Mouse Models of CMML
Abstract
:1. Introduction
2. Pathogenesis and CMML Treatment
3. Genetic Models
3.1. Oncogenes
3.2. Epigenetic Regulators
3.3. Others Regulators of Cell Death (Bid etc.)
4. Patient Derived Xenograft (PDX) Models
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Arber, D.A.; Orazi, A. Update on the Pathologic Diagnosis of Chronic Myelomonocytic Leukemia. Mod. Pathol. 2019, 32, 732–740. [Google Scholar] [CrossRef]
- Phekoo, K.J.; Richards, M.A.; Moller, H.; Schey, S.A. South Thames Haematology Specialist Committee. The Incidence and Outcome of Myeloid Malignancies in 2,112 Adult Patients in Southeast England. Haematologica 2006, 91, 1400–1404. [Google Scholar] [PubMed]
- Rollison, D.E.; Howlader, N.; Smith, M.T.; Strom, S.S.; Merritt, W.D.; Ries, L.A.; Edwards, B.K.; List, A.F. Epidemiology of Myelodysplastic Syndromes and Chronic Myeloproliferative Disorders in the United States, 2001–2004, Using Data from the NAACCR and SEER Programs. Blood 2008, 112, 45–52. [Google Scholar] [CrossRef]
- Benzarti, S.; Daskalakis, M.; Feller, A.; Bacher, V.U.; Schnegg-Kaufmann, A.; Rüfer, A.; Holbro, A.; Schmidt, A.; Benz, R.; Solenthaler, M. Trends of Incidence and Survival of Patients with Chronic Myelomonocytic Leukemia between 1999 and 2014: A Comparison between Swiss and American Population-Based Cancer Registries. Cancer Epidemiol. 2019, 59, 51–57. [Google Scholar] [CrossRef]
- Maynadié, M.; Girodon, F.; Manivet-Janoray, I.; Mounier, M.; Mugneret, F.; Bailly, F.; Favre, B.; Caillot, D.; Petrella, T.; Flesch, M. Twenty-Five Years of Epidemiological Recording on Myeloid Malignancies: Data from the Specialized Registry of Hematologic Malignancies of Cote d’Or (Burgundy, France). Haematologica 2011, 96, 55–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guru Murthy, G.S.; Dhakal, I.; Mehta, P. Incidence and Survival Outcomes of Chronic Myelomonocytic Leukemia in the United States. Leuk. Lymphoma 2017, 58, 1648–1654. [Google Scholar] [CrossRef] [PubMed]
- Such, E.; Cervera, J.; Costa, D.; Solé, F.; Vallespí, T.; Luño, E.; Collado, R.; Calasanz, M.J.; Hernández-Rivas, J.M.; Cigudosa, J.C. Cytogenetic Risk Stratification in Chronic Myelomonocytic Leukemia. Haematologica 2011, 96, 375–383. [Google Scholar] [CrossRef] [Green Version]
- Germing, U.; Kündgen, A.; Gattermann, N. Risk Assessment in Chronic Myelomonocytic Leukemia (CMML). Leuk. Lymphoma 2004, 45, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Onida, F.; Kantarjian, H.M.; Smith, T.L.; Ball, G.; Keating, M.J.; Estey, E.H.; Glassman, A.B.; Albitar, M.; Kwari, M.I.; Beran, M. Prognostic Factors and Scoring Systems in Chronic Myelomonocytic Leukemia: A Retrospective Analysis of 213 Patients. Blood 2002, 99, 840–849. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, M.M.; Wassie, E.A.; Lasho, T.L.; Hanson, C.A.; Ketterling, R.; Tefferi, A. Blast Transformation in Chronic Myelomonocytic Leukemia: Risk Factors, Genetic Features, Survival, and Treatment Outcome. Am. J. Hematol. 2015, 90, 411–416. [Google Scholar] [CrossRef]
- Bennett, J.M.; Catovsky, D.; Daniel, M.T.; Flandrin, G.; Galton, D.A.G.; Gralnick, H.; Sultan, C.; Cox, C. The Chronic Myeloid Leukaemias: Guidelines for Distinguishing Chronic Granulocytic, Atypical Chronic Myeloid, and Chronic Myelomonocytic Leukaemia: Proposals by the French-American-British Cooperative Leukaemia Group. Br. J. Haematol. 1994, 87, 746–754. [Google Scholar] [CrossRef]
- Vardiman, J.W.; Harris, N.L.; Brunning, R.D. The World Health Organization (WHO) Classification of the Myeloid Neoplasms. Blood 2002, 100, 2292–2302. [Google Scholar] [CrossRef]
- Valent, P.; Orazi, A.; Savona, M.R.; Patnaik, M.M.; Onida, F.; van de Loosdrecht, A.A.; Haase, D.; Haferlach, T.; Elena, C.; Pleyer, L. Proposed Diagnostic Criteria for Classical Chronic Myelomonocytic Leukemia (CMML), CMML Variants and Pre-CMML Conditions. Haematologica 2019, 104, 1935–1949. [Google Scholar] [CrossRef]
