–mesenchymal transition in colorectal cancer. Br. J. Cancer 2014, 111, 2082–2090. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Ding, C.; Yang, Z.; Liu, T.; Zhang, X.; Zhao, C.; Wang, J. The long non-coding RNA TUG1 indicates a poor prognosis for colorectal cancer and promotes metastasis by affecting epithelial-mesenchymal transition. J. Transl. Med. 2016, 14, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shioiri, M.; Shida, T.; Koda, K.; Oda, K.; Seike, K.; Nishimura, M.; Takano, S.; Miyazaki, M. Slug expression is an independent prognostic parameter for poor survival in colorectal car-cinoma patients. Br. J. Cancer 2006, 94, 1816–1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrera, A.; Herrera, M.; Guerra-Perez, N.; Galindo-Pumariño, C.; Larriba, M.J.; García-Barberán, V.; Gil, B.; Giménez-Moyano, S.; Ferreiro-Monteagudo, R.; Veguillas, P.; et al. Endothelial cell activation on 3D-matrices derived from PDGF-BB-stimulated fibroblasts is mediated by Snail1. Oncogenesis 2018, 7, 1–15. [Google Scholar] [CrossRef]
- Freihen, V.; Rönsch, K.; Mastroianni, J.; Frey, P.; Rose, K.; Boerries, M.; Zeiser, R.; Busch, H.; Hecht, A. SNAIL1 employs β-Catenin-LEF1 complexes to control colorectal cancer cell invasion and proliferation. Int. J. Cancer 2020, 146, 2229–2242. [Google Scholar] [CrossRef] [Green Version]
- Roepman, P.; Schlicker, A.; Tabernero, J.; Majewski, I.; Tian, S.; Moreno, V.; Snel, M.H.; Chresta, C.M.; Rosenberg, R.; Nitsche, U.; et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mis-match repair and epithelial-to-mesenchymal transition. Int. J. Cancer 2014, 134, 552–562. [Google Scholar] [CrossRef]
- Loboda, A.; Nebozhyn, M.V.; Watters, J.W.; Buser, C.A.; Shaw, P.M.; Huang, P.S.; Van’t Veer, L.; Tollenaar, R.A.E.M.; Jackson, D.B.; Agrawal, D.; et al. EMT is the dominant program in human colon cancer. BMC Med. Genomics 2011, 4, 9. [Google Scholar]
- Cai, R.; Lu, Q.; Wang, D. Construction and prognostic analysis of miRNA-mRNA regulatory network in liver metastasis from colorectal cancer. World J. Surg. Oncol. 2021, 19, 7. [Google Scholar] [CrossRef]
- Liu, J.; Cho, Y.B.; Hong, H.K.; Wu, S.; Ebert, P.J.; Bray, S.M.; Wong, S.S.; Ting, J.C.; Calley, J.N.; Whittington, C.F.; et al. Molecular dissection of CRC primary tumors and their matched liver metastases reveals critical role of immune microenvironment, EMT and angiogenesis in cancer metastasis. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Cohnheim, J. Congenitales, quergestreiftes Muskelsarkom der Nieren. Virchows Archiv 1875, 65, 64–69. [Google Scholar] [CrossRef] [Green Version]
- Mani, S.A.; Guo, W.; Liao, M.-J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsoumas, D.; Nikou, S.; Giannopoulou, E.; Tsaniras, S.C.; Sirinian, C.; Maroulis, I.; Taraviras, S.; Zolota, V.; Kalofonos, H.P.; Bravou, V. ILK Expression in Colorectal Cancer Is Associated with EMT, Cancer Stem Cell Markers and Chemoresistance. Cancer Genom. Proteom. 2018, 15, 127–141. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.; Liu, Q.; Chen, C.; Yu, J.; Wang, J. Landscape perspectives of tumor, EMT, and development. Phys. Biol. 2019, 16, 051003. [Google Scholar] [CrossRef]
