Inhibition of Carbonic Anhydrase IX Suppresses Breast Cancer Cell Motility at the Single-Cell Level
Abstract
1. Introduction
2. Results
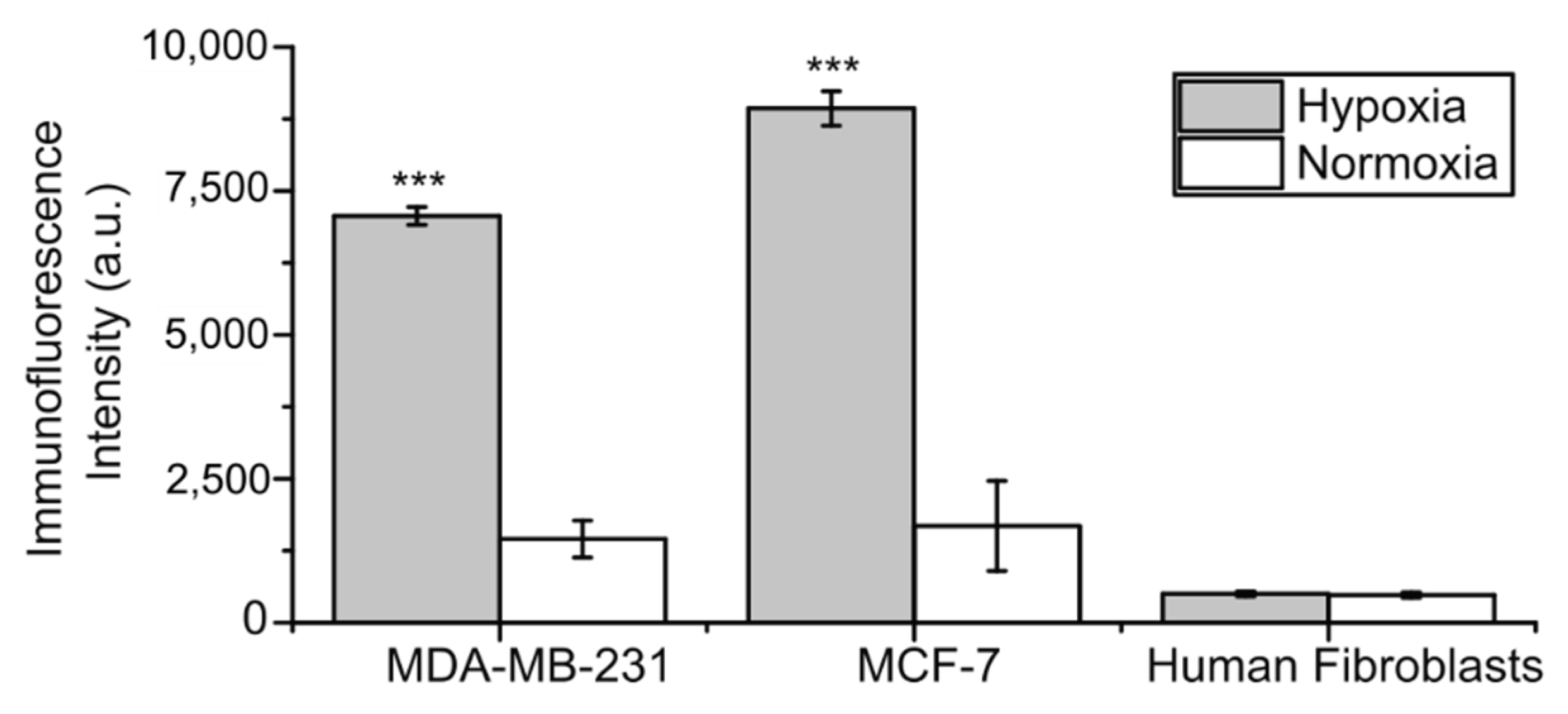
2.1. CA IX Expression in Cell Lines
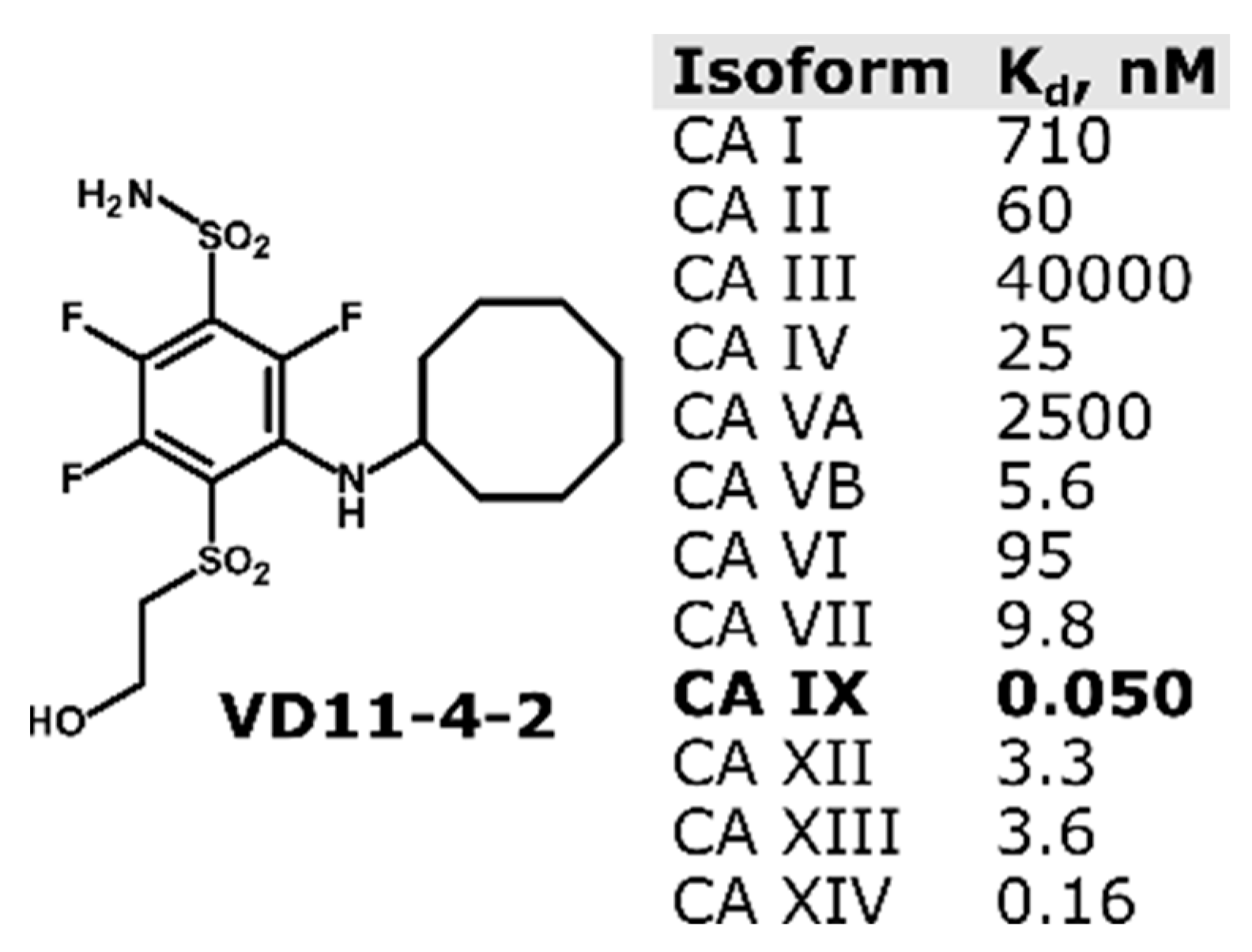
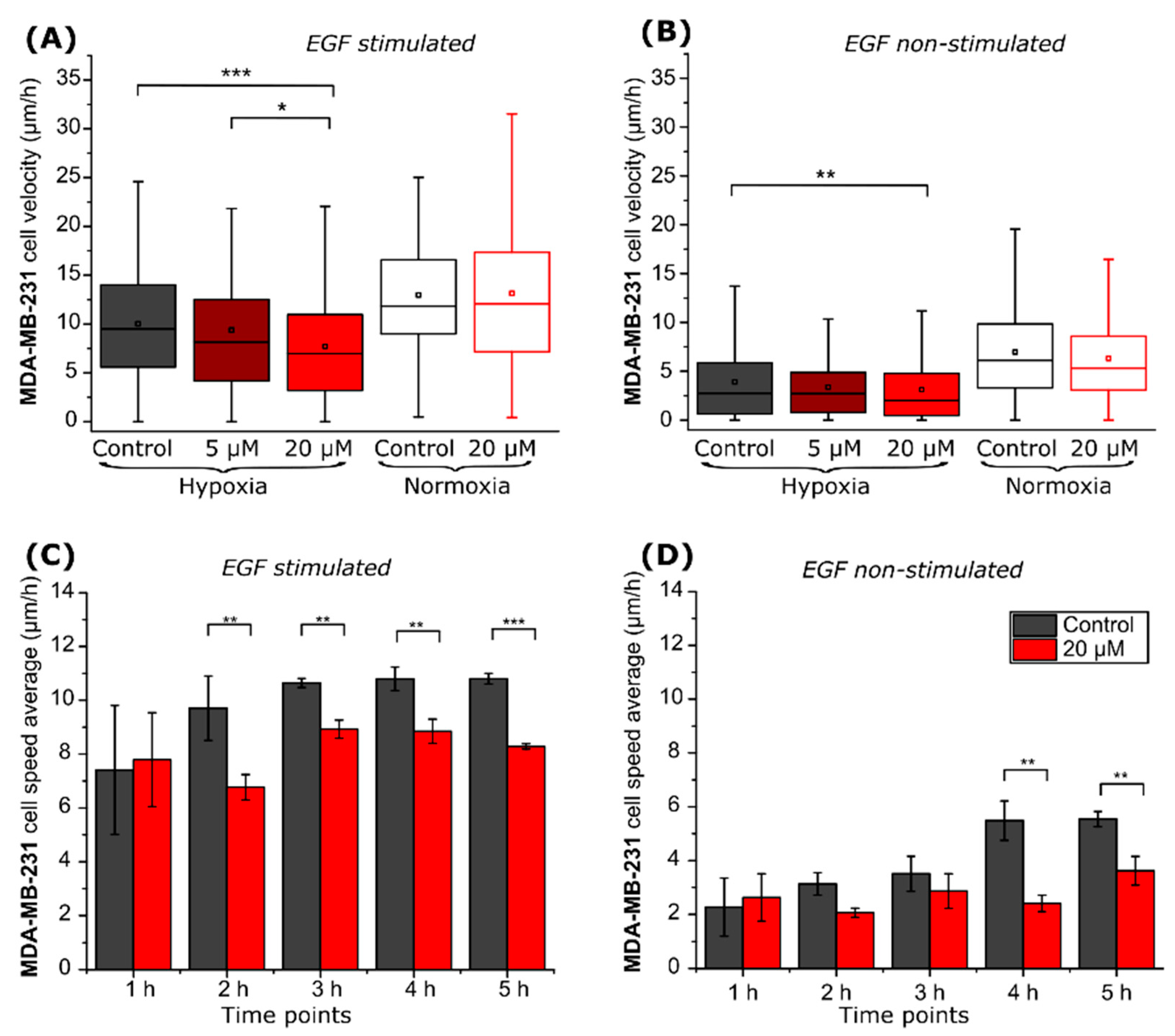
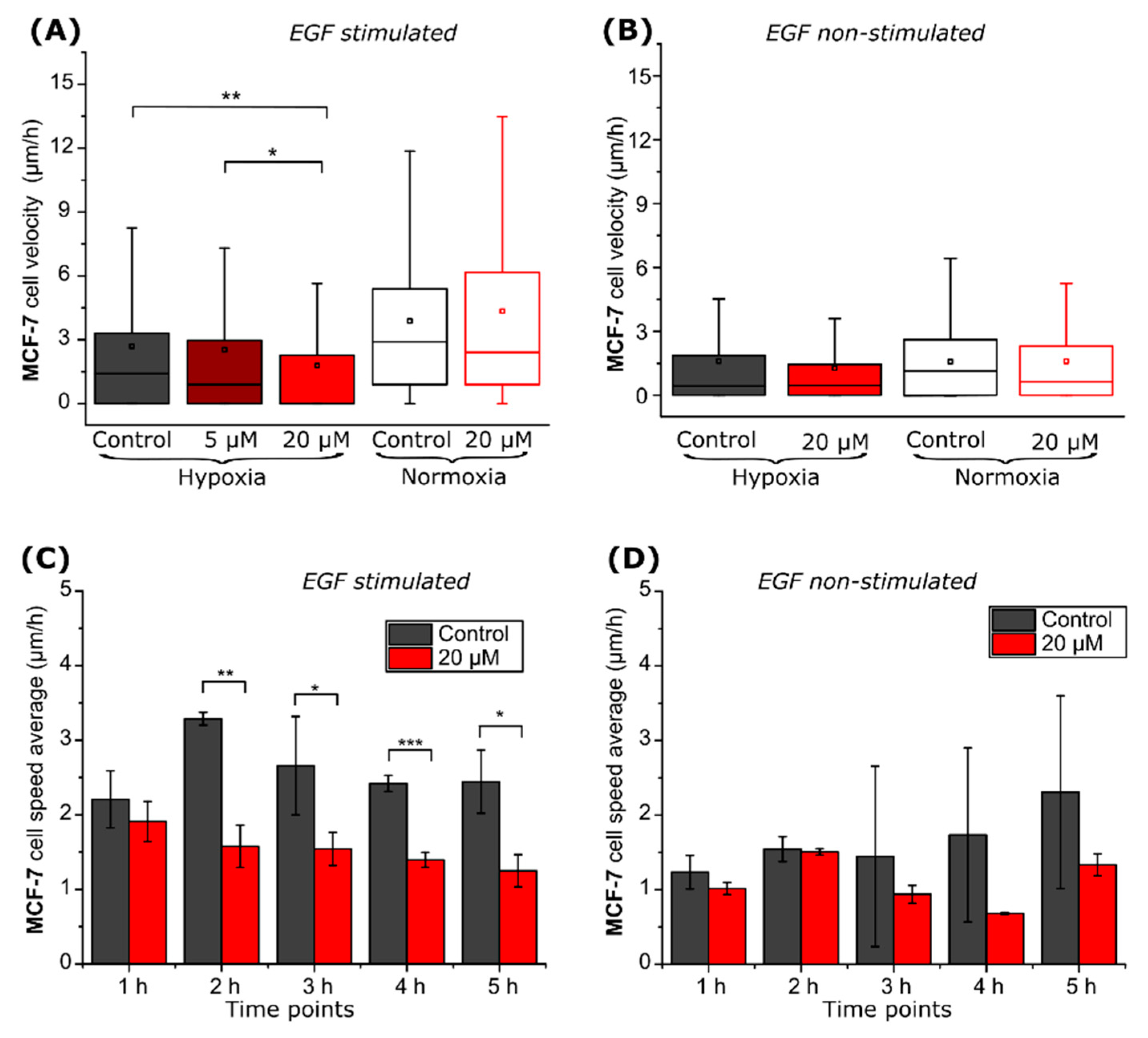
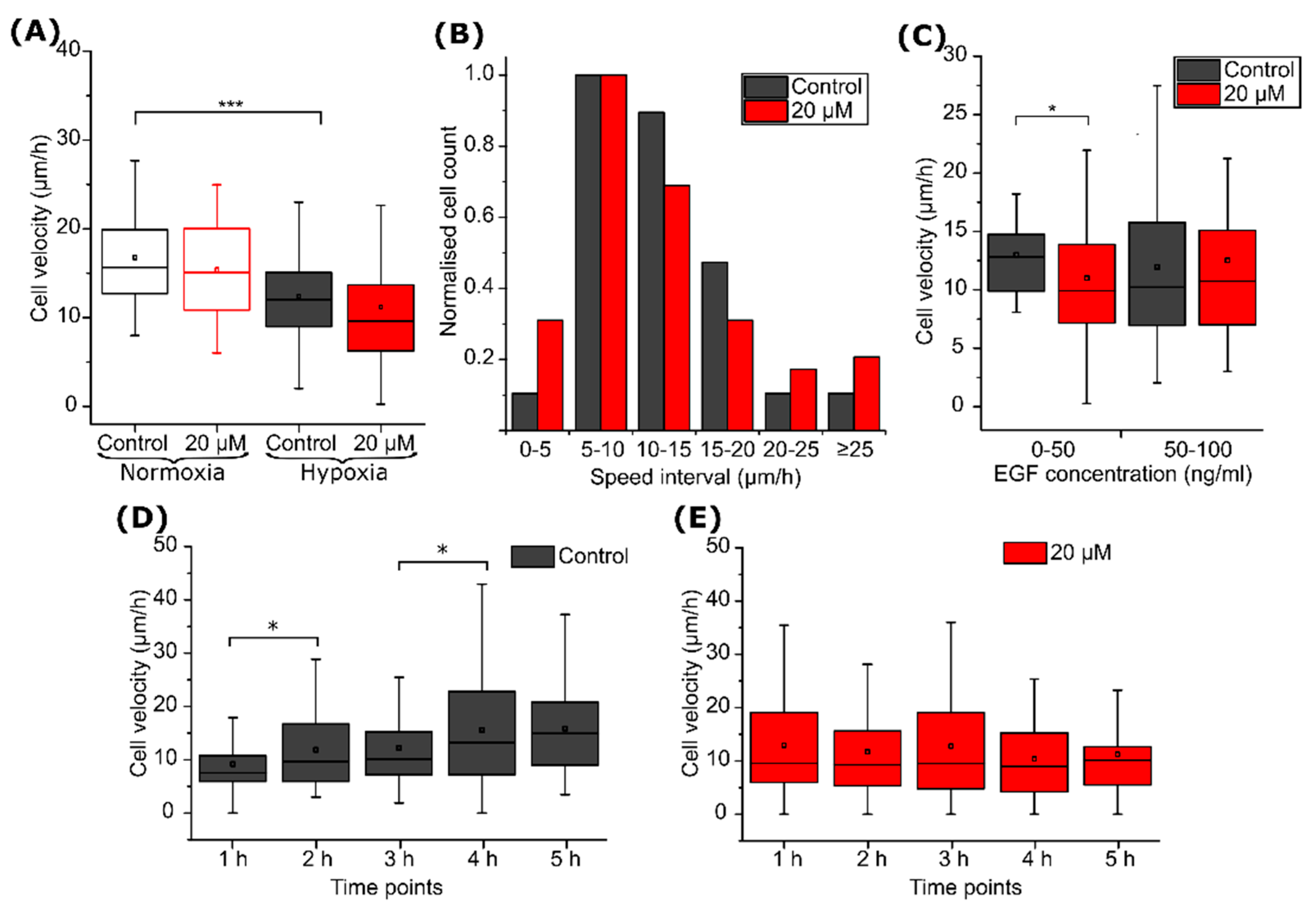
2.2. CA IX Inhibitor Suppresses Cancer Cell Velocity
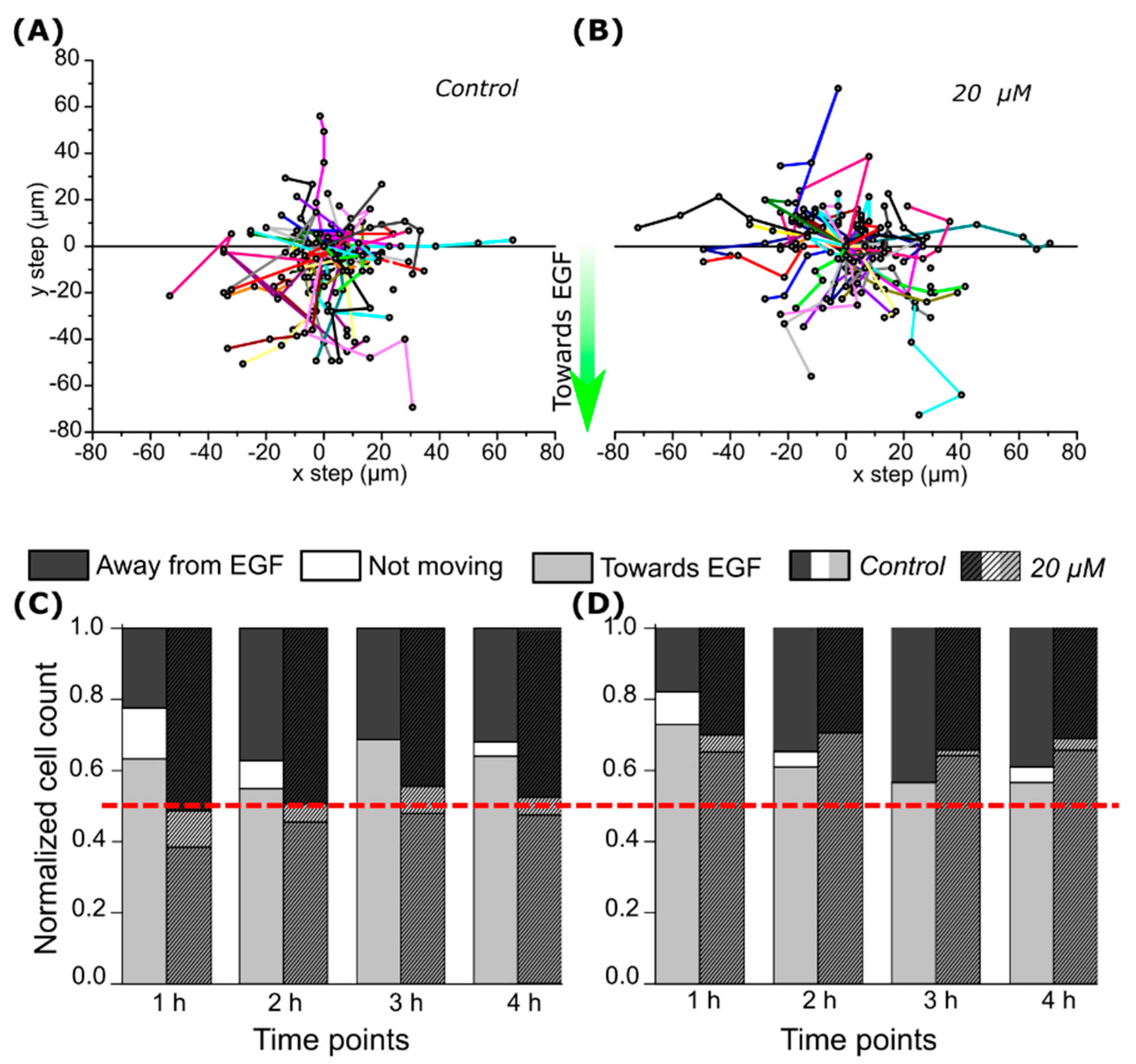
2.3. CA IX Inhibitor Influences Cell Chemotaxis
3. Discussion
4. Materials and Methods
4.1. Cell Culturing
4.2. Immunofluorescence
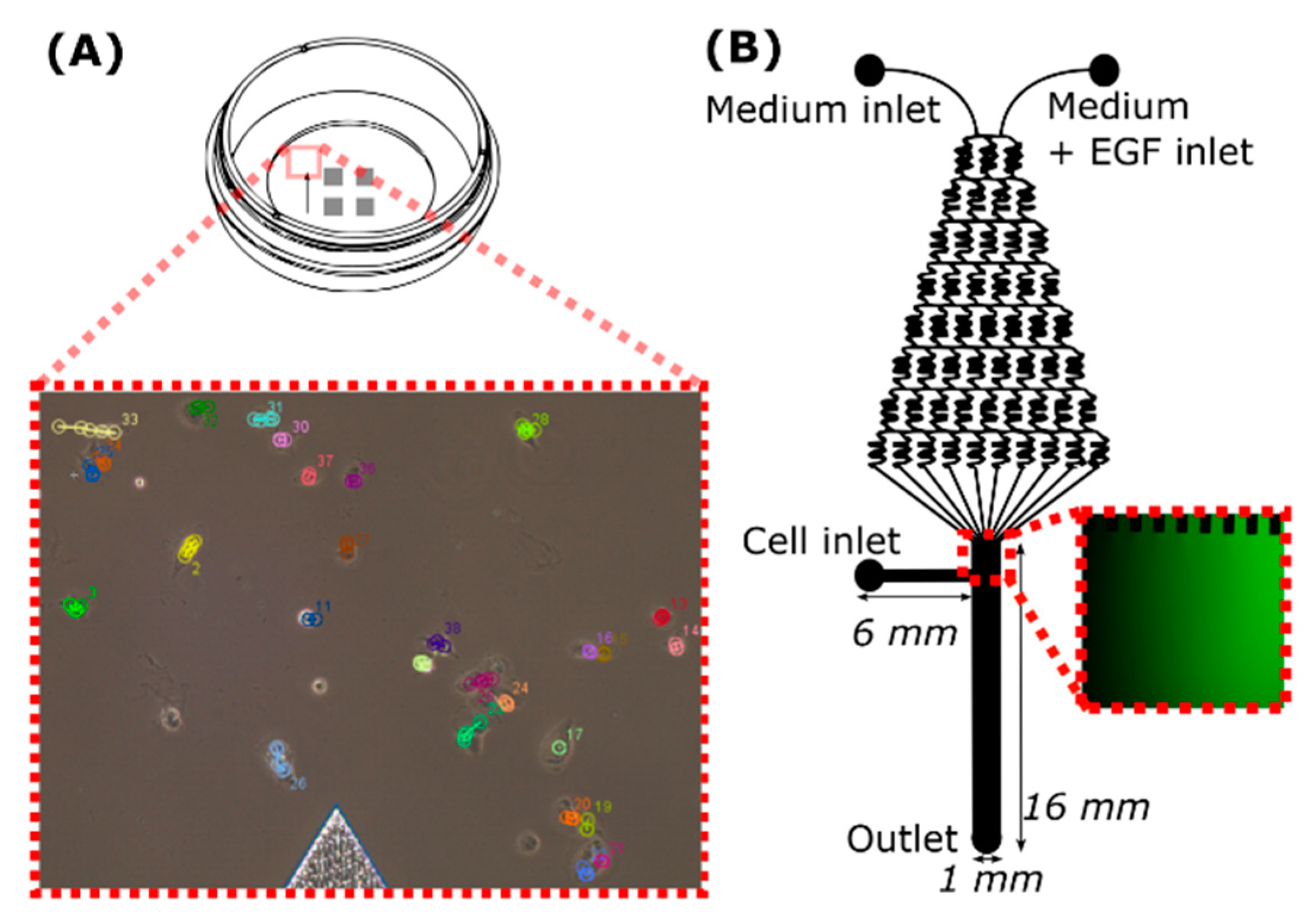
4.3. Migration in µ-Dish
4.4. Migration in the Microfluidic Device
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dillekås, H.; Rogers, M.S.; Straume, O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019, 8, 5574–5576. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Pantel, K.; Kang, Y. Tumor metastasis: Moving new biological insights into the clinic. Nat. Med. 2013, 19, 1450–1464. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.L.; Balasas, T.; Callaghan, J.; Coombes, R.C.; Evans, J.; Hall, J.A.; Kinrade, S.; Jones, D.; Jones, P.S.; Jones, R.; et al. A framework for the development of effective anti-metastatic agents. Nat. Rev. Clin. Oncol. 2019, 16, 185–204. [Google Scholar] [CrossRef]
- Mahon, B.P.; Pinard, M.A.; McKenna, R. Targeting carbonic anhydrase IX activity and expression. Molecules 2015, 20, 2323–2348. [Google Scholar] [CrossRef] [PubMed]