- Vardiman, J.W.; Thiele, J.; Arber, D.A.; Brunning, R.D.; Borowitz, M.J.; Porwit, A.; Harris, N.L.; Le Beau, M.M.; Hellström-Lindberg, E.; Tefferi, A. The 2008 Revision of the World Health Organization (WHO) Classification of Myeloid Neoplasms and Acute Leukemia: Rationale and Important Changes. Blood 2009, 114, 937–951. [Google Scholar] [CrossRef] [Green Version]
- Sangiorgio, V.F.I.; Arber, D.A.; Orazi, A. How I Investigate Chronic Myelomonocytic Leukemia. Int. J. Lab. Hematol. 2020, 42, 101–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patnaik, M.M.; Lasho, T.L.; Finke, C.; Howard, M.T.; Hanson, C.A.; Ketterling, R.P.; Gangat, N.; Tefferi, A. “Proliferative” versus “Dysplastic” Chronic Myelomonocytic Leukemia: Molecular and Prognostic Correlates. Blood 2016, 128, 1987. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Parikh, S.A.; Hanson, C.A.; Tefferi, A. Chronic Myelomonocytic Leukaemia: A Concise Clinical and Pathophysiological Review. Br. J. Haematol. 2014, 165, 273–286. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Wassie, E.A.; Itzykson, R.; Lasho, T.L.; Kosmider, O.; Finke, C.M.; Hanson, C.A.; Ketterling, R.P.; Solary, E.; Tefferi, A.; Patnaik, M.M. Molecular and Prognostic Correlates of Cytogenetic Abnormalities in Chronic Myelomonocytic Leukemia: A Mayo Clinic-French Consortium Study. Am. J. Hematol. 2014, 89, 1111–1115. [Google Scholar] [CrossRef]
- Tang, G.; Zhang, L.; Fu, B.; Hu, J.; Lu, X.; Hu, S.; Patel, A.; Goswami, M.; Khoury, J.D.; Garcia-Manero, G. Cytogenetic Risk Stratification of 417 Patients with Chronic Myelomonocytic Leukemia from a Single Institution. Am. J. Hematol. 2014, 89, 813–818. [Google Scholar] [CrossRef] [Green Version]
- Patnaik, M.M.; Tefferi, A. Cytogenetic and Molecular Abnormalities in Chronic Myelomonocytic Leukemia. Blood Cancer J. 2016, 6, e393. [Google Scholar] [CrossRef] [PubMed]
- Itzykson, R.; Kosmider, O.; Renneville, A.; Gelsi-Boyer, V.; Meggendorfer, M.; Morabito, M.; Berthon, C.; Adès, L.; Fenaux, P.; Beyne-Rauzy, O. Prognostic Score Including Gene Mutations in Chronic Myelomonocytic Leukemia. J. Clin. Oncol. 2013, 31, 2428–2436. [Google Scholar] [CrossRef]
- Jankowska, A.M.; Makishima, H.; Tiu, R.V.; Szpurka, H.; Huang, Y.; Traina, F.; Visconte, V.; Sugimoto, Y.; Prince, C.; O’Keefe, C. Mutational Spectrum Analysis of Chronic Myelomonocytic Leukemia Includes Genes Associated with Epigenetic Regulation: UTX, EZH2, and DNMT3A. Blood 2011, 118, 3932–3941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patnaik, M.M.; Zahid, M.F.; Lasho, T.L.; Finke, C.; Ketterling, R.L.; Gangat, N.; Robertson, K.D.; Hanson, C.A.; Tefferi, A. Number and Type of TET2 Mutations in Chronic Myelomonocytic Leukemia and Their Clinical Relevance. Blood Cancer J. 2016, 6, e472. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, V.; Kohlmann, A.; Eder, C.; Haferlach, C.; Kern, W.; Cross, N.C.P.; Haferlach, T.; Schnittger, S. Molecular Profiling of Chronic Myelomonocytic Leukemia Reveals Diverse Mutations in >80% of Patients with TET2 and EZH2 Being of High Prognostic Relevance. Leukemia 2011, 25, 877–879. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Lasho, T.L.; Finke, C.M.; Hanson, C.A.; Hodnefield, J.M.; Knudson, R.A.; Ketterling, R.P.; Pardanani, A.; Tefferi, A. Spliceosome Mutations Involving SRSF2, SF3B1, and U2AF35 in Chronic Myelomonocytic Leukemia: Prevalence, Clinical Correlates, and Prognostic Relevance. Am. J. Hematol. 2013, 88, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Meggendorfer, M.; Roller, A.; Haferlach, T.; Eder, C.; Dicker, F.; Grossmann, V.; Kohlmann, A.; Alpermann, T.; Yoshida, K.; Ogawa, S. SRSF2 Mutations in 275 Cases with Chronic Myelomonocytic Leukemia (CMML). Blood 2012, 120, 3080–3088. [Google Scholar] [CrossRef]
- Kohlmann, A.; Grossmann, V.; Klein, H.-U.; Schindela, S.; Weiss, T.; Kazak, B.; Dicker, F.; Schnittger, S.; Dugas, M.; Kern, W. Next-Generation Sequencing Technology Reveals a Characteristic Pattern of Molecular Mutations in 72.8% of Chronic Myelomonocytic Leukemia by Detecting Frequent Alterations in TET2, CBL, RAS, and RUNX1. J. Clin. Oncol. 2010, 28, 3858–3865. [Google Scholar] [CrossRef] [PubMed]