- Ocaña, O.H.; Córcoles, R.; Fabra, Á.; Moreno-Bueno, G.; Acloque, H.; Vega, S.; Barrallo-Gimeno, A.; Cano, A.; Nieto, M.A. Metastatic Colonization Requires the Repression of the Epithelial-Mesenchymal Transition Inducer Prrx1. Cancer Cell 2012, 22, 709–724. [Google Scholar] [CrossRef] [Green Version]
- Greaves, M. Cancer stem cells: Back to Darwin? Semin. Cancer Biol. 2010, 20, 65–70. [Google Scholar] [CrossRef]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Brooks, S.A.; Lomax-Browne, H.J.; Carter, T.M.; Kinch, C.E.; Hall, D.M.S. Molecular interactions in cancer cell metastasis. Acta Histochem. 2010. [Google Scholar] [CrossRef]
- Rosen, J.M.; Jordan, C.T. The Increasing Complexity of the Cancer Stem Cell Paradigm. Science 2009, 324, 1670–1673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, P.B.; Chaffer, C.L.; Weinberg, R.A. Cancer stem cells: Mirage or reality? Nat. Med. 2009, 15, 1010–1012. [Google Scholar] [CrossRef]
- Iinuma, H.; Watanabe, T.; Mimori, K.; Adachi, M.; Hayashi, N.; Tamura, J.; Matsuda, K.; Fukushima, R.; Okinaga, K.; Sasako, M.; et al. Clinical Significance of Circulating Tumor Cells, Including Cancer Stem-Like Cells, in Peripheral Blood for Recurrence and Prognosis in Patients with Dukes’ Stage B and C Colorectal Cancer. J. Clin. Oncol. 2011, 29, 1547–1555. [Google Scholar] [CrossRef]
- Gazzaniga, P.; Gradilone, A.; Petracca, A.; Nicolazzo, C.; Raimondi, C.; Iacovelli, R.; Naso, G.; Cortesi, E. Molecular markers in circulating tumour cells from metastatic colorectal cancer patients. J. Cell. Mol. Med. 2010, 14, 2073–2077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Gibson, P.H.; Currle, D.S.; Tong, Y.; Richardson, R.J.; Bayazitov, I.T.; Poppleton, H.; Zakharenko, S.; Ellison, D.W.; Gilbertson, R.J. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nat. Cell Biol. 2008, 457, 603–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shmelkov, S.V.; Butler, J.M.; Hooper, A.T.; Hormigo, A.; Kushner, J.; Milde, T.; Clair, R.S.; Baljevic, M.; White, I.; Jin, D.K.; et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133– metastatic colon cancer cells initiate tumors. J. Clin. Investig. 2008, 118, 2111–2120. [Google Scholar] [CrossRef]
- Dalerba, P.; Dylla, S.J.; Park, I.-K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M.; et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163. [Google Scholar] [CrossRef] [Green Version]
- Vermeulen, L.; Todaro, M.; de Sousa Mello, F.; Sprick, M.R.; Kemper, K.; Perez Alea, M.; Richel, D.J.; Stassi, G.; Medema, J.P. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc. Natl. Acad. Sci. USA 2008, 105, 13427–13432. [Google Scholar] [CrossRef] [Green Version]
- Chu, P.; Clanton, D.J.; Snipas, T.S.; Lee, J.; Mitchell, E.; Nguyen, M.-L.; Hare, E.; Peach, R.J. Characterization of a subpopulation of colon cancer cells with stem cell-like properties. Int. J. Cancer 2009, 124, 1312–1321. [Google Scholar] [CrossRef]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of Pancreatic Cancer Stem Cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermann, P.C.; Huber, S.L.; Herrler, T.; Aicher, A.; Ellwart, J.W.; Guba, M.; Bruns, C.J.; Heeschen, C. Distinct Populations of Cancer Stem Cells Determine Tumor Growth and Metastatic Activity in Human Pancreatic Cancer. Cell Stem Cell 2007, 1, 313–323. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.J.; Dosch, J.; Simeone, D.M. Pancreatic Cancer Stem Cells. J. Clin. Oncol. 2008, 26, 2806–2812. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Cai, Z.; Li, S.; Cheng, Y.; Gao, H.; Liu, F.; Wu, S.; Liu, S.; Dong, Y.; Zheng, L.; et al. Expression and clinical relevance of epithelial and mesenchymal markers in circulating tumor cells from colorectal cancer. Oncotarget 2016, 8, 9293–9302. [Google Scholar] [CrossRef] [Green Version]