- Mboge, M.Y.; Mahon, B.P.; McKenna, R.; Frost, S.C. Carbonic anhydrases: Role in pH control and cancer. Metabolites 2018, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Švastová, E.; Žilka, N.; Zat’ovičová, M.; Gibadulinová, A.; Čiampor, F.; Pastorek, J.; Pastoreková, S. Carbonic anhydrase IX reduces E-cadherin-mediated adhesion of MDCK cells via interaction with β-catenin. Exp. Cell Res. 2003, 290, 332–345. [Google Scholar] [CrossRef]
- Debreova, M.; Csaderova, L.; Burikova, M.; Lukacikova, L.; Kajanova, I.; Sedlakova, O.; Kery, M.; Kopacek, J.; Zatovicova, M.; Bizik, J.; et al. CAIX regulates invadopodia formation through both a pH-dependent mechanism and interplay with actin regulatory proteins. Int. J. Mol. Sci. 2019, 20, 2745. [Google Scholar] [CrossRef]
- Csaderova, L.; Debreova, M.; Radvak, P.; Stano, M.; Vrestiakova, M.; Kopacek, J.; Pastorekova, S.; Svastova, E. The effect of carbonic anhydrase IX on focal contacts during cell spreading and migration. Front. Membr. Physiol. Membr. Biophys. 2013, 4, 271. [Google Scholar] [CrossRef]
- Jiwa, L.S.; Van Diest, P.J.; Hoefnagel, L.D.; Wesseling, J.; Wesseling, P.; Moelans, C.B. Upregulation of Claudin-4, CAIX and GLUT-1 in distant breast cancer metastases. BMC Cancer 2014, 14, 864. [Google Scholar] [CrossRef]
