Characteristics of SlCML39, a Tomato Calmodulin-like Gene, and Its Negative Role in High Temperature Tolerance of Arabidopsis thaliana during Germination and Seedling Growth
Abstract
:1. Introduction
2. Results
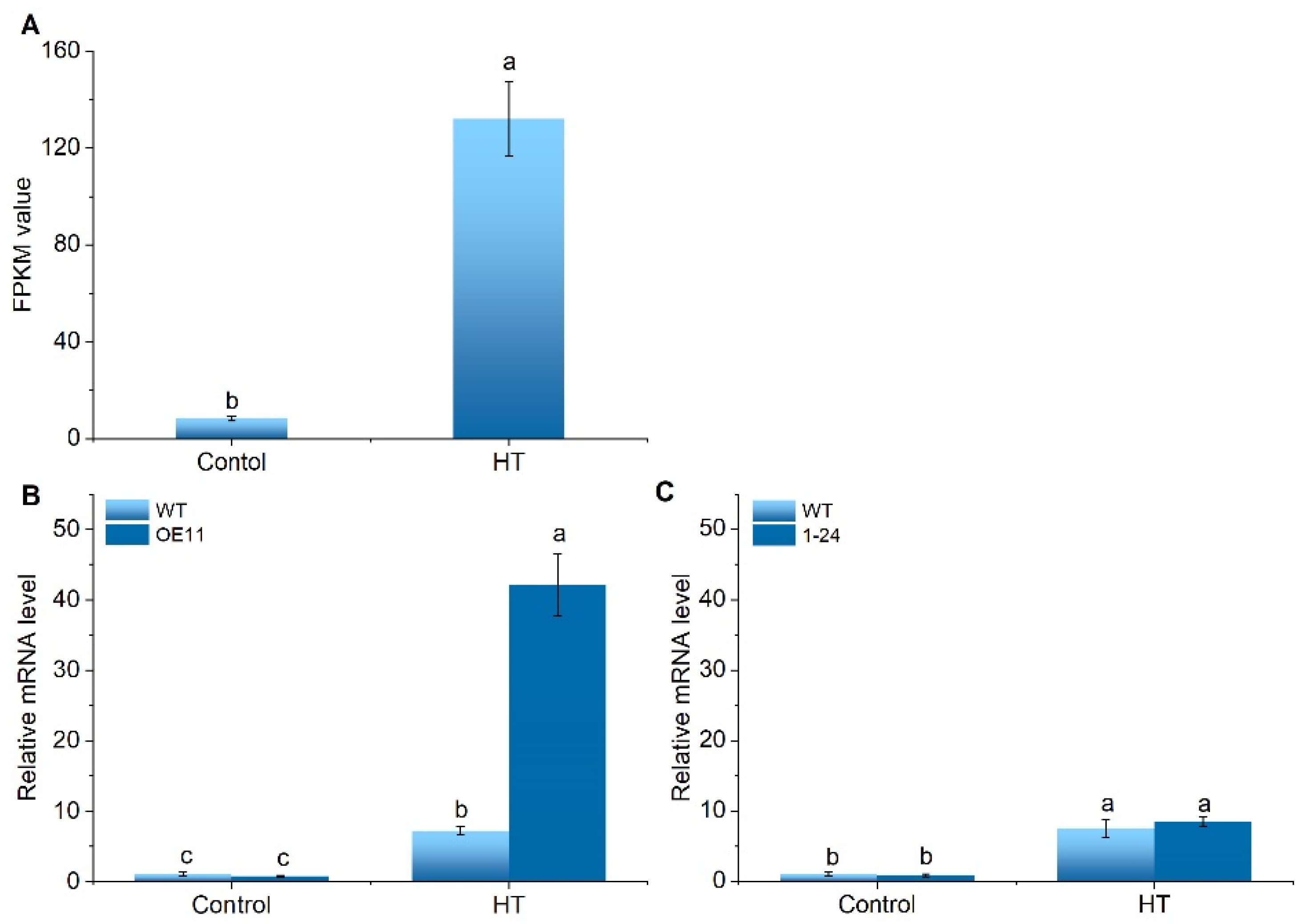
2.1. SlCML39 Is Attractive Enough under HT Stress
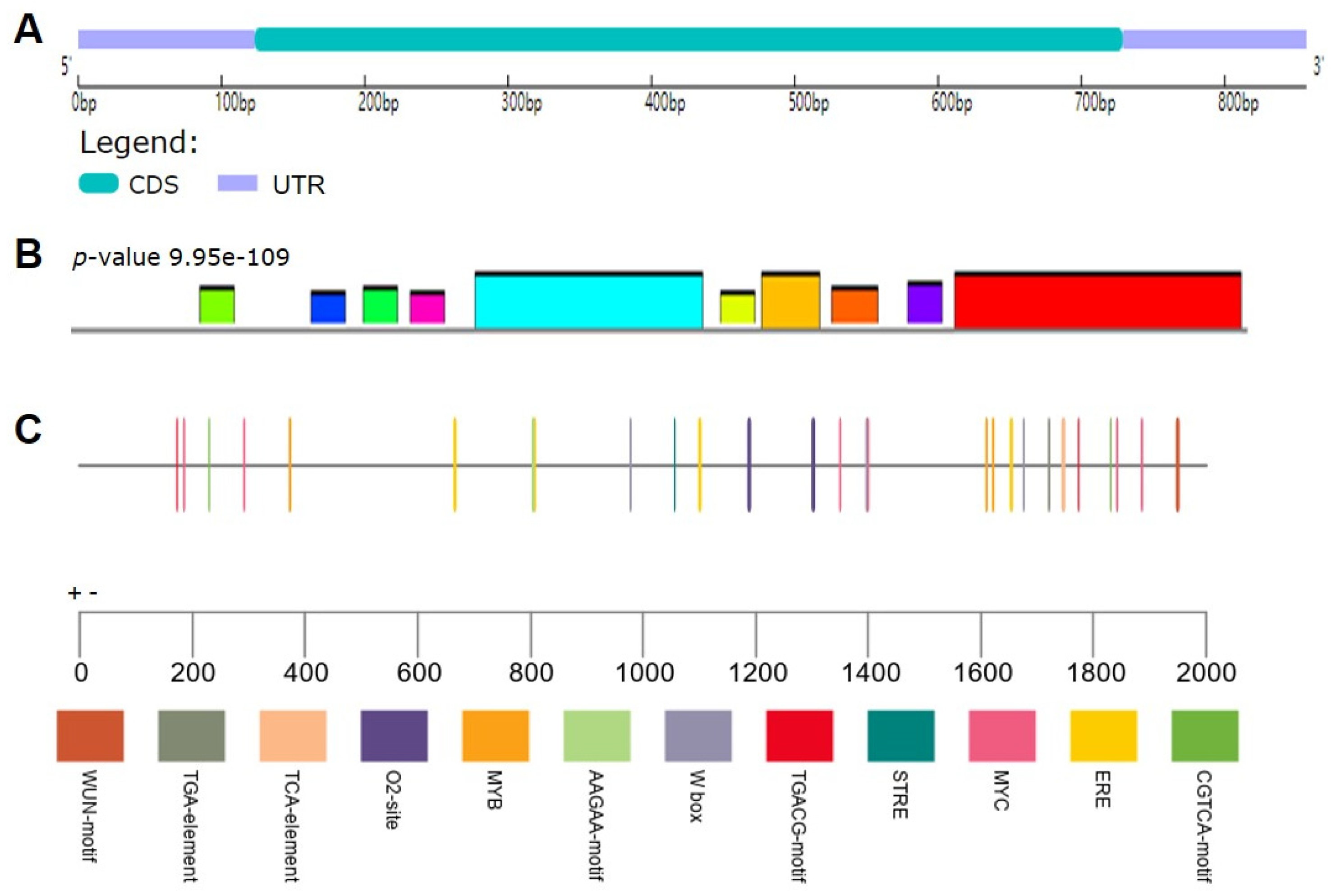
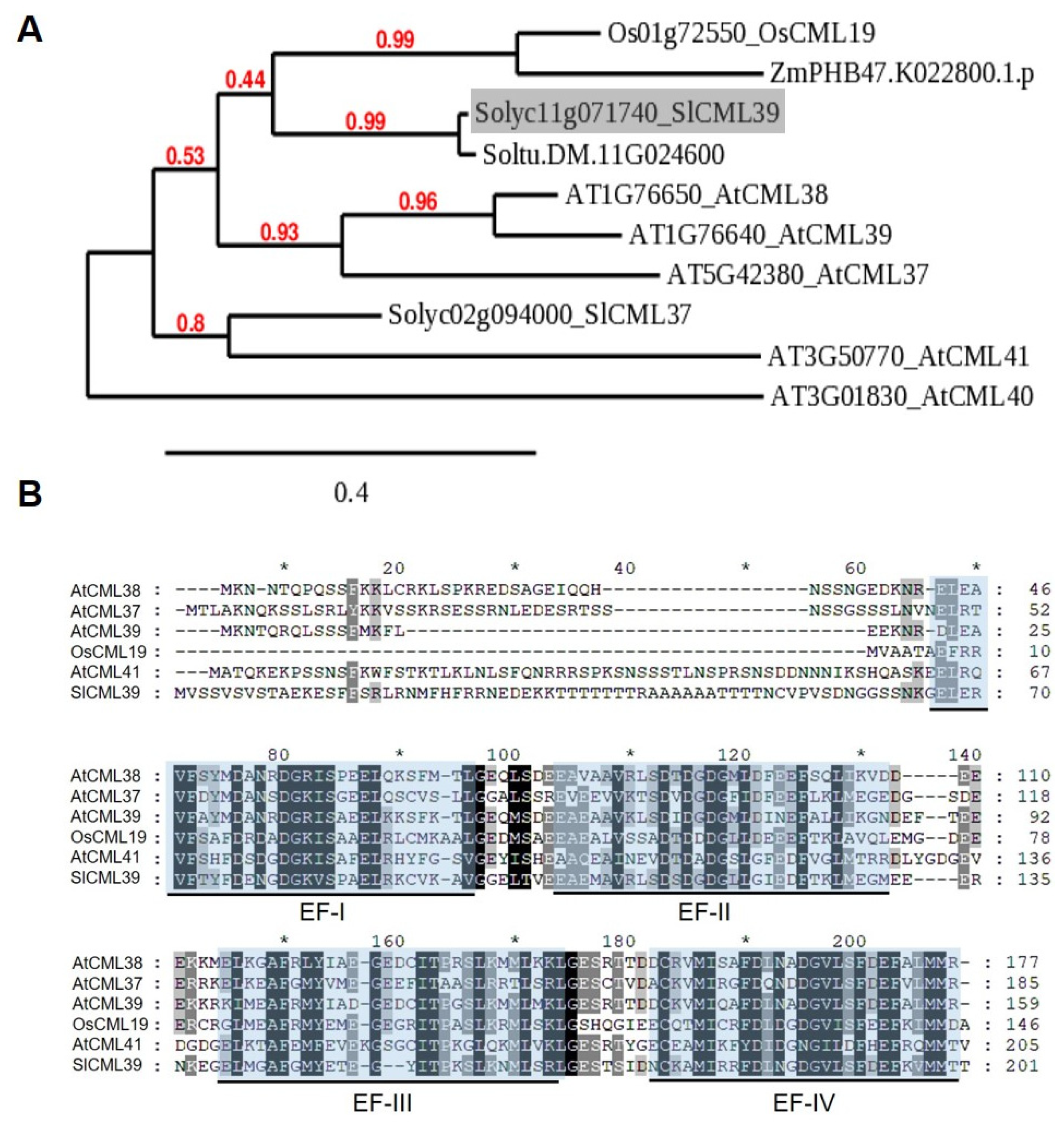
2.2. Characterization of SlCML39
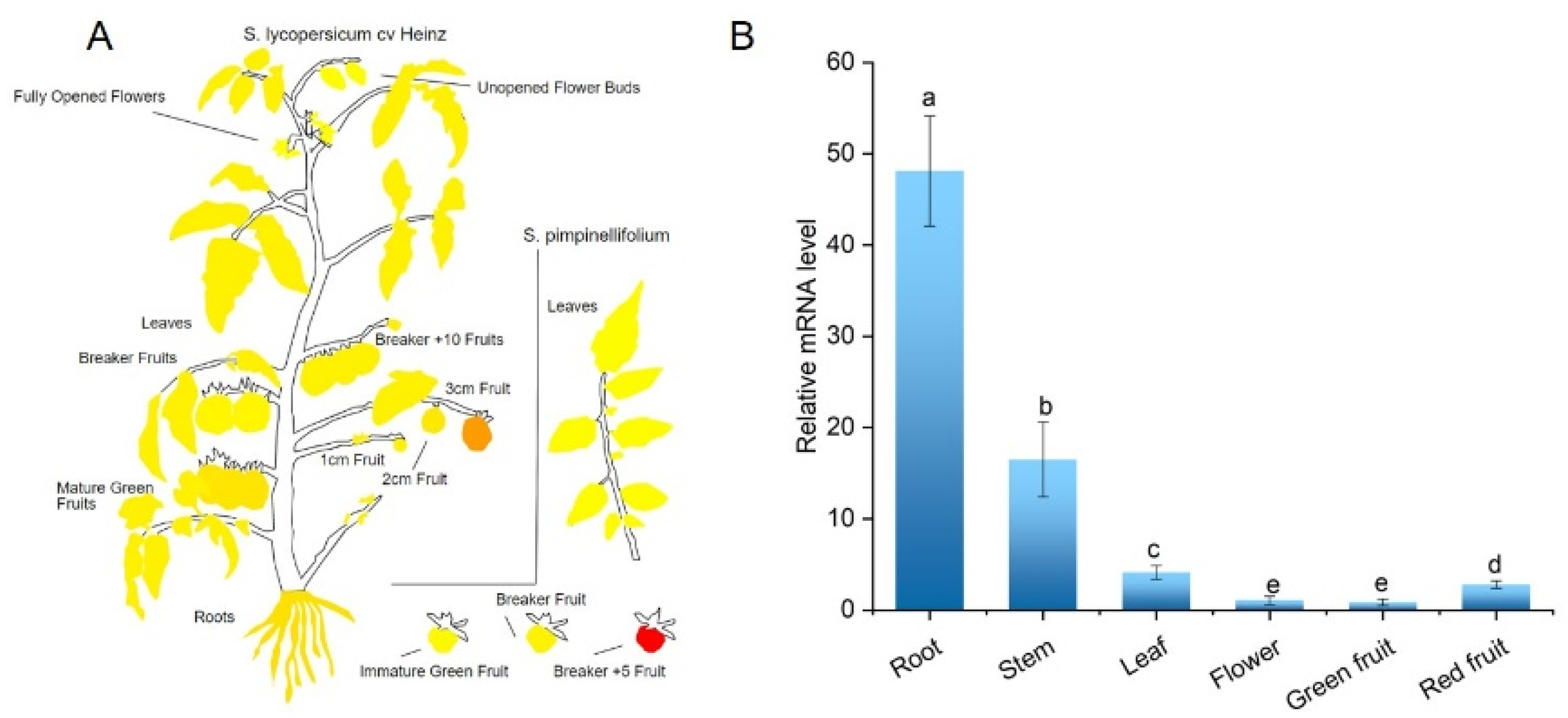
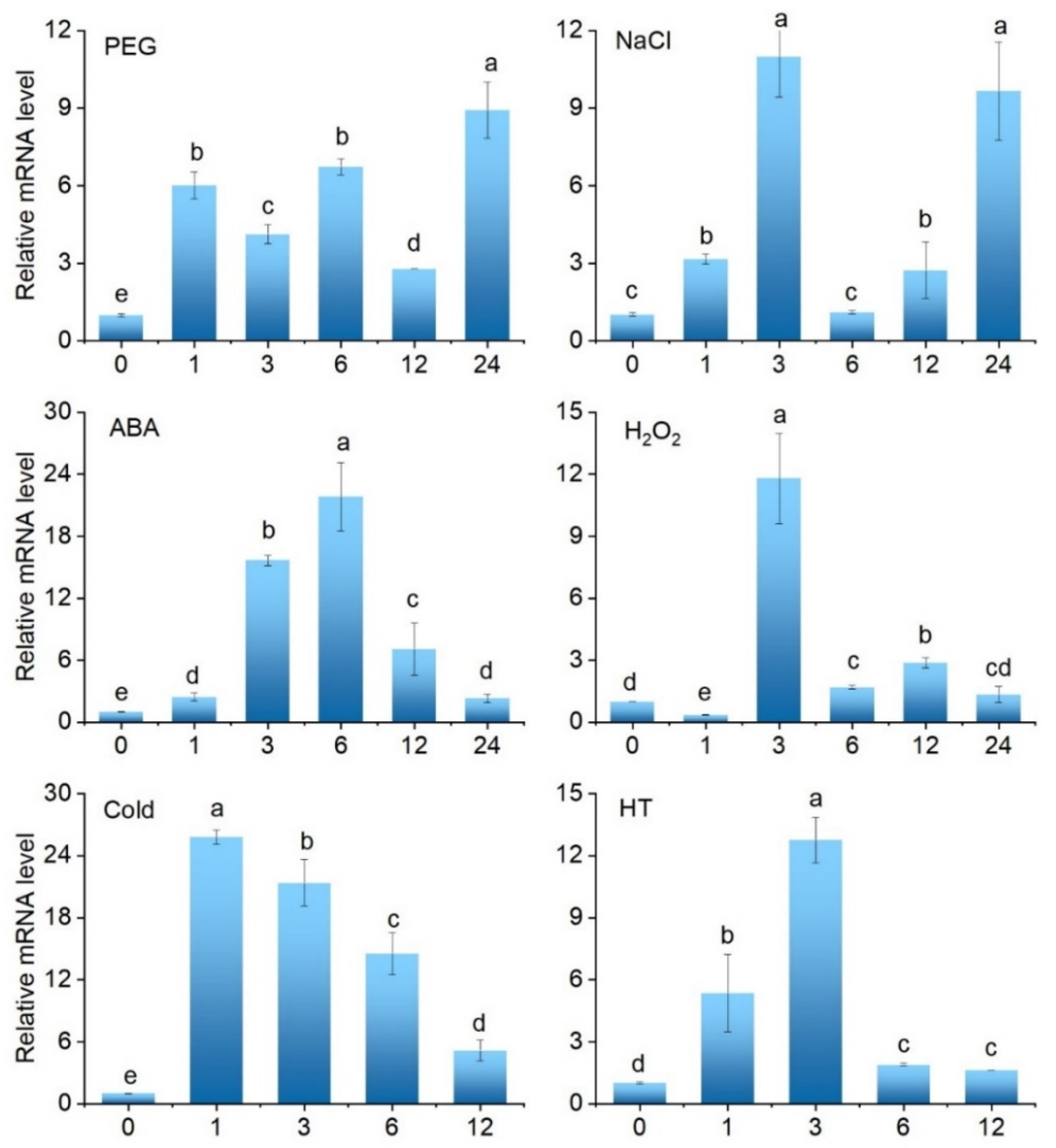
2.3. Expression Profiles of SlCML39
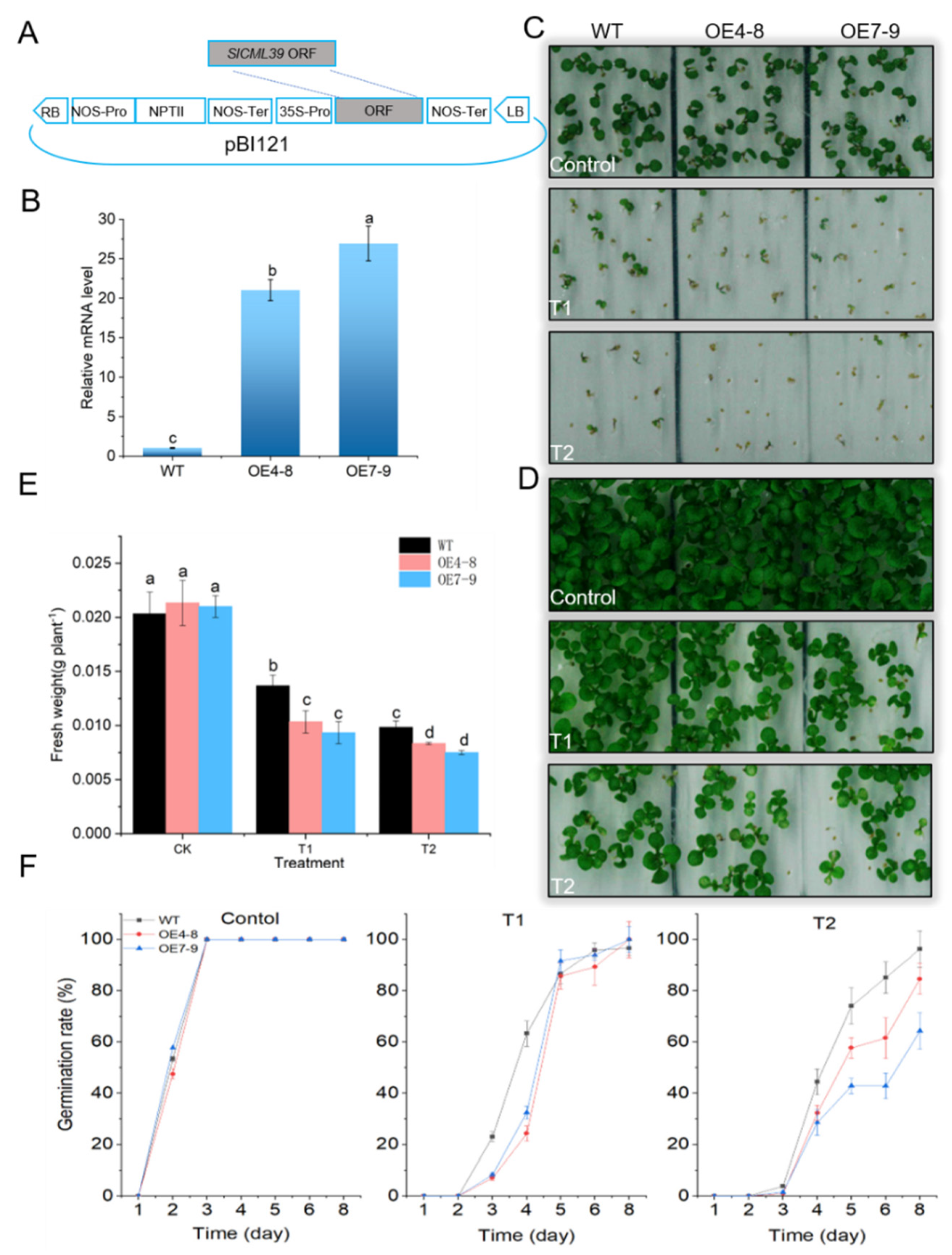
2.4. SlCML39 Negatively Regulated Seed Germination in Arabidopsis under HT Stress
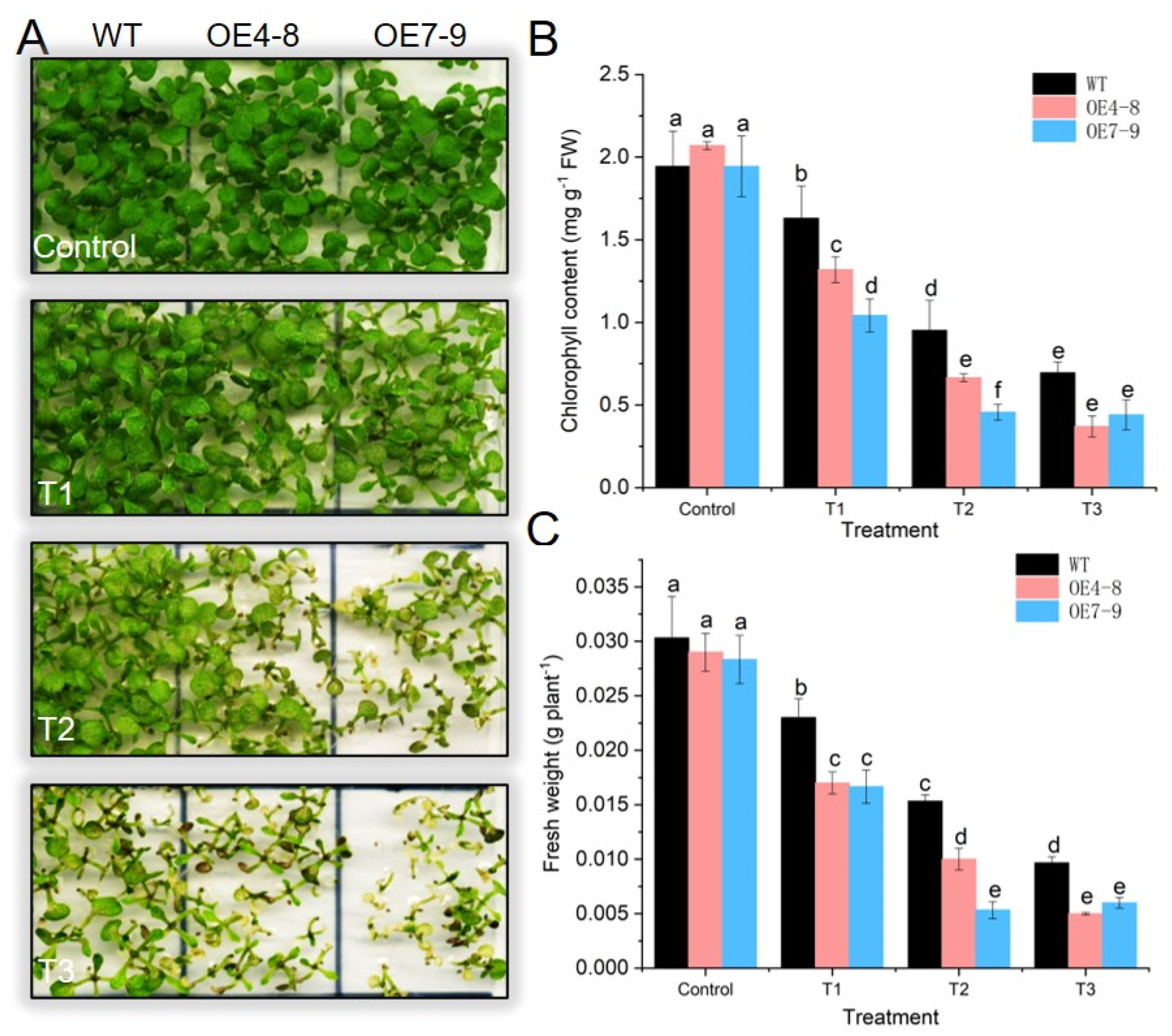
2.5. SlCML39 Overexpression Decreased Arabidopsis Seedling Tolerance to HT Stress
2.6. RNA-Seq of SlCML39-Regulating Gene Network under HT Stress
2.7. SlCML39 Negatively Regulates Stress/ABA-Responsive Genes under HT Stress
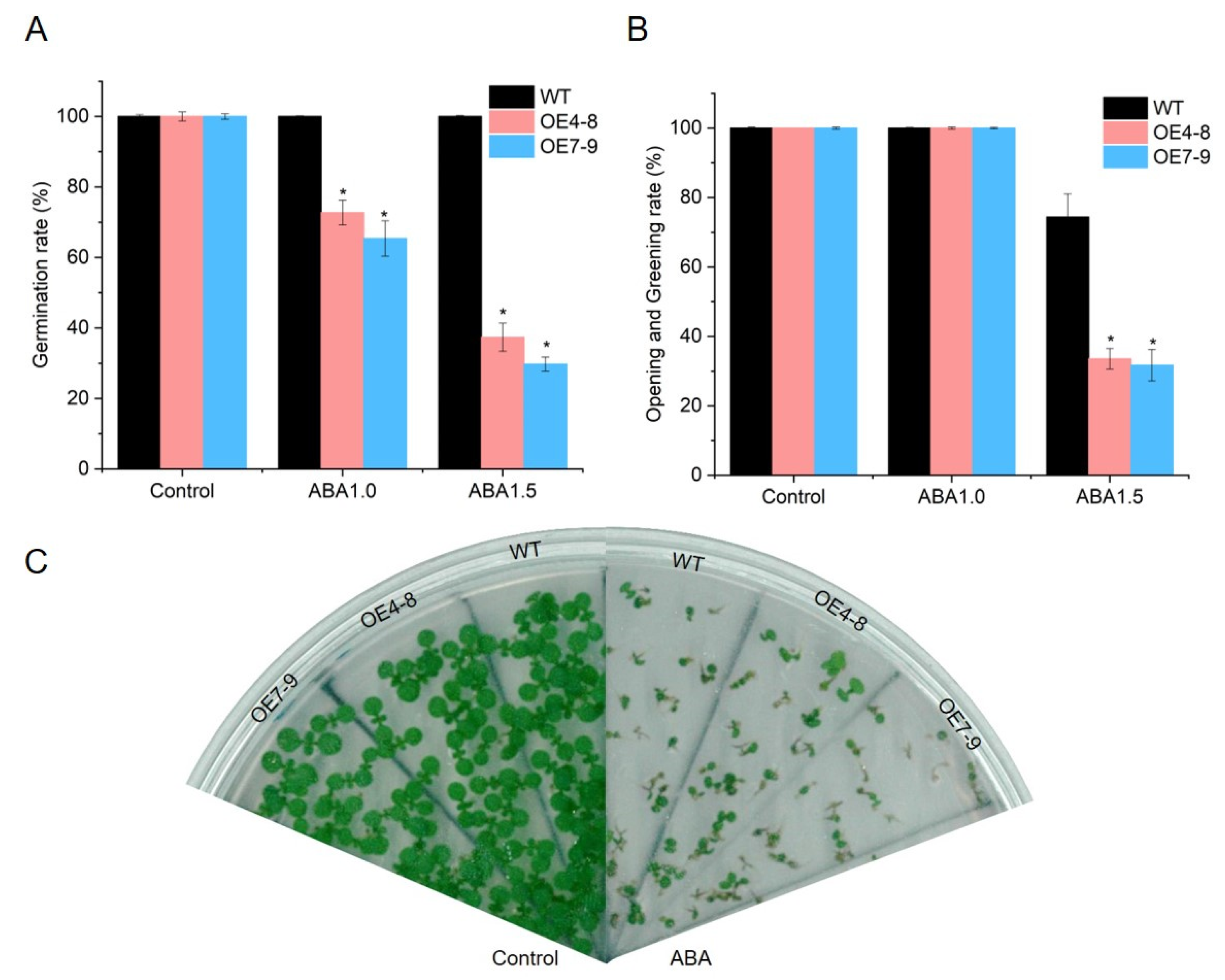
2.8. SlCML39 Is Involved in ABA-Mediated Seed Germination
3. Discussion
3.1. Homologous CMLs of SlCML39
3.2. SlCML39 Expression Is Induced by HT as well as Other Stresses
3.3. SlCML39 Decreased Plant Thermotolerance
3.4. SlCML39 Is Involved in ABA-Regulated Germination
3.5. SlCML39 Regulates Stress/ABA-Responsive Genes under HT
4. Materials and Methods
4.1. Plant Material Growth
4.2. Expression Analysis of SlCML39
4.3. Bioinformatics Analysis of SlCML39
4.4. Heterologous Expression of SlCML39 in Arabidopsis
4.5. HT Tolerance of Arabidopsis
4.6. Seed Germination under ABA Treatment
4.7. ABA Content
4.8. RNA-Seq
4.9. qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Dunand, C.; Snedden, W.; Galaud, J. CaM and CML emergence in the green lineage. Trends Plant Sci. 2015, 20, 483–489. [Google Scholar] [CrossRef]
- DeFalco, T.A.; Bender, K.W.; Snedden, W.A. Breaking the code: Ca2+ sensors in plant signal. Biochem. J. 2010, 425, 27–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gifford, J.L.; Walsh, M.P.; Vogel, H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix—loop—helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, T.K.; Kumar, P.; Bae, H. Genomics and evolutionary aspect of calcium signaling event in calmodulin and calmodulin-like proteins in plants. BMC Plant Biol. 2017, 17, 38. [Google Scholar] [CrossRef] [Green Version]