- Daver, N.; Strati, P.; Jabbour, E.; Kadia, T.; Luthra, R.; Wang, S.; Patel, K.; Ravandi, F.; Cortes, J.; Qin Dong, X. FLT3 Mutations in Myelodysplastic Syndrome and Chronic Myelomonocytic Leukemia. Am. J. Hematol. 2013, 88, 56–59. [Google Scholar] [CrossRef] [Green Version]
- Laborde, R.R.; Patnaik, M.M.; Lasho, T.L.; Finke, C.M.; Hanson, C.A.; Knudson, R.A.; Ketterling, R.P.; Pardanani, A.; Tefferi, A. SETBP1 Mutations in 415 Patients with Primary Myelofibrosis or Chronic Myelomonocytic Leukemia: Independent Prognostic Impact in CMML. Leukemia 2013, 27, 2100–2102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itzykson, R.; Solary, E. An Evolutionary Perspective on Chronic Myelomonocytic Leukemia. Leukemia 2013, 27, 1441–1450. [Google Scholar] [CrossRef]
- Duchmann, M.; Yalniz, F.F.; Sanna, A.; Sallman, D.; Coombs, C.C.; Renneville, A.; Kosmider, O.; Braun, T.; Platzbecker, U.; Willems, L. Prognostic Role of Gene Mutations in Chronic Myelomonocytic Leukemia Patients Treated with Hypomethylating Agents. EBioMedicine 2018, 31, 174–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, C.C.; Khorashad, J.S.; Tantravahi, S.K.; Kelley, T.W.; Zabriskie, M.S.; Yan, D.; Pomicter, A.D.; Reynolds, K.R.; Eiring, A.M.; Kronenberg, Z. Age-Related Mutations and Chronic Myelomonocytic Leukemia. Leukemia 2016, 30, 906–913. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Tong, H.; Du, X.; Li, B.; Gale, R.P.; Qin, T.; Liu, J.; Xu, Z.; Zhang, Y.; Huang, G. Impact of TET2, SRSF2, ASXL1 and SETBP1 Mutations on Survival of Patients with Chronic Myelomonocytic Leukemia. Exp. Hematol. Oncol. 2015, 4, 14. [Google Scholar] [CrossRef] [Green Version]
- Patnaik, M.M.; Itzykson, R.; Lasho, T.L.; Kosmider, O.; Finke, C.M.; Hanson, C.A.; Knudson, R.A.; Ketterling, R.P.; Tefferi, A.; Solary, E. ASXL1 and SETBP1 Mutations and Their Prognostic Contribution in Chronic Myelomonocytic Leukemia: A Two-Center Study of 466 Patients. Leukemia 2014, 28, 2206–2212. [Google Scholar] [CrossRef] [PubMed]
- Sallman, D.A.; Komrokji, R.; Cluzeau, T.; Vaupel, C.; Al Ali, N.H.; Lancet, J.; Hall, J.; List, A.; Padron, E.; Song, J. ASXL1 Frameshift Mutations Drive Inferior Outcomes in CMML without Negative Impact in MDS. Blood Cancer J. 2017, 7, 633. [Google Scholar] [CrossRef] [Green Version]
- Gelsi-Boyer, V.; Trouplin, V.; Roquain, J.; Adélaïde, J.; Carbuccia, N.; Esterni, B.; Finetti, P.; Murati, A.; Arnoulet, C.; Zerazhi, H. ASXL1 Mutation Is Associated with Poor Prognosis and Acute Transformation in Chronic Myelomonocytic Leukaemia. Br. J. Haematol. 2010, 151, 365–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClure, R.F.; Ewalt, M.D.; Crow, J.; Temple-Smolkin, R.L.; Pullambhatla, M.; Sargent, R.; Kim, A.S. Clinical Significance of DNA Variants in Chronic Myeloid Neoplasms: A Report of the Association for Molecular Pathology. J. Mol. Diagn. 2018, 20, 717–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patnaik, M.M.; Wassie, E.A.; Padron, E.; Onida, F.; Itzykson, R.; Lasho, T.L.; Kosmider, O.; Finke, C.M.; Hanson, C.A.; Ketterling, R.P. Chronic Myelomonocytic Leukemia in Younger Patients: Molecular and Cytogenetic Predictors of Survival and Treatment Outcome. Blood Cancer J. 2015, 5, e270. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Itzykson, R.; Renneville, A.; de Renzis, B.; Dreyfus, F.; Laribi, K.; Bouabdallah, K.; Vey, N.; Toma, A.; Recher, C. Molecular Predictors of Response to Decitabine in Advanced Chronic Myelomonocytic Leukemia: A Phase 2 Trial. Blood 2011, 118, 3824–3831. [Google Scholar] [CrossRef] [Green Version]
- Meldi, K.; Qin, T.; Buchi, F.; Droin, N.; Sotzen, J.; Micol, J.-B.; Selimoglu-Buet, D.; Masala, E.; Allione, B.; Gioia, D. Specific Molecular Signatures Predict Decitabine Response in Chronic Myelomonocytic Leukemia. J. Clin. Investig. 2015, 125, 1857–1872. [Google Scholar] [CrossRef] [PubMed]