- Wei, T.; Zhang, X.; Zhang, Q.; Yang, J.; Chen, Q.; Wang, J.; Li, X.; Chen, J.; Ma, T.; Li, G.; et al. Vimentin-positive circulating tumor cells as a biomarker for diagnosis and treatment monitoring in patients with pancreatic cancer. Cancer Lett. 2019, 452, 237–243. [Google Scholar] [CrossRef]
- Poruk, K.E.; Valero, V.; Saunders, T.; Blackford, A.L.; Griffin, J.; Poling, J.; Hruban, R.H.; Anders, R.A.; Herman, J.; Zheng, L.; et al. Circulating Tumor Cell Phenotype Predicts Recurrence and Survival in Pancreatic Adenocarcinoma. Ann. Surg. 2016, 264, 1073–1081. [Google Scholar] [CrossRef] [Green Version]
- Buscail, E.; Chiche, L.; Laurent, C.; Vendrely, V.; Denost, Q.; Denis, J.; Thumerel, M.; Lacorte, J.; Bedel, A.; Moreau-Gaudry, F.; et al. Tumor-proximal liquid biopsy to improve diagnostic and prognostic performances of circulating tumor cells. Mol. Oncol. 2019, 13, 1811–1826. [Google Scholar] [CrossRef]
- Tien, Y.W.; Kuo, H.-C.; Ho, B.-I.; Chang, M.-C.; Chang, Y.-T.; Cheng, M.-F.; Chen, H.-L.; Liang, T.-Y.; Wang, C.-F.; Huang, C.-Y.; et al. A High Circulating Tumor Cell Count in Portal Vein Predicts Liver Metastasis from Periampullary or Pancreatic Cancer. Medicine 2016, 95, e3407. [Google Scholar] [CrossRef]
- Vona, G.; Sabile, A.; Louha, M.; Sitruk, V.; Romana, S.P.; Schütze, K.; Capron, F.; Franco, D.; Pazzagli, M.; Vekemans, M.; et al. Isolation by Size of Epithelial Tumor Cells: A New Method for the Immunomorphological and Molecular Characterization of Circulating Tumor Cells. Am. J. Pathol. 2000, 156, 57–63. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [Green Version]
- Ogle, L.F.; Orr, J.G.; Willoughby, C.; Hutton, C.; McPherson, S.; Plummer, R.; Boddy, A.; Curtin, N.; Jamieson, D.; Reeves, H.L. Imagestream detection and characterisation of circulating tumour cells—A liquid biopsy for hepatocellular carcinoma? J. Hepatol. 2016, 65, 305–313. [Google Scholar] [CrossRef] [Green Version]
- Harouaka, R.; Kang, Z.; Zheng, S.-Y.; Cao, L. Circulating tumor cells: Advances in isolation and analysis, and challenges for clinical applications. Pharmacol. Ther. 2014, 141, 209–221. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Ma, Y.; Zhao, X.; Tian, X.; Sun, Y.; Yang, Y.; Zhao, X. Spatial heterogeneity in epithelial to mesenchymal transition properties of circulating tumor cells associated with distant recurrence in pancreatic cancer patients. Ann. Transl. Med. 2020, 8, 676. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ma, H.; Yan, Y.; Chen, Y.; Fu, S.; Wang, J.; Wang, Y.; Chen, H.; Liu, J. cMET promotes metastasis and epithelial-mesenchymal transition in colorectal carcinoma by re-pressing RKIP. J. Cell. Physiol. 2021, 236, 3963–3978. [Google Scholar] [CrossRef] [PubMed]