- McCormack, E.; Braam, J. Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol. 2003, 159, 585–598. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Liu, W.; Li, W.; Liu, Y.; Wang, W.; Xie, P.; Kang, Y.; Liao, L.; Qian, L.; Liu, Z.; et al. Genome-wide identification and expression analysis of cam/cml genes in brassica napus under abiotic stress. J. Plant Physiol. 2020, 255, 153251. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Xu, C.; Cao, H.; Shi, Y.; Chen, J.; Chai, Y.; Li, Z. Tomato calmodulin-like protein SlCML37 is a calcium (Ca2+) sensor that interacts with proteasome maturation factor SlUMP1 and plays a role in tomato fruit chilling stress tolerance. J. Plant Physiol. 2021, 25, 153373. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, S.S.; Wang, M.; Qiao, Z.; Bao, C.C.; Zhang, W. Arabidopsis thaliana, calmodulin-like protein CML24 regulates pollen tube growth by modulating the actin cytoskeleton and controlling the cytosolic Ca2+ concentration. Plant Mol. Biol. 2014, 86, 225–236. [Google Scholar] [CrossRef]
- Wang, S.S.; Diao, W.Z.; Yang, X.; Qiao, Z.; Wang, M.; Acharya, B.R.; Zhang, W. Arabidopsis thaliana CML25 mediates the Ca2+ regulation of K+ transmembrane trafficking during pollen germination and tube elongation. Plant Cell Environ. 2015, 38, 2372–2386. [Google Scholar] [CrossRef]
- Bender, K.W.; Rosenbaum, D.M.; Vanderbeld, B.; Ubaid, M.; Snedden, W.A. The Arabidopsis calmodulin-like protein, CML39, functions during early seedling establishment. Plant J. 2013, 76, 634–647. [Google Scholar] [CrossRef]
- Dobney, S.; Chiasson, D.; Lam, P.; Smith, S.P.; Snedden, W.A. The calmodulin-related calcium sensor CML42 plays a role in trichome branching. J. Biol. Chem. 2009, 284, 31647–31657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vadassery, J.; Reichelt, M.; Hause, B.; Gershenzon, J.; Boland, W.; Mithofer, A. CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant. Physiol. 2012, 159, 1159–1175. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Cheval, C.; Laohavisit, A.; Hocking, B.; Chiasson, D.; Olsson, T.S.G.; Shirasu, K.; Faulkner, C.; Gilliham, M. A calmodulin-like protein regulates plasmodesmal closure during bacterial immune responses. New Phytol. 2017, 215, 77–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Astegno, A.; Bonza, M.C.; Vallone, R.; La Verde, V.; D’Onofrio, M.; Luoni, L.; Molesini, B.; Dominici, P. Arabidopsis calmodulin-like protein CML36 is a calcium (Ca2+) sensor that interacts with the plasma membrane Ca2+-ATPase isoform ACA8 and stimulates its activity. J. Biol. Chem. 2017, 292, 15049–15061. [Google Scholar] [CrossRef] [Green Version]
- Magnan, F.; Ranty, B.; Charpenteau, M.; Sotta, B.; Galaud, J.P.; Aldon, D. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 2008, 56, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.Y.; Rocha, P.S.; Wang, M.L.; Xu, M.L.; Cui, Y.C.; Li, L.Y.; Zhu, Y.X.; Xia, X. A novel rice calmodulin-like gene, OsMSR2, enhances drought and salt tolerance and increases ABA sensitivity in Arabidopsis. Planta 2011, 234, 47–59. [Google Scholar] [CrossRef]
