Affinity Purification Coupled to Stable Isotope Dilution LC-MS/MS Analysis to Discover IgG4 Glycosylation Profiles for Autoimmune Pancreatitis
Abstract
1. Introduction
2. Results
2.1. Sample Pretreatment Process
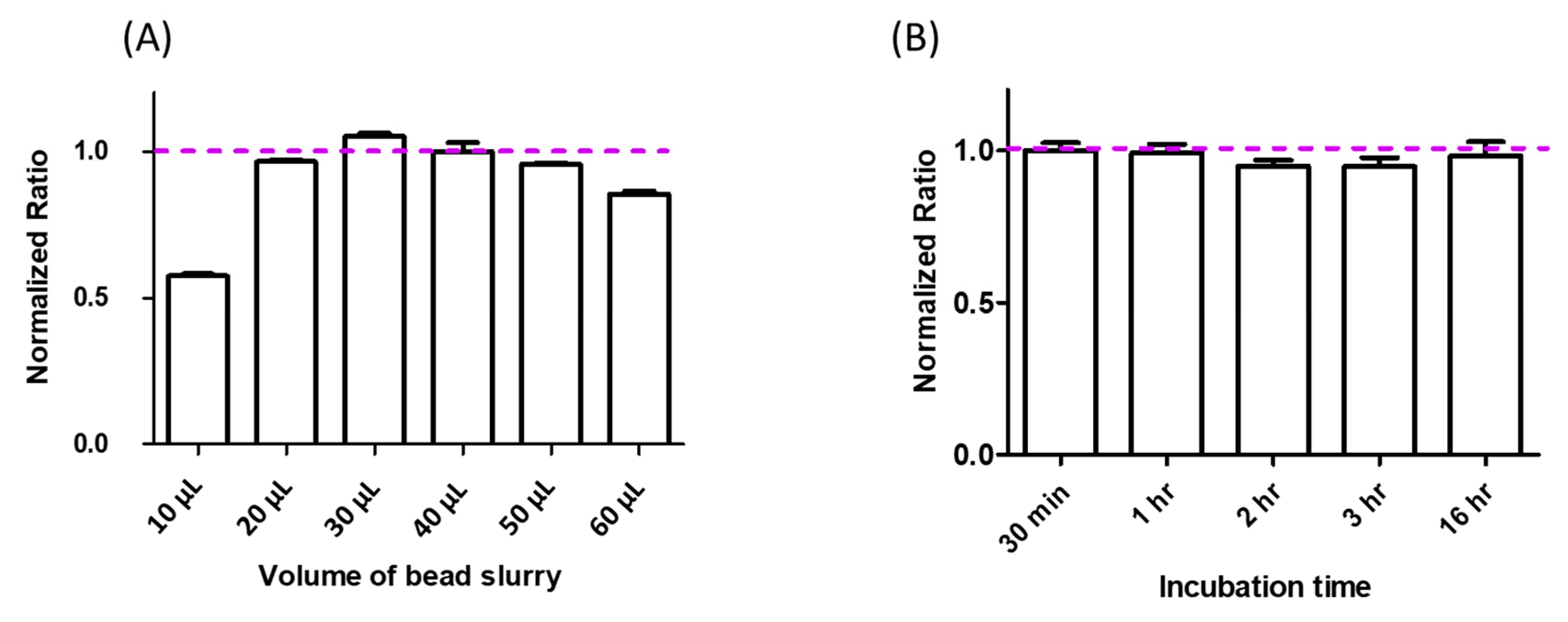
2.1.1. Optimization of Volume of Bead Slurry for Human IgG4 Purification
2.1.2. Optimization of Sample Incubation Time
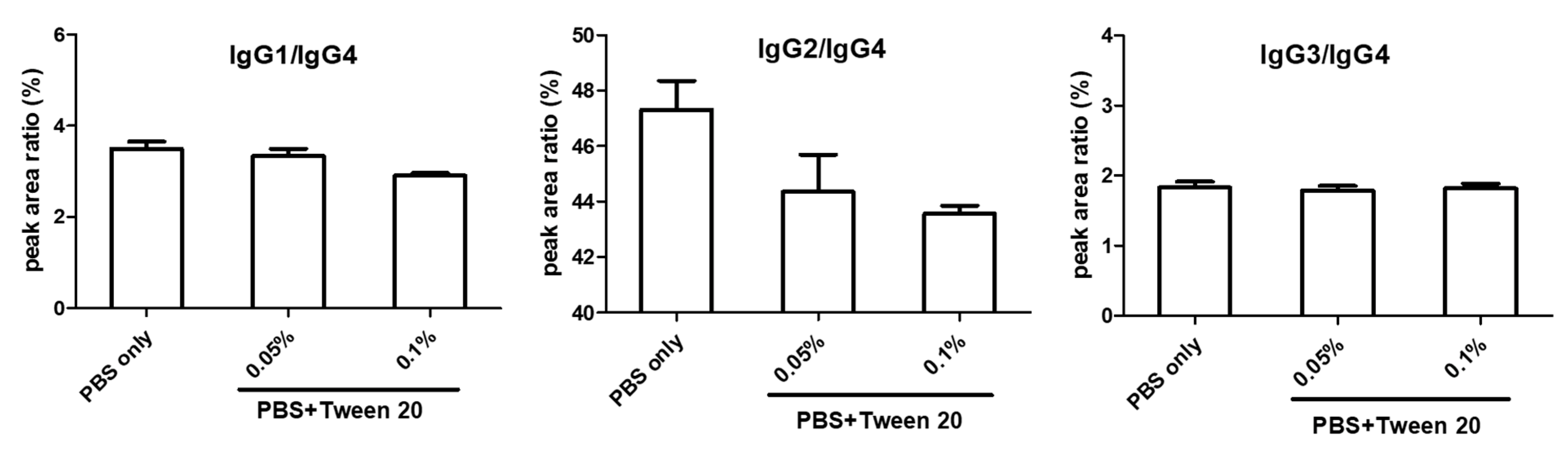
2.1.3. Wash Step and IgG2 Contamination
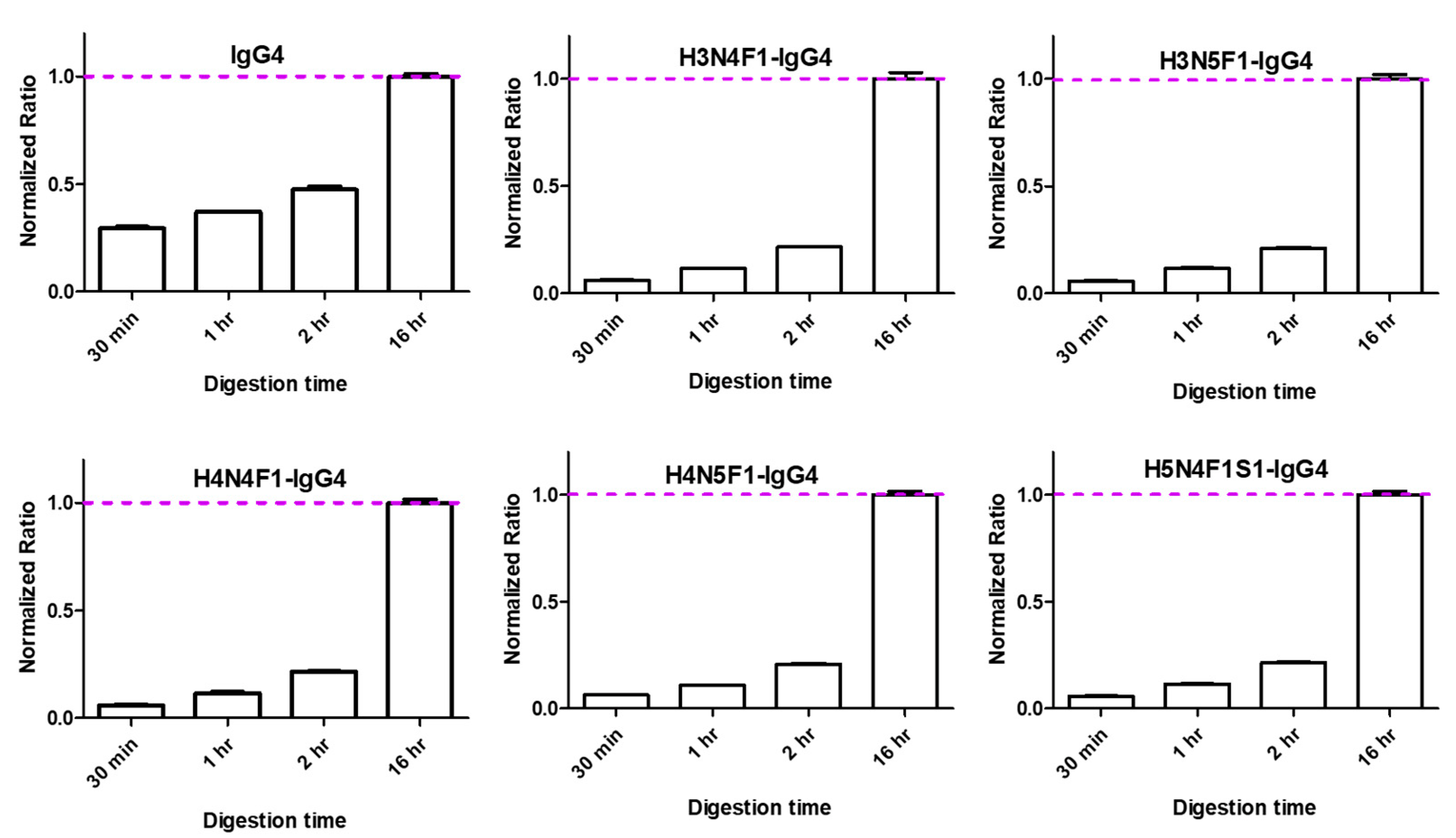
2.1.4. Optimization of on-Bead Digestion Time
2.2. UHPLC-MS/MS Analysis and the General Workflow
2.2.1. UHPLC-MS/MS Condition
2.2.2. Performance of the Analytical Method
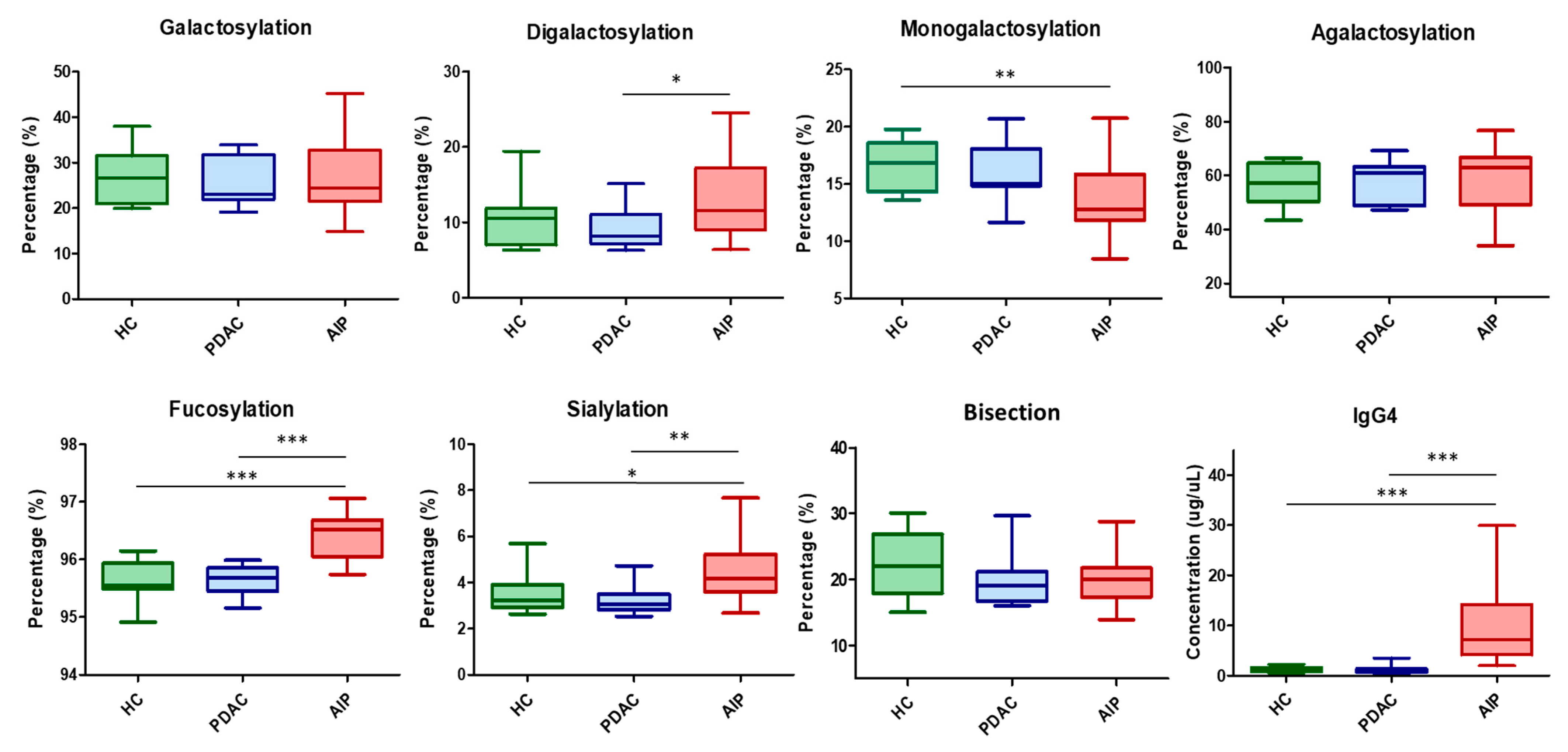
2.3. Application to Clinical Samples
3. Discussion
4. Materials and Methods
4.1. Standards and Reagents
4.2. Sample Preparation Process
4.3. UHPLC-MS/MS Analysis
4.4. Performance of the UHPLC-MS/MS Method
4.5. Analysis of Clinical Samples
4.6. Data analysis and Glycosylation Profiling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maruyama, M.; Watanabe, T.; Kanai, K.; Oguchi, T.; Muraki, T.; Hamano, H.; Arakura, N.; Kawa, S. International Consensus Diagnostic Criteria for Autoimmune Pancreatitis and Its Japanese Amendment Have Improved Diagnostic Ability over Existing Criteria. Gastroenterol. Res. Pract. 2013, 2013, 456965. [Google Scholar] [CrossRef] [PubMed]