- Kosmider, O.; Gelsi-Boyer, V.; Ciudad, M.; Racoeur, C.; Jooste, V.; Vey, N.; Quesnel, B.; Fenaux, P.; Bastie, J.-N.; Beyne-Rauzy, O. TET2 Gene Mutation Is a Frequent and Adverse Event in Chronic Myelomonocytic Leukemia. Haematologica 2009, 94, 1676–1681. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.E.; Mohamedali, A.M.; Kulasekararaj, A.; Lim, Z.; Gäken, J.; Lea, N.C.; Przychodzen, B.; Mian, S.A.; Nasser, E.E.; Shooter, C. Next-Generation Sequencing of the TET2 Gene in 355 MDS and CMML Patients Reveals Low-Abundance Mutant Clones with Early Origins, but Indicates No Definite Prognostic Value. Blood 2010, 116, 3923–3932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoi, K.; Cross, N.C.P. Molecular Pathogenesis of Atypical CML, CMML and MDS/MPN-Unclassifiable. Int. J. Hematol. 2015, 101, 229–242. [Google Scholar] [CrossRef] [Green Version]
- Bejar, R.; Lord, A.; Stevenson, K.; Bar-Natan, M.; Pérez-Ladaga, A.; Zaneveld, J.; Wang, H.; Caughey, B.; Stojanov, P.; Getz, G. TET2 Mutations Predict Response to Hypomethylating Agents in Myelodysplastic Syndrome Patients. Blood 2014, 124, 2705–2712. [Google Scholar] [CrossRef] [PubMed]
- Arbab Jafari, P.; Ayatollahi, H.; Sadeghi, R.; Sheikhi, M.; Asghari, A. Prognostic Significance of SRSF2 Mutations in Myelodysplastic Syndromes and Chronic Myelomonocytic Leukemia: A Meta-Analysis. Hematology 2018, 23, 778–784. [Google Scholar] [CrossRef] [Green Version]
- Kuo, M.-C.; Liang, D.-C.; Huang, C.-F.; Shih, Y.-S.; Wu, J.-H.; Lin, T.-L.; Shih, L.-Y. RUNX1 Mutations Are Frequent in Chronic Myelomonocytic Leukemia and Mutations at the C-Terminal Region Might Predict Acute Myeloid Leukemia Transformation. Leukemia 2009, 23, 1426–1431. [Google Scholar] [CrossRef]
- Carr, R.M.; Vorobyev, D.; Lasho, T.; Marks, D.L.; Tolosa, E.J.; Vedder, A.; Almada, L.L.; Yurcheko, A.; Padioleau, I.; Alver, B. RAS Mutations Drive Proliferative Chronic Myelomonocytic Leukemia via a KMT2A-PLK1 Axis. Nat. Commun. 2021, 12, 2901. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, M.M.; Tefferi, A. Chronic Myelomonocytic Leukemia: 2020 Update on Diagnosis, Risk Stratification and Management. Am. J. Hematol. 2020, 95, 97–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patnaik, M.M.; Barraco, D.; Lasho, T.L.; Finke, C.M.; Hanson, C.A.; Ketterling, R.P.; Gangat, N.; Tefferi, A. DNMT3A Mutations Are Associated with Inferior Overall and Leukemia-Free Survival in Chronic Myelomonocytic Leukemia. Am. J. Hematol. 2017, 92, 56–61. [Google Scholar] [CrossRef] [Green Version]
- Shou, L.-H.; Cao, D.; Dong, X.-H.; Fang, Q.; Wu, Y.; Zhang, Y.; Fei, J.-P.; Xu, B.-L. Prognostic Significance of SETBP1 Mutations in Myelodysplastic Syndromes, Chronic Myelomonocytic Leukemia, and Chronic Neutrophilic Leukemia: A Meta-Analysis. PLoS ONE 2017, 12, e0171608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damm, F.; Itzykson, R.; Kosmider, O.; Droin, N.; Renneville, A.; Chesnais, V.; Gelsi-Boyer, V.; de Botton, S.; Vey, N.; Preudhomme, C. SETBP1 Mutations in 658 Patients with Myelodysplastic Syndromes, Chronic Myelomonocytic Leukemia and Secondary Acute Myeloid Leukemias. Leukemia 2013, 27, 1401–1403. [Google Scholar] [CrossRef] [PubMed]
- Wudhikarn, K.; Loghavi, S.; Mangaonkar, A.A.; Al-Kali, A.; Binder, M.; Carr, R.; Reichard, K.; Finke, C.; Howard, M.; Gangat, N. SF3B1-Mutant CMML Defines a Predominantly Dysplastic CMML Subtype with a Superior Acute Leukemia-Free Survival. Blood Adv. 2020, 4, 5716–5721. [Google Scholar] [PubMed]
- Merlevede, J.; Droin, N.; Qin, T.; Meldi, K.; Yoshida, K.; Morabito, M.; Chautard, E.; Auboeuf, D.; Fenaux, P.; Braun, T. Mutation Allele Burden Remains Unchanged in Chronic Myelomonocytic Leukaemia Responding to Hypomethylating Agents. Nat. Commun. 2016, 7, 10767. [Google Scholar] [CrossRef] [Green Version]
- Patel, B.J.; Przychodzen, B.; Thota, S.; Radivoyevitch, T.; Visconte, V.; Kuzmanovic, T.; Clemente, M.; Hirsch, C.; Morawski, A.; Souaid, R. Genomic Determinants of Chronic Myelomonocytic Leukemia. Leukemia 2017, 31, 2815–2823. [Google Scholar] [CrossRef] [PubMed]