- Chen, C.; Sun, X.; Duanmu, H.; Zhu, D.; Yu, Y.; Cao, L.; Liu, A.; Jia, B.; Xiao, J.; Zhu, Y. GsCML27, a gene encoding a calcium binding EF-hand protein from Glycine soja, plays differential roles in plant responses to Bicarbonate, salt and osmotic stresses. PLoS ONE 2015, 10, e0141888. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Qiao, Z.; Liu, H.; Acharya, B.R.; Li, C.; Zhang, W. CML20, an arabidopsis calmodulin-like protein, negatively regulates guard cell aba signal and drought stress tolerance. Front. Plant Sci. 2017, 8, 824. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Huang, J.; Hou, Q.; Liu, Y.; Wang, J.; Deng, S. Isolation and functional characterization of a Salt-responsive calmodulin-like gene MpCML40 from semi-mangrove Millettia pinnata. Int. J. Mol. Sci. 2021, 22, 3475. [Google Scholar] [CrossRef]
- Munir, S.; Liu, H.; Xing, Y.; Hussain, S.; Ouyang, B.; Zhang, Y.; Li, H.; Ye, Z. Overexpression of calmodulin-like (ShCML44) stress-responsive gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses. Sci. Rep. 2016, 6, 1–20. [Google Scholar] [CrossRef]
- Shi, J.; Du, X. Identification, characterization and expression analysis of calmodulin and calmodulin-like proteins in Solanum pennellii. Sci Rep. 2020, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Chaisson, D.; Ekengren, S.K.; Martin, G.B.; Dobney, S.L.; Snedden, W.A. Calmodulin-like proteins from Arabidopsis and tomato are involved in host defense against Pseudomonas syringae pv. tomato. Plant Mol. Biol. 2005, 58, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Mo, S.; Qian, Y.; Yuan, G.; Wu, X.; Ge, C. Integrated proteome and transcriptome analyses revealed key factors involved in tomato (Solanum lycopersicum) under high temperature stress. Food Energy Sec. 2020, 9, e239. [Google Scholar] [CrossRef]
- Munir, S.; Khan, M.R.; Song, J.; Munir, S.; Zhang, Y.; Ye, Z.; Wang, T. Genome wide identification, characterization and expression analysis of calmodulin-like (CML) proteins in tomato (Solanum lycopersicum). Plant Physiol. Biochem. 2016, 102, 167–179. [Google Scholar] [CrossRef]
- Ding, H.; He, J.; Wu, Y.; Wu, X.; Ge, C.; Wang, Y.; Zhong, S.; Peiter, E.; Liang, J.; Xu, W. The tomato mitogen-activated protein kinase SlMPK1 is as a negative regulator of the high temperature. Plant Physiol. 2018, 177, 633–651. [Google Scholar] [CrossRef] [Green Version]
- Lu, P.P.; Zheng, W.J.; Wang, C.T.; Shi, W.; Fu, J.; Chen, M.; Chen, J.; Zhou, Y.; Xi, Y.; Xu, Z. Wheat Bax Inhibitor-1 interacts with TaFKBP62 and mediates response to heat stress. BMC Plant. Bio. 2018, 18, 259. [Google Scholar] [CrossRef]
