Alternative Splicing of TaGS3 Differentially Regulates Grain Weight and Size in Bread Wheat
Abstract
:1. Introduction
2. Results
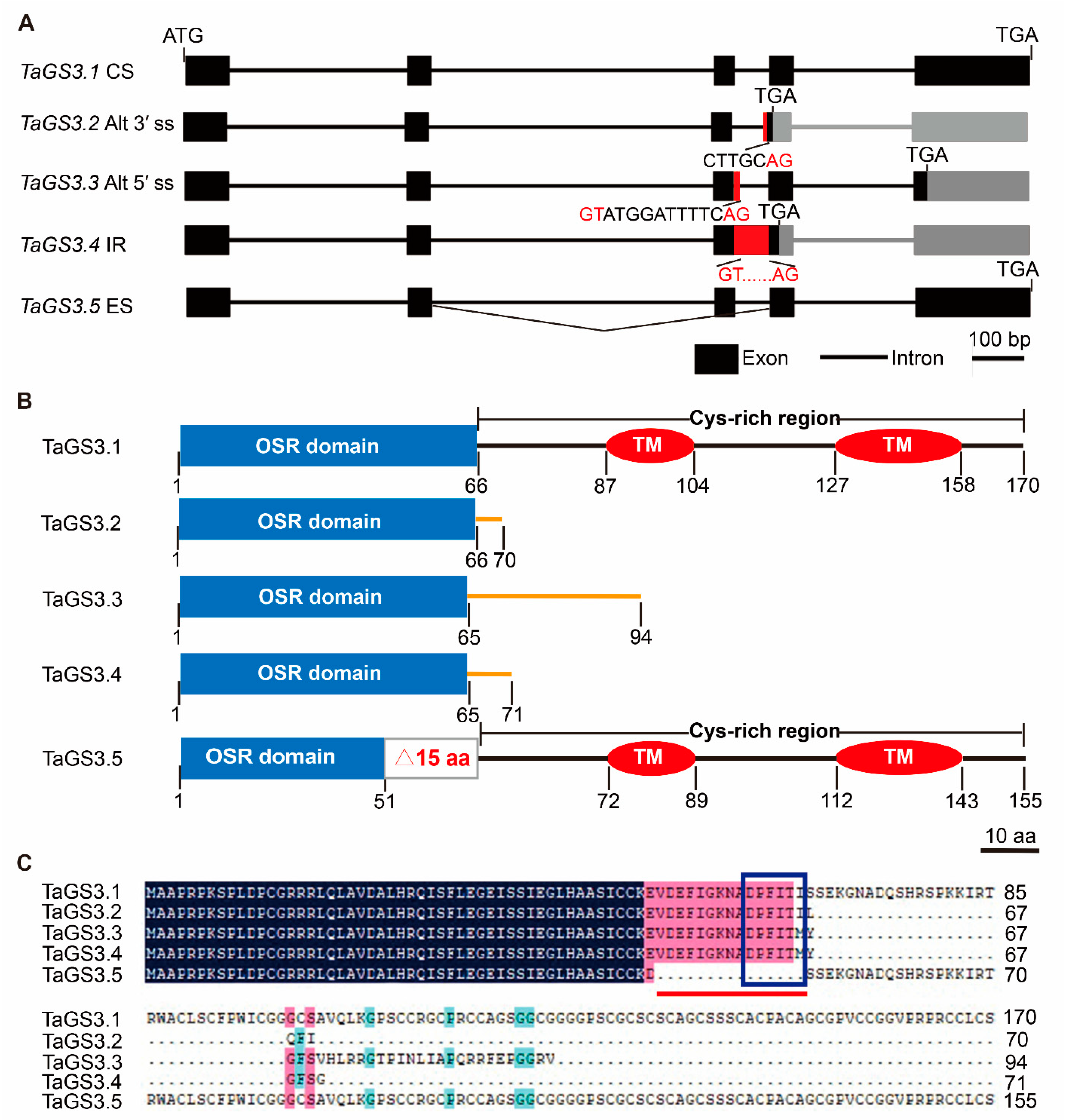
2.1. AS Occurrence of TaGS3 in Wheat
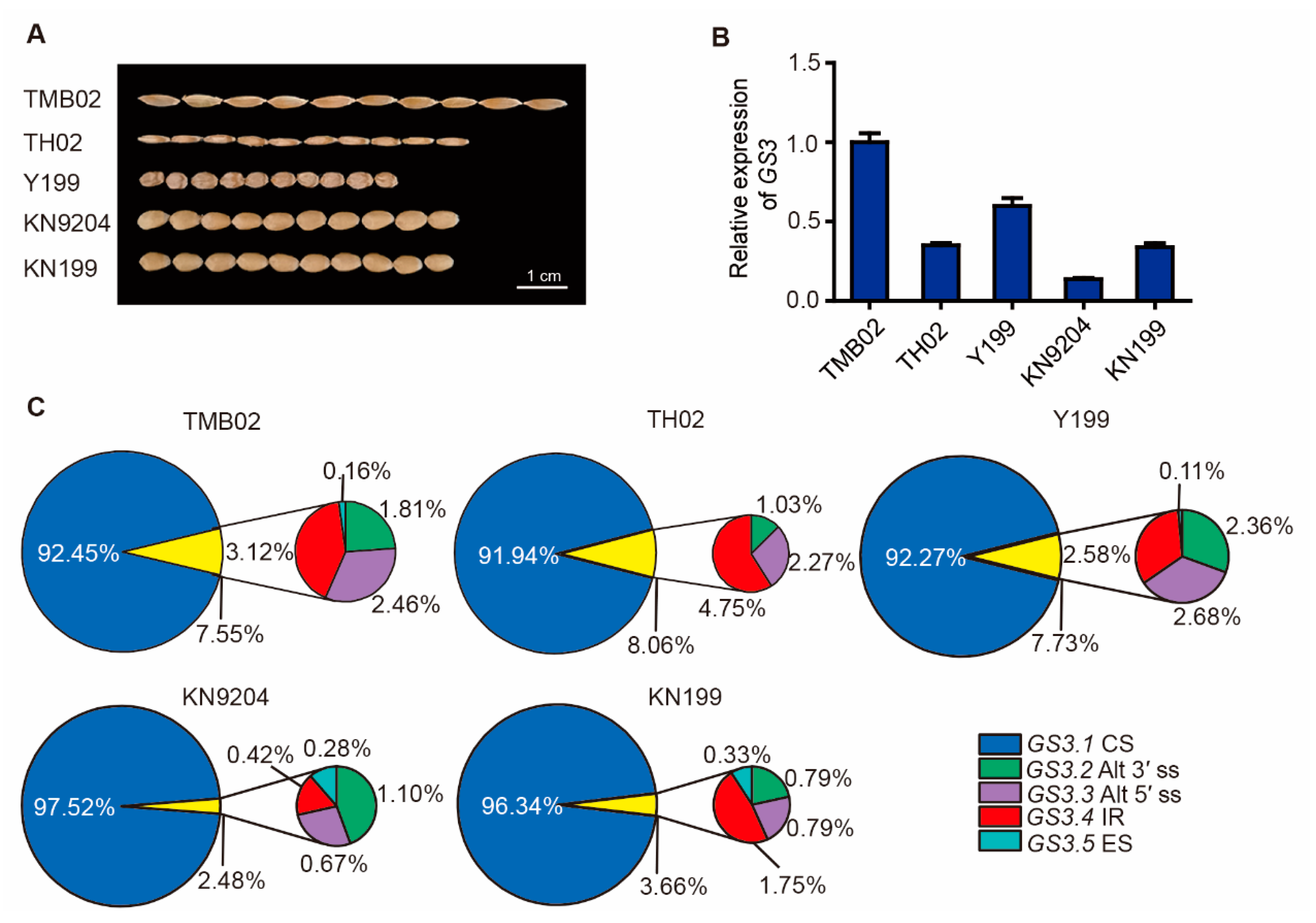
2.2. GS3 Splicing Variants in Wheat Species
2.3. Expression Patterns and Subcellular Localization of TaGS3 Splicing Variants
2.4. Overexpression of TaGS3 Splicing Variants Confers Different Effects on Wheat Grain Weight and Size
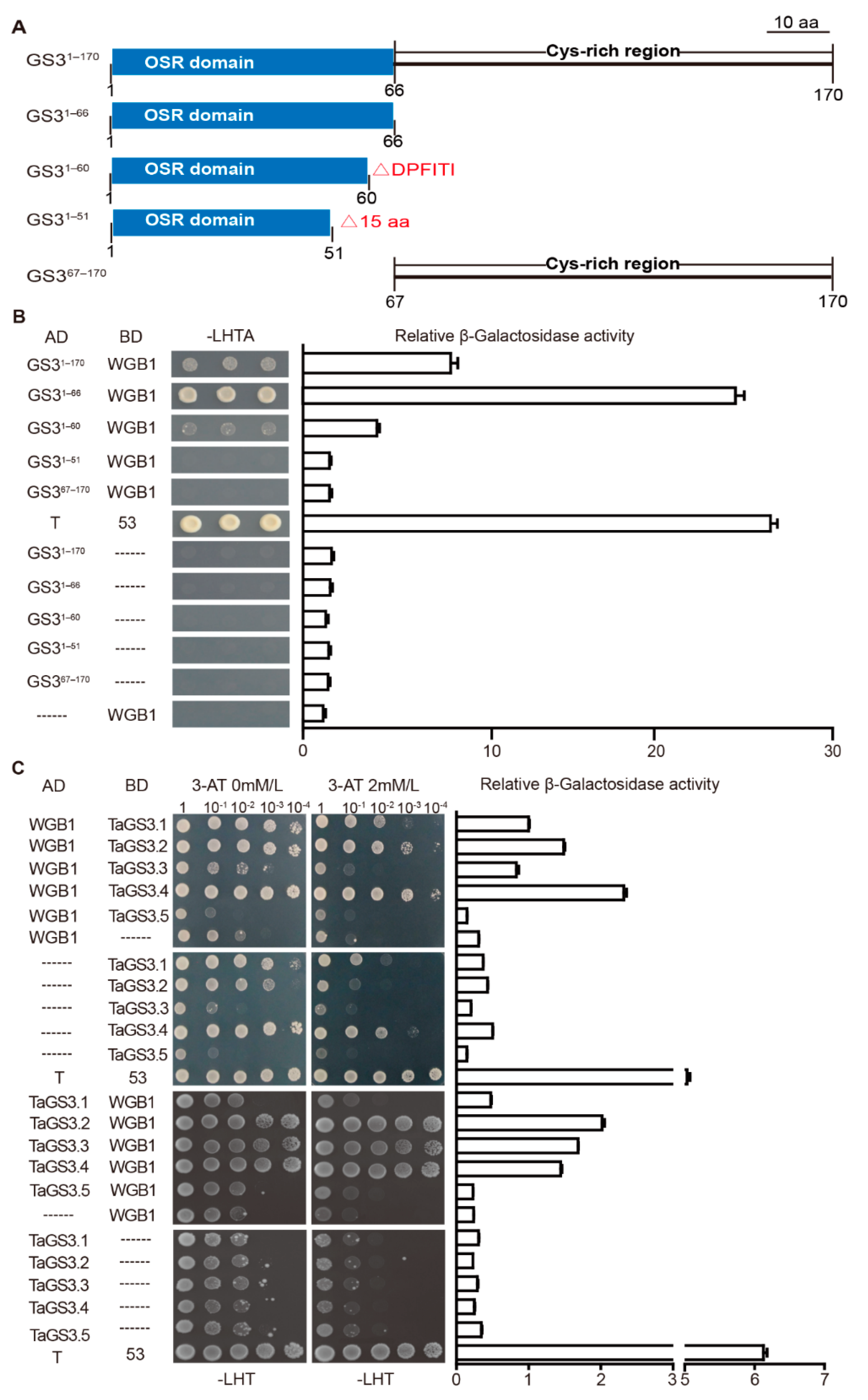
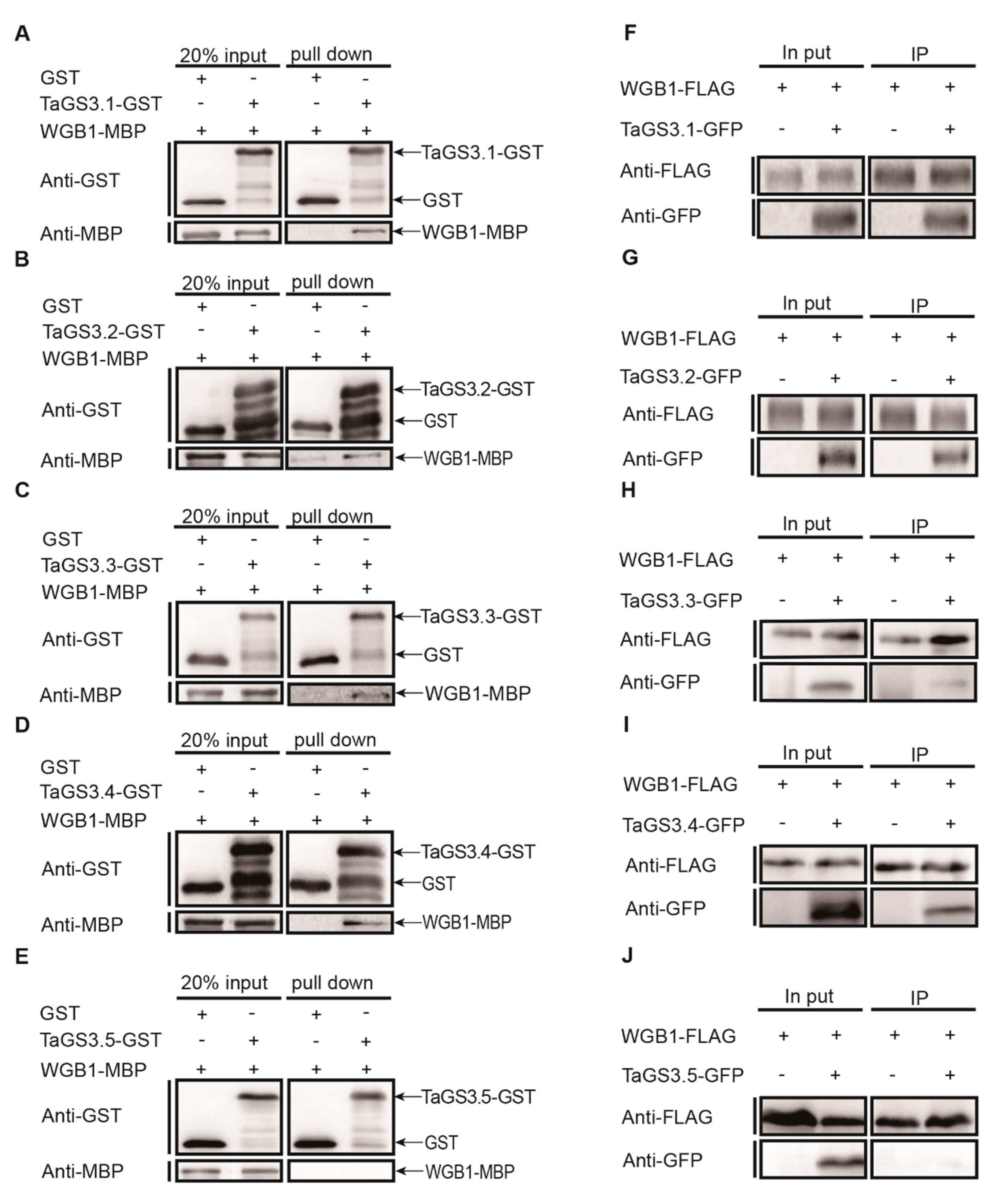
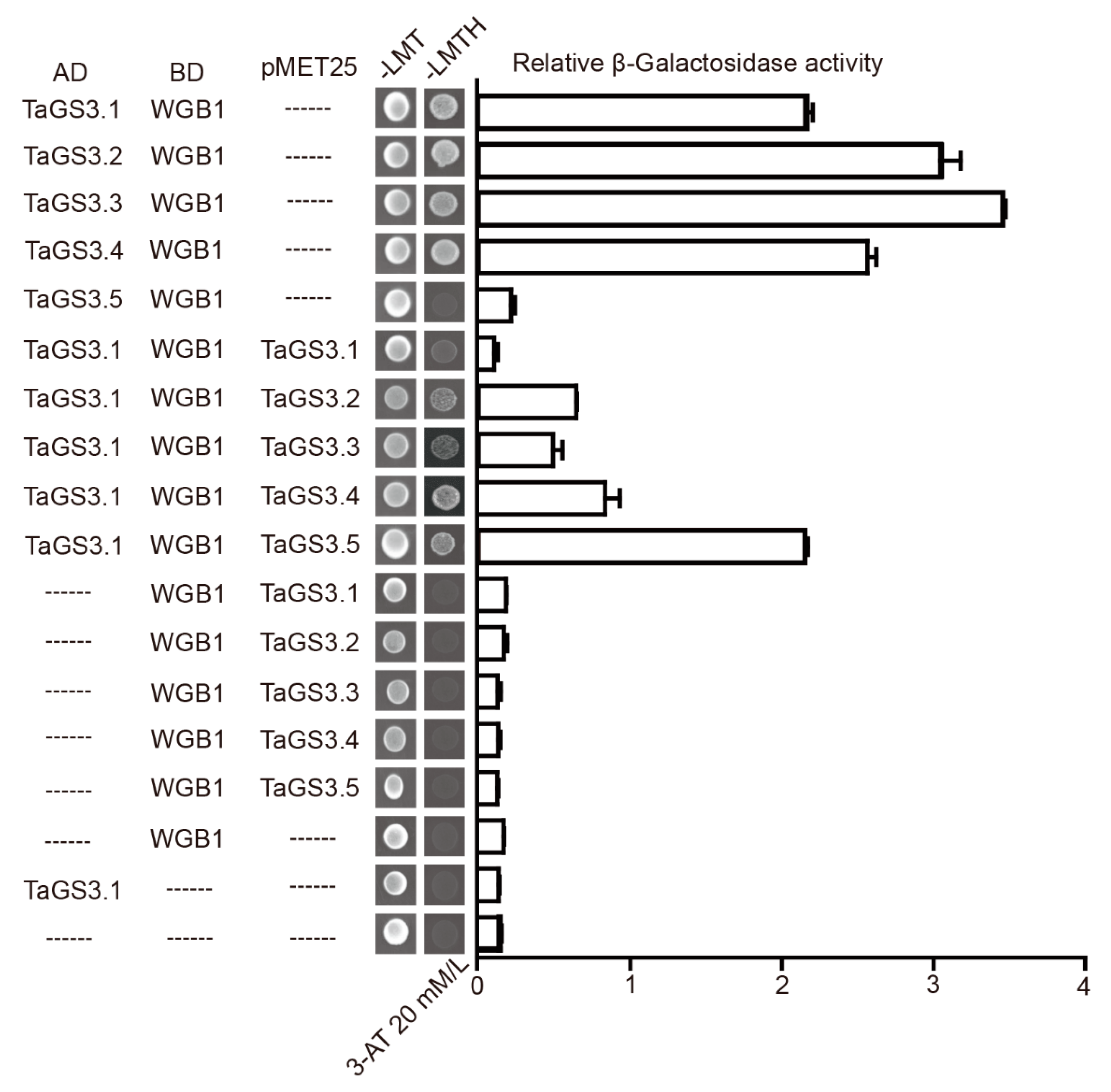
2.5. TaGS3.1, TaGS3.2, TaGS3.3, and TaGS3.4 Physically Interact with WGB1
2.6. Competitive Interactions of TaGS3 Isoforms with WGB1
3. Discussion
3.1. Conservation and Significance of GS3 AS in Gramineae
3.2. Differential Functions and Mechanisms of TaGS3 ASs in Wheat
3.3. Functional Diversification of GS3 in Rice and Wheat
4. Materials and Methods
4.1. Plant Materials
4.2. Isolation of GS3 Splicing Variants
4.3. Phenotype Assessment
4.4. Plasmid Construct
4.5. RNA Extraction and qRT-PCR
4.6. Subcellular Localization Assay
4.7. Yeast Two- and Three-Hybrid Assays