- Swayampakula, M.; McDonald, P.C.; Vallejo, M.; Coyaud, E.; Chafe, S.C.; Westerback, A.; Venkateswaran, G.; Shankar, J.; Gao, G.; Laurent, E.M.N.; et al. The interactome of metabolic enzyme carbonic anhydrase IX reveals novel roles in tumor cell migration and invadopodia/MMP14-mediated invasion. Oncogene 2017, 36, 6244–6261. [Google Scholar] [CrossRef]
- Gieling, R.G.; Babur, M.; Mamnani, L.; Burrows, N.; Telfer, B.A.; Carta, F.; Winum, J.Y.; Scozzafava, A.; Supuran, C.T.; Williams, K.J. Antimetastatic effect of sulfamate carbonic anhydrase IX inhibitors in breast carcinoma xenografts. J. Med. Chem. 2012, 55, 5591–5600. [Google Scholar] [CrossRef] [PubMed]
- Lock, F.E.; McDonald, P.C.; Lou, Y.; Serrano, I.; Chafe, S.C.; Ostlund, C.; Aparicio, S.; Winum, J.-Y.; Supuran, C.T.; Dedhar, S. Targeting carbonic anhydrase IX depletes breast cancer stem cells within the hypoxic niche. Oncogene 2013, 32, 5210–5219. [Google Scholar] [CrossRef]
- Güttler, A.; Theuerkorn, K.; Riemann, A.; Wichmann, H.; Kessler, J.; Thews, O.; Bache, M.; Vordermark, D. Cellular and radiobiological effects of carbonic anhydrase IX in human breast cancer cells. Oncol. Rep. 2019, 41, 2585–2594. [Google Scholar] [CrossRef] [PubMed]
- Ward, C.; Meehan, J.; Mullen, P.; Supuran, C.; Michael Dixon, J.; Thomas, J.S.; Winum, J.Y.; Lambin, P.; Dubois, L.; Pavathaneni, N.K.; et al. Evaluation of carbonic anhydrase IX as a therapeutic target for inhibition of breast cancer invasion and metastasis using a series of in vitro breast cancer models. Oncotarget 2015, 6, 24856–24870. [Google Scholar] [CrossRef] [PubMed]