- Shitara, K.; Kim, T.M.; Yokota, T.; Goto, M.; Satoh, T.; Ahn, J.-H.; Kim, H.S.; Assadourian, S.; Gomez, C.; Harnois, M.; et al. Phase I dose-escalation study of the c-Met tyrosine kinase inhibitor SAR125844 in Asian patients with advanced solid tumors, including patients with MET-amplified gastric cancer. Oncotarget 2017, 8, 79546–79555. [Google Scholar] [CrossRef] [Green Version]
- Oshi, M.; Tokumaru, Y.; Mukhopadhyay, S.; Yan, L.; Matsuyama, R.; Endo, I.; Takabe, K. Annexin A1 Expression Is Associated with Epithelial–Mesenchymal Transition (EMT), Cell Proliferation, Prognosis, and Drug Response in Pancreatic Cancer. Cells 2021, 10, 653. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Tian, X.; Ji, L.; Zhang, X.; Cao, Y.; Shen, C.; Hu, Y.; Wong, J.W.H.; Fang, J.-Y.; Hong, J.; et al. A tumor microenvironment-specific gene expression signature predicts chemotherapy resistance in colorectal cancer patients. NPJ Precis. Oncol. 2021, 5, 1–14. [Google Scholar] [CrossRef]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Wang, G.; Zhu, D.; Huang, Y.; Luo, Y.; Su, P.; Chen, X.; Wang, Q. Epithelial-mesenchymal transition and cancer stem cell-like phenotype induced by Twist1 con-tribute to acquired resistance to irinotecan in colon cancer. Int. J. Oncol. 2017, 51, 515–524. [Google Scholar] [CrossRef]
- Skarkova, V.; Kralova, V.; Krbal, L.; Matouskova, P.; Soukup, J.; Rudolf, E. Oxaliplatin and irinotecan induce heterogenous changes in the EMT markers of metas-tasizing colorectal carcinoma cells. Exp. Cell Res. 2018, 369, 295–303. [Google Scholar] [CrossRef]
- Huang, X.; Liu, J.; Liu, H.; Mo, X.; Meng, Y.; Zhang, L.; Deng, Y.; Zhang, Y.; Tang, W. A Combined Epithelial Mesenchymal Transformation and DNA Repair Gene Panel in Colorectal Cancer with Prognostic and Therapeutic Implication. Front. Oncol. 2021, 10, 2955. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greco, L.; Rubbino, F.; Morelli, A.; Gaiani, F.; Grizzi, F.; de’Angelis, G.L.; Malesci, A.; Laghi, L. Epithelial to Mesenchymal Transition: A Challenging Playground for Translational Research. Current Models and Focus on TWIST1 Relevance and Gastrointestinal Cancers. Int. J. Mol. Sci. 2021, 22, 11469. https://doi.org/10.3390/ijms222111469
Greco L, Rubbino F, Morelli A, Gaiani F, Grizzi F, de’Angelis GL, Malesci A, Laghi L. Epithelial to Mesenchymal Transition: A Challenging Playground for Translational Research. Current Models and Focus on TWIST1 Relevance and Gastrointestinal Cancers. International Journal of Molecular Sciences. 2021; 22(21):11469. https://doi.org/10.3390/ijms222111469
Chicago/Turabian StyleGreco, Luana, Federica Rubbino, Alessandra Morelli, Federica Gaiani, Fabio Grizzi, Gian Luigi de’Angelis, Alberto Malesci, and Luigi Laghi. 2021. "Epithelial to Mesenchymal Transition: A Challenging Playground for Translational Research. Current Models and Focus on TWIST1 Relevance and Gastrointestinal Cancers" International Journal of Molecular Sciences 22, no. 21: 11469. https://doi.org/10.3390/ijms222111469
APA StyleGreco, L., Rubbino, F., Morelli, A., Gaiani, F., Grizzi, F., de’Angelis, G. L., Malesci, A., & Laghi, L. (2021). Epithelial to Mesenchymal Transition: A Challenging Playground for Translational Research. Current Models and Focus on TWIST1 Relevance and Gastrointestinal Cancers. International Journal of Molecular Sciences, 22(21), 11469. https://doi.org/10.3390/ijms222111469