- Zhu, F.Y.; Chen, M.X.; Ye, N.H.; Qiao, W.M.; Gao, B.; Law, W.K.; Tian, Y.; Zhang, D.; Zhang, D.; Liu, T.Y.; et al. Comparative performance of the BGISEQ-500 and Illumina HiSeq4000 sequencing platforms for transcriptome analysis in plants. Plant Methods 2018, 14, 69. [Google Scholar] [CrossRef] [Green Version]
- Chao, L.M.; Liu, Y.Q.; Chen, D.Y.; Xue, X.Y.; Mao, Y.B.; Chen, X.Y. Arabidopsis transcription factors SPL1 and SPL12 confer plant thermotolerance at reproductive stage. Mol. Plant 2017, 10, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.H.; Matsui, A.; Tanaka, M.; Mizunashi, K.; Nakaminami, K.; Hayashi, M.; Iida, K.; Toyoda, T.; Nguyen, D.V.; Seki, M. Loss of Arabidopsis 5′–3′ exoribonuclease AtXRN4 function enhances heat stress tolerance of plants subjected to severe heat stress. Plant Cell Physiol. 2015, 56, 1762–1772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Song, N.; Zheng, M.; Liu, X.; Liu, Z.; Xing, J.; Ma, J.; Guo, W.; Yao, Y.; Peng, H.; et al. Histone acetyltransferase GCN 5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J. 2015, 84, 1178–1191. [Google Scholar] [CrossRef] [PubMed]
- Wiese, A.J.; Steinbachová, L.; Timofejeva, L.; Čermák, V.; Klodová, B.; Ganji, R.S.; Limones-Mendez, M.; Bokvaj, P.; Hafidh, S.; Potešil, D.; et al. Arabidopsis bZIP18 and bZIP52 accumulate in nuclei following heat stress where they regulate the expression of a similar set of genes. Int. J. Mol. Sci. 2021, 22, 530. [Google Scholar] [CrossRef]
- Weng, J.K.; Ye, M.; Li, B.; Noel, J.P. Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity. Cell 2016, 166, 881–893. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Wang, B.; Tang, K.; Hsu, C.C.; Xie, S.; Du, H.; Yang, Y.; Tao, W.A.; Zhu, J.K. An arabidopsis nucleoporin NUP85 modulates plant responses to ABA and salt stress. PLoS Genet. 2017, 13, e1007124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Cao, J.; He, J.; Chen, Q.; Li, X.; Yang, Y. Molecular mechanism for the regulation of ABA homeostasis during plant development and stress responses. Int. J. Mol. Sci. 2018, 19, 3643. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.C.; Niu, C.Y.; Yang, C.R.; Jinn, T.L. The heat stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol. 2016, 172, 1182–1199. [Google Scholar] [CrossRef]
- Suzuki, N.; Bassil, E.; Hamilton, J.S.; Inupakutika, M.A.; Zandalinas, S.I.; Tripathy, D.; Luo, Y.; Dion, E.; Fukui, G.; Kumazaki, A.; et al. ABA is required for plant acclimation to a combination of salt and heat stress. PLoS ONE 2016, 11, e0147625. [Google Scholar] [CrossRef] [Green Version]