- Kai, F.; Drain, A.P.; Weaver, V.M. The extracellular matrix modulates the metastatic journey. Dev. Cell 2019, 49, 332–346. [Google Scholar] [CrossRef] [PubMed]
- Decaestecker, C.; Debeir, O.; Van Ham, P.; Kiss, R. Can anti-migratory drugs be screened in vitro? A review of 2D and 3D assays for the quantitative analysis of cell migration. Med. Res. Rev. 2007, 27, 149–176. [Google Scholar] [CrossRef]
- Insua-Rodríguez, J.; Oskarsson, T. The extracellular matrix in breast cancer. Adv. Drug Deliv. Rev. 2016, 97, 41–55. [Google Scholar] [CrossRef]
- Wolf, K.; Alexander, S.; Schacht, V.; Coussens, L.M.; Von Andrian, U.H.; Van Rheenen, J.; Deryugina, E.; Friedl, P. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 2009, 20, 931–941. [Google Scholar] [CrossRef]
- Dudutiene, V.; Matuliene, J.; Smirnov, A.; Timm, D.D.; Zubriene, A.; Baranauskiene, L.; Morkunaite, V.; Smirnoviene, J.; Michailoviene, V.; Juozapaitiene, V.; et al. Discovery and Characterization of Novel Selective Inhibitors of Carbonic Anhydrase IX. J. Med. Chem. 2014, 57, 9435–9446. [Google Scholar] [CrossRef]
- Kazokaitė, J.; Ames, S.; Becker, H.M.; Deitmer, J.W.; Matulis, D. Selective inhibition of human carbonic anhydrase IX in Xenopus oocytes and MDA-MB-231 breast cancer cells. J. Enzyme Inhib. Med. Chem. 2016, 31, 38–44. [Google Scholar] [CrossRef][Green Version]
- Kazokaitė, J.; Aspatwar, A.; Kairys, V.; Parkkila, S.; Matulis, D. Fluorinated benzenesulfonamide anticancer inhibitors of carbonic anhydrase IX exhibit lower toxic effects on zebrafish embryonic development than ethoxzolamide. Drug Chem. Toxicol. 2017, 40, 309. [Google Scholar] [CrossRef] [PubMed]