4.8. Pull-Down Assay
4.9. Coimmunoprecipitation Assay
4.10. Protein Extraction and Immunoblotting Assay
4.11. Statistical Analyses
4.12. Accession Numbers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Brisson, N.; Gate, P.; Gouache, D.; Charmet, G.; Oury, F.-X.; Huard, F. Why are wheat yields stagnating in Europe? A comprehensive data analysis for France. Field Crops Res. 2010, 119, 201–212. [Google Scholar] [CrossRef]
- Li, N.; Xu, R.; Li, Y. Molecular Networks of Seed Size Control in Plants. Annu. Rev. Plant Biol. 2019, 70, 435–463. [Google Scholar] [CrossRef]
- Schmidt, C.J.; Thomas, T.C.; Levine, M.A.; Neer, E.J. Specificity of G-protein β and γ subunit interaction. J. Biol. Chem. 1992, 267, 13807–13810. [Google Scholar] [CrossRef]
- Trusov, Y.; Rookes, J.E.; Tilbrook, K.; Chakravorty, D.; Mason, M.G.; Anderson, D.; Chen, J.G.; Jones, A.M.; Botella, J.R. Heterotrimeric G protein gamma subunits provide functional selectivity in G beta gamma dimer signaling in Arabidopsis. Plant Cell 2007, 19, 1235–1250. [Google Scholar] [CrossRef] [Green Version]