- Chari, S.T.; Kloeppel, G.; Zhang, L.; Notohara, K.; Lerch, M.M.; Shimosegawa, T. Histopathologic and clinical subtypes of autoimmune pancreatitis: The honolulu consensus document. Pancreatology 2010, 10, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Kamisawa, T.; Zen, Y.; Pillai, S.; Stone, J.H. IgG4-related disease. Lancet 2015, 385, 1460–1471. [Google Scholar] [CrossRef]
- Kamisawa, T.; Takuma, K.; Tabata, T.; Inaba, Y.; Egawa, N.; Tsuruta, K.; Hishima, T.; Sasaki, T.; Itoi, T. Serum IgG4-negative autoimmune pancreatitis. J. Gastroenterol. 2011, 46, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.H.; Horie, R.; Sekiguchi, H.; Peikert, T.; Yi, E.S. Spectrum of Disorders Associated with Elevated Serum IgG4 Levels Encountered in Clinical Practice. Int. J. Rheumatol. 2012, 2012, 232960. [Google Scholar] [CrossRef]
- Ghazale, A.; Chari, S.T.; Smyrk, T.C.; Levy, M.J.; Topazian, M.D.; Takahashi, N.; Clain, J.E.; Pearson, R.K.; Pelaez-Luna, M.; Petersen, B.T.; et al. Value of serum IgG4 in the diagnosis of autoimmune pancreatitis and in distinguishing it from pancreatic cancer. Am. J. Gastroenterol. 