- Janoniene, A.; Liu, Z.; Baranauskiene, L.; Mäkilä, E.; Ma, M.; Salonen, J.; Hirvonen, J.; Zhang, H.; Petrikaite, V.; Santos, H.A. A Versatile Carbonic Anhydrase IX Targeting Ligand-Functionalized Porous Silicon Nanoplatform for Dual Hypoxia Cancer Therapy and Imaging. ACS Appl. Mater. Interfaces 2017, 9, 13976–13987. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, D.; Liang, W.; Zehua, L.; Wu, R.; Janoniene, A.; Ma, M.; Pan, G.; Baranauskiene, L.; Zhang, L.; et al. Microfluidic Encapsulation of Prickly Zinc-doped Copper Oxide Nanoparticles with VD1142 Modified Spermine Acetalated Dextran for Efficient Cancer Therapy. Adv. Healthc. Mater. 2017, 6, 1601406. [Google Scholar] [CrossRef]
- Chu, C.Y.; Jin, Y.T.; Zhang, W.; Yu, J.; Yang, H.P.; Wang, H.Y.; Zhang, Z.J.; Liu, X.P.; Zou, Q. CA IX is upregulated in CoCl2-induced hypoxia and associated with cell invasive potential and a poor prognosis of breast cancer. Int. J. Oncol. 2016, 48, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Ciccone, V.; Filippelli, A.; Angeli, A.; Supuran, C.T.; Morbidelli, L. Pharmacological inhibition of CA-IX impairs tumor cell proliferation, migration and invasiveness. Int. J. Mol. Sci. 2020, 21, 2983. [Google Scholar] [CrossRef]
- Liao, S.Y.; Lerman, M.I.; Stanbridge, E.J. Expression of transmembrane carbonic anhydrases, CAIX and CAXII, in human development. BMC Dev. Biol. 2009, 9, 22. [Google Scholar] [CrossRef]