- Mo, S.; Qian, Y.; Zhang, W.; Qian, L.; Wang, Y.; Cailin, G.; Ding, H. Mitogen-activated protein kinase action in plant response to high-temperature stress: A mini review. Protoplasma 2021, 258, 477–482. [Google Scholar] [CrossRef]
- Farooq, M.A.; Niazi, A.K.; Akhtar, J.; Farooq, M.; Souri, Z.; Karimi, N.; Rengel, Z. Acquiring control: The evolution of ROS-Induced oxidative stress and redox signal pathways in plant stress responses. Plant Physiol. Bioch. 2019, 141, 353–369. [Google Scholar] [CrossRef]
- Vanderbeld, B.; Snedden, W.A. Developmental and stimulus-induced expression patterns of Arabidopsis calmodulin-like genes CML37, CML38 and CML39. Plant Mol. Biol. 2007, 64, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Scholz, S.S.; Reichelt, M.; Vadassery, J.; Mithöfer, A. Calmodulin-like protein CML37 is a positive regulator of ABA during drought stress in Arabidopsis. Plant Signal. Behav. 2015, 10, e1011951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lokdarshi, A.; Conner, W.C.; McClintock, C.; Li, T.; Roberts, D.M. Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol. 2016, 170, 1046–1059. [Google Scholar] [CrossRef] [Green Version]
- Campos, W.F.; Dressano, K.; Ceciliato, P.H.; Guerrero-Abad, J.C.; Silva, A.L.; Fiori, C.S.; Amanda, M.D.C.; Bergonci, T.; Claus, L.A.N.; Silva-Filho, M.C.; et al. Arabidopsis thaliana rapid alkalinization factor 1–mediated root growth inhibition is dependent on calmodulin-like protein 38. J. Biol. Chem. 2018, 293, 2159–2171. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Li, J.; Lyu, M.; Kong, X.; Hu, S.; Song, Q.; Zuo, K. CALMODULIN-LIKE-38 and PEP1 RECEPTOR 2 integrate nitrate and brassinosteroid signals to regulate root growth. Plant Physiol. 2021. [Google Scholar] [CrossRef]
- Midhat, U.; Ting, M.K.; Teresinski, H.J.; Snedden, W.A. The calmodulin-like protein, CML39, is involved in regulating seed development, germination, and fruit development in Arabidopsis. Plant Mol. Biol. 2018, 96, 375–392. [Google Scholar] [CrossRef]
- Zeng, H.; Xu, L.; Singh, A.; Wang, H.; Du, L.; Poovaiah, B.W. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 2015, 6, 600. [Google Scholar] [CrossRef] [Green Version]
- Dubrovina, A.S.; Aleynova, O.A.; Ogneva, Z.V.; Suprun, A.R.; Ananev, A.A.; Kiselev, K.V. The effect of abiotic stress conditions on expression of calmodulin (CaM) and calmodulin-like (CML) genes in wild-growing grapevine Vitis amurensis. Plants 2019, 8, 602. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Shen, X.J.; Yuan, H.Z.; Liu, Y.; Liao, X.; Wang, Q.; Liu, L.L.; Li, F.; Li, T.H. Isolation and characterization of dehydration-responsive element-binding factor 2C (MsDREB2C) from Malus sieversii Roem. Plant Cell Physiol. 2013, 54, 1415–1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar]