- Trusov, Y.; Chakravorty, D.; Botella, J.R. Diversity of heterotrimeric G-protein gamma subunits in plants. BMC Res. Notes 2012, 5, 608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Liu, Y.; Zheng, L.; Chen, L.; Li, N.; Corke, F.; Lu, Y.; Fu, X.; Zhu, Z.; Bevan, M.W.; et al. The plant-specific G protein gamma subunit AGG3 influences organ size and shape in Arabidopsis thaliana. New Phytol. 2012, 194, 690–703. [Google Scholar] [CrossRef]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lin, T.; Meng, X.; Du, H.; Zhang, J.; Liu, G.; Chen, M.; Jing, Y.; Kou, L.; Li, X.; et al. A route to de novo domestication of wild allotetraploid rice. Cell 2021, 184, 1156–1170.e14. [Google Scholar] [CrossRef]
- Fan, C.; Yu, S.; Wang, C.; Xing, Y. A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor. Appl. Genet. 2009, 118, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Li, H.F.; Zhi, L.Y.; Su, Q.N.; Liu, J.J.; Ren, X.L.; Meng, D.Y.; Zhang, N.; Ji, J.; Zhang, X.Y.; et al. Functional markers developed from TaGS3, a negative regulator of grain weight and size, for marker-assisted selection in wheat. Crop J. 2020, 8, 943–952. [Google