- Hussain, S.A.; Ganesan, R.; Reynolds, G.; Gross, L.; Stevens, A.; Pastorek, J.; Murray, P.G.; Perunovic, B.; Anwar, M.S.; Billingham, L.; et al. Hypoxia-regulated carbonic anhydrase IX expression is associated with poor survival in patients with invasive breast cancer. Br. J. Cancer 2007, 96, 104–109. [Google Scholar] [CrossRef]
- Müller, V.; Riethdorf, S.; Rack, B.; Janni, W.; Fasching, P.A.; Solomayer, E.; Aktas, B.; Kasimir-Bauer, S.; Zeitz, J.; Pantel, K.; et al. Prospective evaluation of serum tissue inhibitor of metalloproteinase 1 and carbonic anhydrase IX in correlation to circulating tumor cells in patients with metastatic breast cancer. Breast Cancer Res. 2011, 13, R71. [Google Scholar] [CrossRef]
- Svastova, E.; Pastorekova, S. Carbonic anhydrase IX: A hypoxia-controlled “catalyst” of cell migration. Cell Adhes. Migr. 2013, 7, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Kazokaite, J.; Niemans, R.; Dudutiene, V.; Becker, H.M.; Leitans, J.; Zubriene, A.; Baranauskiene, L.; Gondi, G.; Zeidler, R.; Matuliene, J.; et al. Novel fluorinated carbonic anhydrase IX inhibitors reduce hypoxia-induced acidification and clonogenic survival of cancer cells. Oncotarget 2018, 9, 26800–26816. [Google Scholar] [CrossRef]
- Comşa, Ş.; Cîmpean, A.M.; Raica, M. The story of MCF-7 breast cancer cell line: 40 Years of experience in research. Anticancer Res. 2015, 35, 3147–3154. [Google Scholar]