2007, 102, 1646–1653. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, X.; Joseph, L.; Zhao, L.; Hart, J.; Xiao, S.Y. Pancreatic ductal adenocarcinoma with autoimmune pancreatitis-like histologic and immunohistochemical features. Hum. Pathol. 2014, 45, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Kamisawa, T.; Shimosegawa, T.; Okazaki, K.; Nishino, T.; Watanabe, H.; Kanno, A.; Okumura, F.; Nishikawa, T.; Kobayashi, K.; Ichiya, T.; et al. Standard steroid treatment for autoimmune pancreatitis. Gut 2009, 58, 1504–1507. [Google Scholar] [CrossRef]
- Khosroshahi, A.; Wallace, Z.S.; Crowe, J.L.; Akamizu, T.; Azumi, A.; Carruthers, M.N.; Chari, S.T.; Della-Torre, E.; Frulloni, L.; Goto, H.; et al. International Consensus Guidance Statement on the Management and Treatment of IgG4-Related Disease. Arthritis Rheumatol. 2015, 67, 1688–1699. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef] [PubMed]
- Yau, L.F.; Liu, J.; Jiang, M.; Bai, G.; Wang, J.R.; Jiang, Z.H. An integrated approach for comprehensive profiling and quantitation of IgG-Fc glycopeptides with application to rheumatoid