- Delk, N.A.; Johnson, K.A.; Chowdhury, N.I.; Braam, J. CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 2005, 139, 240–253. [Google Scholar] [CrossRef] [Green Version]
- Sreenivasulu, N.; Harshavardhan, V.T.; Govind, G.; Seiler, C.; Kohli, A. Contrapuntal role of ABA: Does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 2012, 506, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Zang, X.; Li, H.; Liu, Z.; Zhao, A.; Liu, J.; Peng, H.; Yao, Y.; Hu, Z.; Ni, Z. Unconventional splicing of wheat TabZIP60 confers heat tolerance in transgenic Arabidopsis. Plant Sci. 2018, 274, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Brinker, M.; Brosché, M.; Vinocur, B.; Abo-Ogiala, A.; Fayyaz, P.; Janz, D.; Ottow, E.A.; Cullmann, A.D.; Saborowski, J.; Kangasjärvi, J.; et al. Linking the salt transcriptome with physiological responses of a salt-resistant Populus species as a strategy to identify genes important for stress acclimation. Plant Physiol. 2010, 154, 1697–1709. [Google Scholar] [CrossRef] [Green Version]
- Koops, P.; Pelser, S.; Ignatz, M.; Klose, C.; Marrocco-Selden, K.; Kretsch, T. EDL3 is an F-box protein involved in the regulation of abscisic acid signal in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 5547–5560. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.A.; Plattner, S.; Radakovic, Z.; Wieczorek, K.; Elashry, A.; Grundler, F.M.; Ammelburg, M.; Siddique, S.; Bohlmann, H. An Arabidopsis ATPase gene involved in nematode-induced syncytium development and abiotic stress responses. Plant J. 2013, 74, 852–866. [Google Scholar] [CrossRef] [Green Version]
- Hsu, K.H.; Liu, C.C.; Wu, S.J.; Ku, O.Y.Y.; Lu, C.A.; Wu, C.R.; Lian, P.J.; Hong, C.Y.; Ke, Y.T.; Huang, J.H. Expression of a gene encoding a rice RING zinc-finger protein, OsRZFP34, enhances stomata opening. Plant Mol. Biol. 2014, 86, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, Y.; Kieffer, M.; Yu, H.; Kepinski, S.; Estelle, M. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat. Commun. 2016, 7, 10269. [Google Scholar] [CrossRef]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Kang, L.; Li, Y.; Wu, C.; Zheng, C.; Liu, P.; Huang, J. RING finger protein RGLG1 and RGLG2 negatively modulate MAPKKK18 mediated drought stress tolerance in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 484–493. [Google Scholar] [CrossRef]