- Malcovati, L.; Papaemmanuil, E.; Ambaglio, I.; Elena, C.; Gallì, A.; Della Porta, M.G.; Travaglino, E.; Pietra, D.; Pascutto, C.; Ubezio, M. Driver Somatic Mutations Identify Distinct Disease Entities within Myeloid Neoplasms with Myelodysplasia. Blood 2014, 124, 1513–1521. [Google Scholar] [CrossRef]
- Steensma, D.P.; Bejar, R.; Jaiswal, S.; Lindsley, R.C.; Sekeres, M.A.; Hasserjian, R.P.; Ebert, B.L. Clonal Hematopoiesis of Indeterminate Potential and Its Distinction from Myelodysplastic Syndromes. Blood 2015, 126, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Mangaonkar, A.A.; Patnaik, M.M. Advances in Chronic Myelomonocytic Leukemia and Future Prospects: Lessons Learned from Precision Genomics. Adv. Cell Gene Ther. 2019, 2, e48. [Google Scholar] [CrossRef] [PubMed]
- Cervera, N.; Itzykson, R.; Coppin, E.; Prebet, T.; Murati, A.; Legall, S.; Vey, N.; Solary, E.; Birnbaum, D.; Gelsi-Boyer, V. Gene Mutations Differently Impact the Prognosis of the Myelodysplastic and Myeloproliferative Classes of Chronic Myelomonocytic Leukemia. Am. J. Hematol. 2014, 89, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Itzykson, R.; Fenaux, P.; Bowen, D.; Cross, N.C.P.; Cortes, J.; De Witte, T.; Germing, U.; Onida, F.; Padron, E.; Platzbecker, U. Diagnosis and Treatment of Chronic Myelomonocytic Leukemias in Adults: Recommendations from the European Hematology Association and the European LeukemiaNet. HemaSphere 2018, 2, e150. [Google Scholar] [CrossRef] [PubMed]
- Elmariah, H.; DeZern, A.E. Chronic Myelomonocytic Leukemia: 2018 Update to Prognosis and Treatment. Curr. Hematol. Malig. Rep. 2019, 14, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Eissa, H.; Gooley, T.A.; Sorror, M.L.; Nguyen, F.; Scott, B.L.; Doney, K.; Loeb, K.R.; Martin, P.J.; Pagel, J.M.; Radich, J.P. Allogeneic Hematopoietic Cell Transplantation for Chronic Myelomonocytic Leukemia: Relapse-Free Survival Is Determined by Karyotype and Comorbidities. Biol. Blood Marrow Transplant. 2011, 17, 908–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Witte, T.; Bowen, D.; Robin, M.; Malcovati, L.; Niederwieser, D.; Yakoub-Agha, I.; Mufti, G.J.; Fenaux, P.; Sanz, G.; Martino, R. Allogeneic Hematopoietic Stem Cell Transplantation for MDS and CMML: Recommendations from an International Expert Panel. Blood 2017, 129, 1753–1762. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, M.M.; Tefferi, A. Chronic Myelomonocytic Leukemia: 2016 Update on Diagnosis, Risk Stratification, and Management. Am. J. Hematol. 2016, 91, 631–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfonso, A.; Montalban-Bravo, G.; Takahashi, K.; Jabbour, E.J.; Kadia, T.; Ravandi, F.; Cortes, J.; Estrov, Z.; Borthakur, G.; Pemmaraju, N. Natural History of Chronic Myelomonocytic Leukemia Treated with Hypomethylating Agents. Am. J. Hematol. 2017, 92, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Moyo, T.K.; Savona, M.R. Therapy for Chronic Myelomonocytic Leukemia in a New Era. Curr. Hematol. Malig. Rep. 2017, 12, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Padron, E.; DeZern, A.E.; Niyongere, S.; Ball, M.C.; Balasis, M.; Ramadan, H.; Lancet, J.E.; List, A.F.; Mesa, R.A.; Roboz, G.J. Promising Results of a Phase 1/2 Clinical Trial of Ruxolitinib in Patients with Chronic Myelomonocytic Leukemia. Blood 2017, 130, 162. [Google Scholar]
- Fenaux, P.; Raza, A.; Mufti, G.J.; Aul, C.; Germing, U.; Kantarjian, H.; Cripe, L.; Kerstens, R.; De Porre, P.; Kurzrock, R. A Multicenter Phase 2 Study of the Farnesyltransferase Inhibitor Tipifarnib in Intermediate- to High-Risk Myelodysplastic Syndrome. Blood 2007, 109, 4158–4163. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Tefferi, A. Chronic Myelomonocytic Leukemia: 2018 Update on Diagnosis, Risk Stratification and Management. Am. J. Hematol. 2018, 93, 824–840. [Google Scholar] [CrossRef]
- Burgstaller, S.; Stauder, R.; Kuehr, T.; Lang, A.; Machherndl-Spandl, S.; Mayrbaeurl, B.; Noesslinger, T.; Petzer, A.; Valent, P.; Greil, R. A Phase I Study of Lenalidomide in Patients with Chronic Myelomonocytic Leukemia (CMML)—AGMT_CMML-1. Leuk. Lymphoma 2018, 59, 1121–1126. [Google Scholar] [CrossRef]