arthritis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1122–1123, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Chen, B.; Wang, Y.; Xia, P.; He, C.; Liu, Y.; Zhang, R.; Zhang, M.; Li, Z. Disease-specific IgG Fc N-glycosylation as personalized biomarkers to differentiate gastric cancer from benign gastric diseases. Sci. Rep. 2016, 6, 25957. [Google Scholar] [CrossRef]
- Simurina, M.; de Haan, N.; Vuckovic, F.; Kennedy, N.A.; Stambuk, J.; Falck, D.; Trbojevic-Akmacic, I.; Clerc, F.; Razdorov, G.; Khon, A.; et al. Glycosylation of Immunoglobulin G Associates With Clinical Features of Inflammatory Bowel Diseases. Gastroenterology 2018, 154, 1320–1333.e10. [Google Scholar] [CrossRef]
- Kawaguchi-Sakita, N.; Kaneshiro-Nakagawa, K.; Kawashima, M.; Sugimoto, M.; Tokiwa, M.; Suzuki, E.; Kajihara, S.; Fujita, Y.; Iwamoto, S.; Tanaka, K.; et al. Serum immunoglobulin G Fc region N-glycosylation profiling by matrix-assisted laser desorption/ionization mass spectrometry can distinguish breast cancer patients from cancer-free controls. Biochem. Biophys Res. Commun. 2016, 469, 1140–1145. [Google Scholar] [CrossRef]
- Shiao, J.Y.; Chang, Y.T.; Chang, M.C.; Chen, M.X.; Liu, L.W.; Wang, X.Y.; Tsai, Y.J.; Kuo, T.C.; Tsai, I.L. Development of efficient on-bead protein elution process coupled to ultra-high performance liquid