Scholar] [CrossRef]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, P.; Quilichini, T.D.; Zhai, C.; Qin, L.; Nilsen, K.T.; Li, Q.; Sharpe, A.G.; Kochian, L.V.; Zou, J.; Reddy, A.S.; et al. Alternative splicing dynamics and evolutionary divergence during embryogenesis in wheat species. Plant Biotechnol. J. 2021. [CrossRef]
- Syed, N.H.; Kalyna, M.; Marquez, Y.; Barta, A.; Brown, J.W. Alternative splicing in plants-coming of age. Trends Plant Sci. 2012, 17, 616–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Z.; Wu, J.; Geng, S.; Feng, N.; Chen, F.; Kong, X.; Song, G.; Chen, K.; Li, A.; Mao, L.; et al. Regulation of FT splicing by an endogenous cue in temperate grasses. Nat. Commun. 2017, 8, 14320. [Google Scholar] [CrossRef] [Green Version]
- Quesada, V.; Macknight, R.; Dean, C.; Simpson, G.G. Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J. 2003, 22, 3142–3152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pose, D.; Verhage, L.; Ott, F.; Yant, L.; Mathieu, J.; Angenent, G.C.; Immink, R.G.; Schmid, M. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 2013, 503, 414–417. [Google Scholar] [CrossRef]