- Komrokji, R.; Garcia-Manero, G.; Ades, L.; Prebet, T.; Steensma, D.P.; Jurcic, J.G.; Sekeres, M.A.; Berdeja, J.; Savona, M.R.; Beyne-Rauzy, O. Sotatercept with Long-Term Extension for the Treatment of Anaemia in Patients with Lower-Risk Myelodysplastic Syndromes: A Phase 2, Dose-Ranging Trial. Lancet Haematol. 2018, 5, e63–e72. [Google Scholar] [CrossRef]
- Komrokji, R.S.; Garcia-Manero, G.; Ades, L.; Laadem, A.; Vo, B.; Prebet, T.; Stamatoullas, A.; Boyd, T.; Delaunay, J.; Steensma, D.P. An Open-Label, Phase 2, Dose-Finding Study of Sotatercept (ACE-011) in Patients with Low or Intermediate-1 (Int-1)-Risk Myelodysplastic Syndromes (MDS) or Non-Proliferative Chronic Myelomonocytic Leukemia (CMML) and Anemia Requiring Transfusion. Blood 2014, 124, 3251. [Google Scholar] [CrossRef]
- Bagley, C.J.; Woodcock, J.M.; Stomski, F.C.; Lopez, A.F. The Structural and Functional Basis of Cytokine Receptor Activation: Lessons from the Common β Subunit of the Granulocyte-Macrophage Colony-Stimulating Factor, Interleukin-3 (IL-3), and IL-5 Receptors. Blood 1997, 89, 1471–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, S.E.; Gasson, J.C. Characterization of the Role of the Human Granulocyte-Macrophage Colony-Stimulating Factor Receptor α Subunit in the Activation of JAK2 and STAT5. Blood 1998, 92, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Largaespada, D.A.; Brannan, C.I.; Jenkins, N.A.; Copeland, N.G. Nf1 Deficiency Causes Ras-Dediated Granulocyte/Macrophage Colony Stimulating Factor Hypersensitivity and Chronic Myeloid Leukaemia. Nat. Genet. 1996, 12, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, R.A.; O’Marcaigh, A.; Wardak, Z.; Zhang, Y.-Y.; Dranoff, G.; Jacks, T.; Clapp, D.W.; Shannon, K.M. Nf1 and Gmcsf Interact in Myeloid Leukemogenesis. Mol. Cell 2000, 5, 189–195. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Y.; Li, Z.; Du, J.; Ryu, M.-J.; Taylor, P.R.; Fleming, M.D.; Young, K.H.; Pitot, H.; Zhang, J. Endogenous Oncogenic Nras Mutation Promotes Aberrant GM-CSF Signaling in Granulocytic/Monocytic Precursors in a Murine Model of Chronic Myelomonocytic Leukemia. Blood 2010, 116, 5991–6002. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Ranheim, E.A.; Du, J.; Liu, Y.; Wang, J.; Kong, G.; Zhang, J. Deficiency of β Common Receptor Moderately Attenuates the Progression of Myeloproliferative Neoplasm in NrasG12D/+ Mice. J. Biol. Chem. 2015, 290, 19093–19103. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Liu, Y.; Li, Z.; Wang, Z.; Tan, L.X.; Ryu, M.-J.; Meline, B.; Du, J.; Young, K.H.; Ranheim, E. Endogenous Oncogenic Nras Mutation Initiates Hematopoietic Malignancies in a Dose- and Cell Type-Dependent Manner. Blood 2011, 118, 368–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, I.T.; Kutok, J.L.; Williams, I.R.; Cohen, S.; Kelly, L.; Shigematsu, H.; Johnson, L.; Akashi, K.; Tuveson, D.A.; Jacks, T. Conditional Expression of Oncogenic K-Ras from Its Endogenous Promoter Induces a Myeloproliferative Disease. J. Clin. Investig. 2004, 113, 528–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.-I.; You, X.; Kong, G.; Ranheim, E.A.; Wang, J.; Du, J.; Liu, Y.; Zhou, Y.; Ryu, M.-J.; Zhang, J. Loss of Dnmt3a and Endogenous Kras(G12D/+) Cooperate to Regulate Hematopoietic Stem and Progenitor Cell Functions in Leukemogenesis. Leukemia 2015, 29, 1847–1856. [Google Scholar] [CrossRef] [Green Version]
- Melo-Cardenas, J.; Xu, Y.; Wei, J.; Tan, C.; Kong, S.; Gao, B.; Montauti, E.; Kirsammer, G.; Licht, J.D.; Yu, J. USP22 Deficiency Leads to Myeloid Leukemia upon Oncogenic Kras Activation through a PU.1-Dependent Mechanism. Blood 2018, 132, 423–434. [Google Scholar] [CrossRef] [Green Version]
- Emanuel, P.D. Juvenile Myelomonocytic Leukemia and Chronic Myelomonocytic Leukemia. Leukemia 2008, 22, 1335–1342. [Google Scholar] [CrossRef]