chromatography-tandem mass spectrometry to determine immunoglobulin G subclass and glycosylation for discovery of bio-signatures in pancreatic disease. J. Chromatogr. A 2020, 1621, 461039. [Google Scholar] [CrossRef] [PubMed]
- Shih, H.C.; Chang, M.C.; Chen, C.H.; Tsai, I.L.; Wang, S.Y.; Kuo, Y.P.; Chen, C.H.; Chang, Y.T. High accuracy differentiating autoimmune pancreatitis from pancreatic ductal adenocarcinoma by immunoglobulin G glycosylation. Clin. Proteom. 2019, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Zauner, G.; Selman, M.H.; Bondt, A.; Rombouts, Y.; Blank, D.; Deelder, A.M.; Wuhrer, M. Glycoproteomic analysis of antibodies. Mol. Cell. Proteom. 2013, 12, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.T.; Chang, M.C.; Tsai, Y.J.; Ferng, C.; Shih, H.C.; Kuo, Y.P.; Chen, C.H.; Tsai, I.L. Method development of immunoglobulin G purification from micro-volumes of human serum for untargeted and targeted proteomics-based antibody repertoire studies. J. Food Drug Anal. 2019, 27, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Lorna De Leoz, M.; Stein, S. NIST Mass Spectrometry Data Center, Gaithersburg, Maryland USA. Available online: https://www.nist.gov/static/glyco-mass-calc/#/ (accessed on 1 September 2021).