- Holliday, D.L.; Speirs, V. Choosing correct breast cancer cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kong, J.; Chang, H.; Kim, H.; Kim, A. EGF induces epithelial-mesenchymal transition through phospho-Smad2/3-Snail signaling pathway in breast cancer cells. Oncotarget 2016, 7, 85021–85032. [Google Scholar] [CrossRef]
- Patel, R.A.; Liu, Y.; Wang, B.; Li, R.; Sebti, S.M. Identification of novel ROCK inhibitors with anti-migratory and anti-invasive activities. Oncogene 2014, 33, 550–555. [Google Scholar] [CrossRef]
- Unbekandt, M.; Croft, D.R.; Crighton, D.; Mezna, M.; McArthur, D.; McConnell, P.; Schüttelkopf, A.W.; Belshaw, S.; Pannifer, A.; Sime, M.; et al. A novel small-molecule MRCK inhibitor blocks cancer cell invasion. Cell Commun. Signal. 2014, 12, 54. [Google Scholar] [CrossRef]
- Svitkina, T. The actin cytoskeleton and actin-based motility. Cold Spring Harb. Perspect. Biol. 2018, 10, a018267. [Google Scholar] [CrossRef]
- Lim, S.; Nam, H.; Jeon, J.S. Chemotaxis Model for Breast Cancer Cells Based on Signal/Noise Ratio. Biophys. J. 2018, 115, 2034–2043. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.J.; Saadi, W.; Lin, F.; Nguyen, C.M.-C.; Jeon, N.L. Differential effects of EGF gradient profiles on MDA-MB-231 breast cancer cell chemotaxis. Exp. Cell Res. 2004, 300, 180–189. [Google Scholar] [CrossRef]
- Biswenger, V.; Baumann, N.; Jürschick, J.; Häckl, M.; Battle, C.; Schwarz, J.; Horn, E.; Zantl, R. Characterization of EGF-guided MDA-MB-231 cell chemotaxis in vitro using a physiological and highly sensitive assay system. PLoS ONE 2018, 13, e0203040. [Google Scholar] [CrossRef] [PubMed]