- Li, K.; Yang, F.; Zhang, G.; Song, S.; Li, Y.; Ren, D.; Miao, Y.; Song, C.P. AIK1, a mitogen-activated protein kinase, modulates abscisic acid responses through the MKK5-MPK6 kinase cascade. Plant Physiol. 2017, 173, 1391–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miles, G.P.; Samuel, M.A.; Ellis, B.E. Suppression of MKK5 reduces ozone-induced signal transmission to both MPK3 and MPK6 and confers increased ozone sensitivity in Arabidopsis thaliana. Plant Signal. Behav. 2009, 4, 687–692. [Google Scholar] [CrossRef] [Green Version]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny. fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M.; Oda, K. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 2008, 227, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.L.; Fu, X.M.; Liu, J.Q.; Ye, T.T.; Hou, S.Y.; Huang, Y.Q.; Yuan, B.F.; Wu, Y.; Feng, Y.Q. Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2012, 905, 67–74. [Google Scholar] [CrossRef]
- Ding, H.; Yuan, G.; Mo, S.; Qian, Y.; Wu, Y.; Chen, Q.; Xu, X.X.; Wu, X.X.; Ge, C. Genome-wide analysis of the plant-specific VQ motif-containing proteins in tomato (Solanum lycopersicum) and characterization of SlVQ6 in thermotolerance. Plant Physiol. Biochem. 2019, 143, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]


| GeneID | Fold Change | padj | Symbol | Description |
|---|---|---|---|---|
| AT3G09940 ** | −1.96 | 0.000 | MDAR3 | Monodehydroascorbate reductase 3 |
| AT4G21830 *,** | −1.92 | 0.000 | MSRB7 | Methionine sulfoxide reductase B7 |
| AT1G02920 | −1.55 | 0.005 | GSTF7 | Glutathione S-transferase 7 |
| AT1G53903 * | −1.47 | 0.015 | Linoleate 9S-lipoxygenase-4 protein | |
| AT1G53885 * | −1.47 | 0.015 | Linoleate 9S-lipoxygenase-4 protein | |
| AT3G50770 * | −1.39 | 0.010 | CML41 | Calmodulin-like 41 |
| AT3G44006 * | −1.37 | 0.038 | unknown protein | |
| AT3G15450 *,** | −1.36 | 0.000 | Aluminum-induced protein | |
| AT5G52300 *,** | −1.36 | 0.029 | RD29B | Low-temperature 65 |
| AT1G24793 * | −1.35 | 0.019 | AtLpxC1 | lipid X C1 |
| AT1G64110 ** | −1.34 | 0.010 | DAA1 | DUO1-activated ATPase 1 |
| AT2G43820 ** | −1.34 | 0.000 | SAGT1 | UDP-glucosyltransferase 74F2 |
| AT4G12530 *,** | −1.32 | 0.012 | AZI7 | Bifunctional inhibitor |
| AT4G27410 *,** | −1.24 | 0.047 | RD26 | Responsive to desiccation 26 |
| AT3G44860 *,** | −1.22 | 0.001 | FAMT | Farnesoic acid carboxyl-O-methyltransferase |
| AT5G59220 *,** | −1.21 | 0.000 | SAG113 | Highly ABA-induced PP2C gene 1/HAI1 |
| AT3G63060 *,** | −1.21 | 0.020 | EDL3 | EID1-like 3 |
| AT5G02020 *,** | −1.19 | 0.000 | SIS | Salt-Induced Serine-rich |
| AT3G48360 *,** | −1.18 | 0.039 | ATBT2 | BTB and TAZ domain protein 2 |
| AT1G05100 *,** | −1.15 | 0.021 | MAPKKK18 | MAP kinase kinase kinase 18 |