- Konno, N.; Sugimoto, M.; Takagi, T.; Furuya, M.; Asano, T.; Sato, S.; Kobayashi, H.; Migita, K.; Miura, Y.; Aihara, T.; et al. Changes in N-glycans of IgG4 and its relationship with the existence of hypocomplementemia and individual organ involvement in patients with IgG4-related disease. PLoS ONE 2018, 13, e0196163. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Zhang, P.; Li, L.; Liu, C.; Li, J.; Zhang, W.; Li, Y. Assessing serum IgG4 glycosylation profiles of IgG4-related disease using lectin microarray. Clin. Exp. Rheumatol. 2021, 39, 393–402. [Google Scholar] [PubMed]
- Culver, E.L.; van de Bovenkamp, F.S.; Derksen, N.I.L.; Koers, J.; Cargill, T.; Barnes, E.; de Neef, L.A.; Koeleman, C.A.M.; Aalberse, R.C.; Wuhrer, M.; et al. Unique patterns of glycosylation in immunoglobulin subclass G4-related disease and primary sclerosing cholangitis. J. Gastroenterol. Hepatol. 2019, 34, 1878–1886. [Google Scholar] [CrossRef]
- Maverakis, E.; Kim, K.; Shimoda, M.; Gershwin, M.E.; Patel, F.; Wilken, R.; Raychaudhuri, S.; Ruhaak, L.R.; Lebrilla, C.B. Glycans in the immune system and The Altered Glycan Theory of Autoimmunity: A critical review. J. Autoimmun. 2015, 57, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Larson, N.R.; White, D.R.; Middaugh, C.R.; Tolbert, T.; Schöneich, C. Effects of Glycan Structure on the Stability and Receptor Binding of an IgG4-Fc. J. Pharm. Sci. 2020, 109, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Hazen, M.; Marshall, B.; Crowell, S.R.; Ou, Q.; Wong, A.W.; Phung, W.; Vernes, J.M.; Meng, Y.G.; Tejada, M.; et al. Increased in vivo effector function of human IgG4 isotype antibodies through afucosylation. MAbs 2016, 8, 1098–1106. [Google Scholar] [CrossRef] [PubMed]

| Compound Name | RT (min) | Precursor (m/z) | Product (m/z) | Cone (V) | Collision (eV) |
|---|---|---|---|---|---|
| IgG4: TTPPVLDSDGSFFLYSR | 4.77 | 951.5 | 850.4/1293.6 | 35 | 34 |
| H3N3F1-IgG4 a | 2.38 | 805.7 | 204.1/366.1 | 35 | 25 |
| H3N4F1-IgG4 | 2.38 | 873.4 | 204.1/366.1 | 35 | 25 |
| H3N5-IgG4 | 2.40 | 892.4 | 204.1/366.1 | 35 | 25 |
| H3N5F1-IgG4 | 2.39 | 941.0 | 204.1/366.1 | 35 | 25 |
| H4N4F1-IgG4 | 2.36 | 927.4 | 204.1/366.1 | 35 | 25 |
| H4N4F1S1-IgG4 | 2.46 | 1024.4 | 204.1/366.1 | 35 | 25 |
| H4N5-IgG4 | 2.40 | 946.4 | 204.1/366.1 | 35 | 25 |
| H4N5F1-IgG4 | 2.38 | 995.1 | 204.1/366.1 | 35 | 25 |
| H4N5F1S1-IgG4 | 2.48 | 1092.1 | 204.1/366.1 | 35 | 25 |
| H4N5S1-IgG4 | 2.43 | 1043.4 | 204.1/366.1 | 35 | 25 |
| H5N4F1-IgG4 | 2.35 | 981.4 | 204.1/366.1 | 35 | 25 |
| H5N4F1S1-IgG4 | 2.44 | 1078.4 | 204.1/366.1 | 35 | 25 |
| H5N4S2-IgG4 | 2.38 | 1126.8 | 204.1/366.1 | 35 | 25 |