- Seo, P.J.; Park, M.J.; Lim, M.H.; Kim, S.G.; Lee, M.; Baldwin, I.T.; Park, C.M. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 2012, 24, 2427–2442. [Google Scholar] [CrossRef] [Green Version]
- Howitt, C.A.; Cavanagh, C.R.; Bowerman, A.F.; Cazzonelli, C.; Rampling, L.; Mimica, J.L.; Pogson, B.J. Alternative splicing activation of cryptic exons and amino acid substitutions in carotenoid biosynthetic genes are associated with lutein accumulation in wheat endosperm. Funct. Integr. Genomics 2009, 9, 363–376. [Google Scholar] [CrossRef]
- Cyrek, M.; Fedak, H.; Ciesielski, A.; Guo, Y.; Sliwa, A.; Brzezniak, L.; Krzyczmonik, K.; Pietras, Z.; Kaczanowski, S.; Liu, F.; et al. Seed dormancy in Arabidopsis is controlled by alternative polyadenylation of DOG1. Plant Physiol. 2016, 170, 947–955. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Miao, J.; Zhang, Z.; Xiong, H.; Zhu, X.; Sun, X.; Pan, Y.; Liang, Y.; Zhang, Q.; Abdul Rehman, R.M.; et al. Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol. J. 2018, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Y.H.; Gao, L.J.; Liang, Y.T.; Zhao, Y.; Liang, H.F.; Chen, W.W.; Yang, X.H.; Qing, D.J.; Gao, J.; Wu, H.; et al. OrMKK3 Influences Morphology and Grain Size in Rice. J. Plant Biol. 2021, 1–14. [Google Scholar] [CrossRef]