| AT1G23390 *,** | −1.12 | 0.000 | KFB | A kelch domain-containing F-box protein |
| AT3G21220 | −1.11 | 0.000 | MKK5 | MAP kinase kinase 5 |
| AT5G15960 *,** | −1.10 | 0.001 | KIN1 | Stress-responsive protein (KIN1) |
| AT5G15500 *,** | −1.07 | 0.000 | Ankyrin repeat family protein | |
| AT5G67480 *,** | −1.07 | 0.001 | ATBT4 | BTB and TAZ domain protein 4 |
| AT3G27250 *,** | −1.06 | 0.009 | DIL4 | ABA-induced transcription repressor |
| AT5G22920 *,** | −1.01 | 0.004 | AtRZPF34 | RING-type Zinc finger protein |
| AT1G24260 * | −1.00 | 0.008 | SEP3 | MADs box transcription factor |
| AT3G29030 *,** | 1.01 | 0.000 | ATEXP5 | Expansin A5 |
| AT3G56360 * | 1.02 | 0.000 | Hypothetical protein | |
| AT1G35260 * | 1.04 | 0.033 | MLP165 | MLP-like protein 165 |
| AT3G27650 * | 1.13 | 0.024 | LBD25 | LOB domain-containing protein 25 |
| AT2G43870 * | 1.21 | 0.001 | Pectin lyase-like superfamily protein | |
| AT2G41510 * | 1.21 | 0.010 | ATCKX1 | Cytokinin oxidase |
| AT2G38530 * | 1.24 | 0.000 | ATLTPI-5 | Lipid transfer protein 2 |
| AT1G13740 *,** | 1.26 | 0.001 | AFP2 | ABI five binding protein 2 |
| AT1G44800 *,** | 1.30 | 0.000 | SIAR1 | Siliques Are Red 1 |
| AT2G32990 *,** | 1.30 | 0.000 | GH9B8 | Glycosyl hydrolase 9B8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, H.; Qian, Y.; Fang, Y.; Ji, Y.; Sheng, J.; Ge, C. Characteristics of SlCML39, a Tomato Calmodulin-like Gene, and Its Negative Role in High Temperature Tolerance of Arabidopsis thaliana during Germination and Seedling Growth. Int. J. Mol. Sci. 2021, 22, 11479. https://doi.org/10.3390/ijms222111479
Ding H, Qian Y, Fang Y, Ji Y, Sheng J, Ge C. Characteristics of SlCML39, a Tomato Calmodulin-like Gene, and Its Negative Role in High Temperature Tolerance of Arabidopsis thaliana during Germination and Seedling Growth. International Journal of Molecular Sciences. 2021; 22(21):11479. https://doi.org/10.3390/ijms222111479
Chicago/Turabian StyleDing, Haidong, Ying Qian, Yifang Fang, Yurong Ji, Jiarong Sheng, and Cailin Ge. 2021. "Characteristics of SlCML39, a Tomato Calmodulin-like Gene, and Its Negative Role in High Temperature Tolerance of Arabidopsis thaliana during Germination and Seedling Growth" International Journal of Molecular Sciences 22, no. 21: 11479. https://doi.org/10.3390/ijms222111479
APA StyleDing, H., Qian, Y., Fang, Y., Ji, Y., Sheng, J., & Ge, C. (2021). Characteristics of SlCML39, a Tomato Calmodulin-like Gene, and Its Negative Role in High Temperature Tolerance of Arabidopsis thaliana during Germination and Seedling Growth. International Journal of Molecular Sciences, 22(21), 11479. https://doi.org/10.3390/ijms222111479