- Stravinskiene, D.; Imbrasaite, A.; Petrikaite, V.; Matulis, D.; Matuliene, J.; Zvirbliene, A. New monoclonal antibodies for a selective detection of membrane-associated and soluble forms of carbonic anhydrase ix in human cell lines and biological samples. Biomolecules 2019, 9, 304. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janoniene, A.; Mazutis, L.; Matulis, D.; Petrikaite, V. Inhibition of Carbonic Anhydrase IX Suppresses Breast Cancer Cell Motility at the Single-Cell Level. Int. J. Mol. Sci. 2021, 22, 11571. https://doi.org/10.3390/ijms222111571
Janoniene A, Mazutis L, Matulis D, Petrikaite V. Inhibition of Carbonic Anhydrase IX Suppresses Breast Cancer Cell Motility at the Single-Cell Level. International Journal of Molecular Sciences. 2021; 22(21):11571. https://doi.org/10.3390/ijms222111571
Chicago/Turabian StyleJanoniene, Agne, Linas Mazutis, Daumantas Matulis, and Vilma Petrikaite. 2021. "Inhibition of Carbonic Anhydrase IX Suppresses Breast Cancer Cell Motility at the Single-Cell Level" International Journal of Molecular Sciences 22, no. 21: 11571. https://doi.org/10.3390/ijms222111571
APA StyleJanoniene, A., Mazutis, L., Matulis, D., & Petrikaite, V. (2021). Inhibition of Carbonic Anhydrase IX Suppresses Breast Cancer Cell Motility at the Single-Cell Level. International Journal of Molecular Sciences, 22(21), 11571. https://doi.org/10.3390/ijms222111571