- Nakata, Y.; Ueda, T.; Nagamachi, A.; Yamasaki, N.; Ikeda, K.-I.; Sera, Y.; Takubo, K.; Kanai, A.; Oda, H.; Sanada, M. Acquired Expression of Cbl(Q367P) in Mice Induces Dysplastic Myelopoiesis Mimicking Chronic Myelomonocytic Leukemia. Blood 2017, 129, 2148–2160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, M.L.; Sakai, D.S.; Flotho, C.; Kang, M.; Fliegauf, M.; Archambeault, S.; Mullighan, C.G.; Chen, L.; Bergstraesser, E.; Bueso-Ramos, C.E. Mutations in CBL Occur Frequently in Juvenile Myelomonocytic Leukemia. Blood 2009, 114, 1859–1863. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Cai, X.; Cai, C.-L.; Wang, J.; Zhang, W.; Petersen, B.E.; Yang, F.-C.; Xu, M. Deletion of Tet2 in Mice Leads to Dysregulated Hematopoietic Stem Cells and Subsequent Development of Myeloid Malignancies. Blood 2011, 118, 4509–4518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selimoglu-Buet, D.; Rivière, J.; Ghamlouch, H.; Bencheikh, L.; Lacout, C.; Morabito, M.; Diop, M.; Meurice, G.; Breckler, M.; Chauveau, A. A MiR-150/TET3 Pathway Regulates the Generation of Mouse and Human Non-Classical Monocyte Subset. Nat. Commun. 2018, 9, 5455. [Google Scholar] [CrossRef] [Green Version]
- Bera, R.; Chiu, M.-C.; Huang, Y.-J.; Lin, T.-H.; Kuo, M.-C.; Shih, L.-Y. RUNX1 Mutations Promote Leukemogenesis of Myeloid Malignancies in ASXL1-Mutated Leukemia. J. Hematol. Oncol. 2019, 12, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Zheng, H.; Bao, N.; Jiang, S.; Bueso-Ramos, C.E.; Khoury, J.; Class, C.; Lu, Y.; Lin, K.; Yang, H. KDM6B Overexpression Activates Innate Immune Signaling and Impairs Hematopoiesis in Mice. Blood Adv. 2018, 2, 2491–2504. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Xu, L.; Xu, Q.; Yu, L.; Zhao, D.; Chen, P.; Wang, W.; Wang, Y.; Han, G.; Chen, C.D. Utx Loss Causes Myeloid Transformation. Leukemia 2018, 32, 1458–1465. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Harada, Y.; Kagiyama, Y.; Nishikawa, S.; Ding, Y.; Imagawa, J.; Shingai, N.; Kato, N.; Kitaura, J.; Hokaiwado, S. NUP98-HBO1-Fusion Generates Phenotypically and Genetically Relevant Chronic Myelomonocytic Leukemia Pathogenesis. Blood Adv. 2019, 3, 1047–1060. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.-Y.; Eldin, K.W.; Beaudet, A.L. Identification of Chromatin Remodeling Genes Arid4a and Arid4b as Leukemia Suppressor Genes. J. Natl. Cancer Inst. 2008, 100, 1247–1259. [Google Scholar] [CrossRef] [PubMed]
- Aucagne, R.; Droin, N.; Paggetti, J.; Lagrange, B.; Largeot, A.; Hammann, A.; Bataille, A.; Martin, L.; Yan, K.-P.; Fenaux, P. Transcription Intermediary Factor 1γ Is a Tumor Suppressor in Mouse and Human Chronic Myelomonocytic Leukemia. J. Clin. Investig. 2011, 121, 2361–2370. [Google Scholar] [CrossRef] [Green Version]
- Quéré, R.; Saint-Paul, L.; Carmignac, V.; Martin, R.Z.; Chrétien, M.-L.; Largeot, A.; Hammann, A.; Pais de Barros, J.-P.; Bastie, J.-N.; Delva, L. Tif1γ Regulates the TGF-Β1 Receptor and Promotes Physiological Aging of Hematopoietic Stem Cells. Proc. Natl. Acad. Sci. USA 2014, 111, 10592–10597. [Google Scholar] [CrossRef] [Green Version]
- Zinkel, S.S.; Ong, C.C.; Ferguson, D.O.; Iwasaki, H.; Akashi, K.; Bronson, R.T.; Kutok, J.L.; Alt, F.W.; Korsmeyer, S.J. Proapoptotic BID Is Required for Myeloid Homeostasis and Tumor Suppression. Genes Dev. 2003, 17, 229–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mambet, C.; Chivu-Economescu, M.; Matei, L.; Necula, L.G.; Dragu, D.L.; Bleotu, C.; Diaconu, C.C. Murine Models Based on Acute Myeloid Leukemia-Initiating Stem Cells Xenografting. World J. Stem Cells 2018, 10, 57–65. [Google Scholar] [CrossRef]
- Yoshimi, A.; Balasis, M.E.; Vedder, A.; Feldman, K.; Ma, Y.; Zhang, H.; Lee, S.C.W.; Letson, C.; Niyongere, S.; Lu, S.X. Robust Patient-Derived Xenografts of MDS/MPN Overlap Syndromes Capture the Unique Characteristics of CMML and JMML. Blood 2017, 130, 397–407. [Google Scholar] [CrossRef] [Green Version]