| H5N5F1-IgG4 | 2.37 | 1049.1 | 204.1/366.1 | 35 | 25 |
| H5N5S1-IgG4 | 2.44 | 1097.4 | 204.1/366.1 | 35 | 25 |
| IS: TTPPVLDSDGSFFLYSR{13C6,15N4} | 4.78 | 956.5 | 806.9/855.4/303.6 | 35 | 34 |
| Analytes | Calibration Curves (y = ax + b) | |||
|---|---|---|---|---|
| Spiked Protein Concentration(μg μL−1) | a | b | r | |
| IgG4 | 0.14~8.80 | 7.392 | 0.0836 | 0.999 |
| H3N3F1-IgG4 | 0.14~8.80 | 0.017 | −0.0001 | 0.992 |
| H3N4F1-IgG4 | 0.14~8.80 | 0.813 | 0.0392 | 0.995 |
| H3N5-IgG4 a | 0.28~8.80 | 0.024 | 0.0063 | 0.966 |
| H3N5F1-IgG4 | 0.14~8.80 | 0.113 | 0.0024 | 0.995 |
| H4N4F1-IgG4 | 0.14~8.80 | 0.577 | 0.0228 | 0.996 |
| H4N4F1S1-IgG4 | 0.14~8.80 | 0.051 | 0.0002 | 0.995 |
| H4N5-IgG4 a | 0.28~8.80 | 0.019 | 0.0041 | 0.968 |
| H4N5F1-IgG4 | 0.14~8.80 | 0.098 | 0.0020 | 0.995 |
| H4N5F1S1-IgG4 a | 0.55~8.80 | 0.004 | −0.0007 | 0.979 |
| H4N5S1-IgG4 a | 0.55~8.80 | 0.002 | −0.0003 | 0.917 |
| H5N4F1-IgG4 | 0.14~8.80 | 0.071 | 0.0008 | 0.995 |
| H5N4F1S1-IgG4 a | 0.28~8.80 | 0.029 | −0.0003 | 0.994 |
| H5N4S2-IgG4 a | 0.55~8.80 | 0.003 | −0.0005 | 0.980 |
| H5N5F1-IgG4 a | 0.55~8.80 | 0.007 | −0.0009 | 0.984 |
| H5N5S1-IgG4 a | 0.55~8.80 | 0.001 | −0.0003 | 0.948 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.X.; Su, H.-H.; Shiao, C.-Y.; Chang, Y.-T.; Chang, M.-C.; Kao, C.-C.; Wang, S.-Y.; Shih, H.-C.; Tsai, I.-L. Affinity Purification Coupled to Stable Isotope Dilution LC-MS/MS Analysis to Discover IgG4 Glycosylation Profiles for Autoimmune Pancreatitis. Int. J. Mol. Sci. 2021, 22, 11527. https://doi.org/10.3390/ijms222111527
Chen MX, Su H-H, Shiao C-Y, Chang Y-T, Chang M-C, Kao C-C, Wang S-Y, Shih H-C, Tsai I-L. Affinity Purification Coupled to Stable Isotope Dilution LC-MS/MS Analysis to Discover IgG4 Glycosylation Profiles for Autoimmune Pancreatitis. International Journal of Molecular Sciences. 2021; 22(21):11527. https://doi.org/10.3390/ijms222111527
Chicago/Turabian StyleChen, Michael X., Ho-Hsuan Su, Ching-Ya Shiao, Yu-Ting Chang, Ming-Chu Chang, Chih-Chin Kao, San-Yuan Wang, Hsi-Chang Shih, and I-Lin Tsai. 2021. "Affinity Purification Coupled to Stable Isotope Dilution LC-MS/MS Analysis to Discover IgG4 Glycosylation Profiles for Autoimmune Pancreatitis" International Journal of Molecular Sciences 22, no. 21: 11527. https://doi.org/10.3390/ijms222111527
APA StyleChen, M. X., Su, H.-H., Shiao, C.-Y., Chang, Y.-T., Chang, M.-C., Kao, C.-C., Wang, S.-Y., Shih, H.-C., & Tsai, I.-L. (2021). Affinity Purification Coupled to Stable Isotope Dilution LC-MS/MS Analysis to Discover IgG4 Glycosylation Profiles for Autoimmune Pancreatitis. International Journal of Molecular Sciences, 22(21), 11527. https://doi.org/10.3390/ijms222111527