- Taoka, K.; Arai, S.; Kataoka, K.; Hosoi, M.; Miyauchi, M.; Yamazaki, S.; Honda, A.; Aixinjueluo, W.; Kobayashi, T.; Kumano, K. Using Patient-Derived IPSCs to Develop Humanized Mouse Models for Chronic Myelomonocytic Leukemia and Therapeutic Drug Identification, Including Liposomal Clodronate. Sci. Rep. 2018, 8, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Sevin, M.; Debeurme, F.; Laplane, L.; Badel, S.; Morabito, M.; Newman, H.L.; Torres-Martin, M.; Yang, Q.; Badaoui, B.; Wagner-Ballon, O. Cytokine-like Protein 1-Induced Survival of Monocytes Suggests a Combined Strategy Targeting MCL1 and MAPK in CMML. Blood 2021, 137, 3390–3402. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, L.; Selimoglu-Buet, D.; Jego, C.; Morabito, M.; Willekens, C.; Diop, M.K.; Gonin, P.; Lapierre, V.; Droin, N. Engraftment of Chronic Myelomonocytic Leukemia Cells in Immunocompromised Mice Supports Disease Dependency on Cytokines. Blood Adv. 2017, 1, 972–979. [Google Scholar] [CrossRef] [Green Version]
- Kloos, A.; Mintzas, K.; Winckler, L.; Gabdoulline, R.; Alwie, Y.; Jyotsana, N.; Kattre, N.; Schottmann, R.; Scherr, M.; Gupta, C. Effective Drug Treatment Identified by in Vivo Screening in a Transplantable Patient-Derived Xenograft Model of Chronic Myelomonocytic Leukemia. Leukemia 2020, 34, 2951–2963. [Google Scholar] [CrossRef] [PubMed]
- Hunter, A.M.; Newman, H.; DeZern, A.E.; Steensma, D.P.; Niyongere, S.; Roboz, G.J.; Mo, Q.; Chan, O.; Gerds, A.; Sallman, D.A. Integrated Human and Murine Clinical Study Establishes Clinical Efficacy of Ruxolitinib in Chronic Myelomonocytic Leukemia. Clin. Cancer Res. 2021. [Google Scholar] [CrossRef] [PubMed]
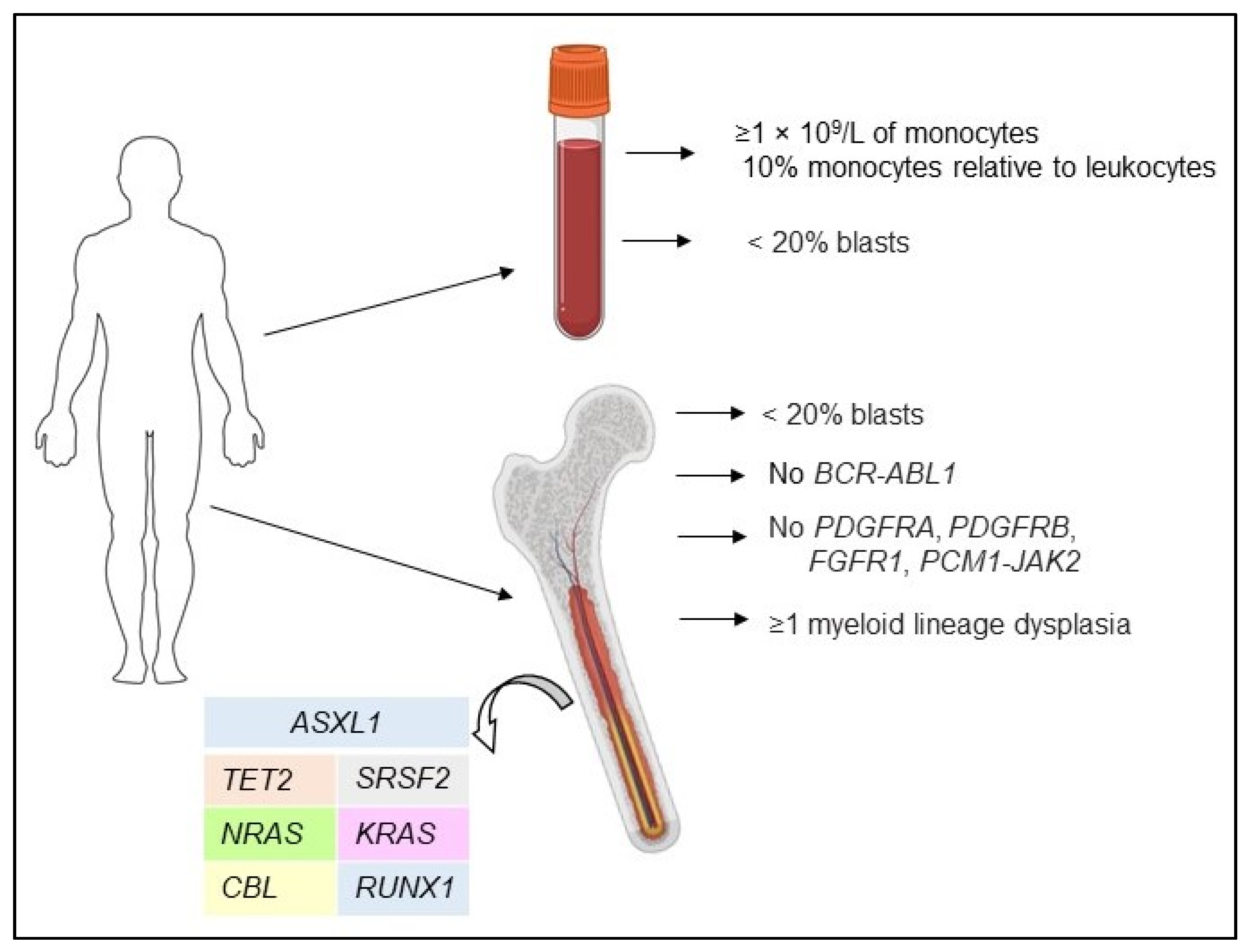
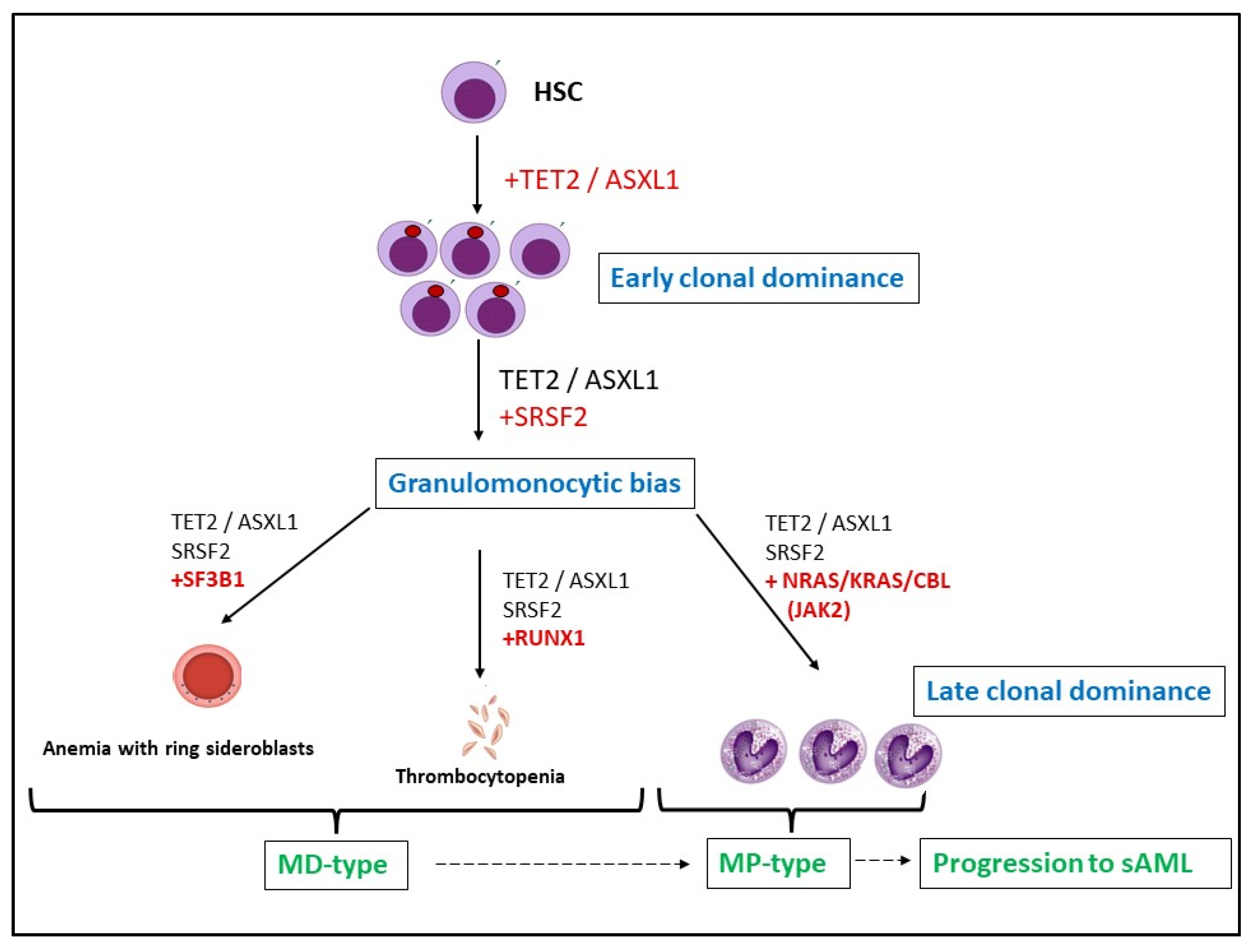
| Gene Name | Mutation Frequency in CMML, % | Prognostic Significance | Treatment Response to HMA |
|---|---|---|---|
| ASXL1 | 34–46 [22,23,24,25,32,33,34] | Marker of poor prognosis, decreased OS [22,32,33,34,35,36,37] Increased progression to AML [37] Controversial data concerning leukemia-free survival [22,38,39] | Controversial data about response to HMA [22,32,39,40,41] |
| TET2 | 32–61 [22,23,24,25,27,32,33,34,42] | Controversial data about prognostic impact [22,25,42,43,44,45] Genotype ASXL1wt/TET2muthad a favorable impact on OS [4,21] | No impact on response or survival on decitabine [40,41,45] TET2mut/ASXL1wt–higher CR rate and ORR to HMA, prolonged OS after treatment with HMA [32] |
| SRSF2 | 29–52 [22,24,27,32,33,34,46] | Controversial data about prognostic impact [22,27,46] | No impact on response to HMAs [22,32,39,41] |
| RUNX1 | 6–22 [22,24,25,27,32] | Controversial data about OS [32,47] Trend towards increased progression to AML [47] | No impact on response to HMAs [22,32,41] |
| NRAS | 2–22 [22,23,24,25,27,32] | Decreased OS [33,48] | No impact on response to HMAs [32,40,41] |
| KRAS | 3–12 [23,24,25,27] | Unclear impact on prognosis [38] | No impact on response or survival on decitabine [40,41] |
| CBL | 10–22 [22,23,24,25,27,32] | Decreased OS [22,32] | No impact on response [32,40,41] Controversial data about OS after therapy with HMAs [22,40] |
| U2AF1 | 5–10 [22,24,32] | No impact on prognosis [49] | No impact on response to HMAs [32,41] |
| DNMT3A | 2–9 [22,24,32] | Decreased overall survival [50] Decreased leukemia-free survival [50] | No impact on response to decitabine [41] |
| SETBP1 | 4–18 [13,24,30,34] | Controversial data about OS and its impact on progression to AML [13,30,35,39,51,52,53] | Unclear impact |
| IDH2 | 4–6 [22,24,25] | Controversial data about prognosis [22,25,38] | Controversial data [22,41] |
| EZH2 | 5–11 [22,25,27] | Decreased OS, increased progression [25,33,38] | Unclear impact |
| FLT3 | <5 [29,38,49] | No impact on prognosis [29,40] | No impact on response to decitabine [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belotserkovskaya, E.; Demidov, O. Mouse Models of CMML. Int. J. Mol. Sci. 2021, 22, 11510. https://doi.org/10.3390/ijms222111510
Belotserkovskaya E, Demidov O. Mouse Models of CMML. International Journal of Molecular Sciences. 2021; 22(21):11510. https://doi.org/10.3390/ijms222111510
Chicago/Turabian StyleBelotserkovskaya, Ekaterina, and Oleg Demidov. 2021. "Mouse Models of CMML" International Journal of Molecular Sciences 22, no. 21: 11510. https://doi.org/10.3390/ijms222111510