- Brenchley, R.; Spannagl, M.; Pfeifer, M.; Barker, G.L.; D’Amore, R.; Allen, A.M.; McKenzie, N.; Kramer, M.; Kerhornou, A.; Bolser, D.; et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 2012, 491, 705–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, K.; Feng, M.; Yang, G.; Sun, L.; Qin, Z.; Cao, J.; Wen, J.; Li, H.; Zhou, Y.; Chen, X.; et al. Changes in alternative splicing in response to domestication and polyploidization in Wheat. Plant Physiol. 2020, 184, 1955–1968. [Google Scholar] [CrossRef]
- Sun, Y.; He, Z.; Ma, W.; Xia, X. Alternative splicing in the coding region of Ppo-A1 directly influences the polyphenol oxidase activity in common wheat (Triticum aestivum L.). Funct. Integr. Genomics 2011, 11, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.J.; Yang, Q.; Zhong, X.J.; Tang, H.P.; Deng, M.; Ma, J.; Qi, P.F.; Wang, J.R.; Chen, G.Y.; Liu, Y.X.; et al. Alternative splicing results in a lack of starch synthase IIa-D in Chinese wheat landrace. Genome 2018, 61, 201–208. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Mao, R.; Wang, Y.; Zhang, L.; Wang, C.; Lv, S.; Liu, X.; Wang, Y.; Ji, W. Transcriptome-wide alternative splicing modulation during plant-pathogen interactions in wheat. Plant Sci. 2019, 288, 110160. [Google Scholar] [CrossRef]
- Egawa, C.; Kobayashi, F.; Ishibashi, M.; Nakamura, T.; Nakamura, C.; Takumi, S. Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes Genet. Syst. 2006, 81, 77–91. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Qin, J.; Tian, X.; Xu, S.; Wang, Y.; Li, H.; Wang, X.; Peng, H.; Yao, Y.; Hu, Z.; et al. Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.). Plant Biotechnol. J. 2018, 16, 714–726. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Zhao, P.; Zhang, A.; Ma, L.; Xu, S.; Wang, X. Alternative splicing diversified the heat response and evolutionary strategy of conserved heat shock protein 90s in Hexaploid Wheat (Triticum aestivum L.). Front Genet. 2020, 11, 577897. [Google Scholar] [CrossRef]
- Gracheva, E.O.; Cordero-Morales, J.F.; Gonzalez-Carcacia, J.A.; Ingolia, N.T.; Manno, C.; Aranguren, C.I.; Weissman, J.S.; Julius, D. Ganglion-specific splicing of TRPV1 underlies infrared sensation in vampire bats. Nature 2011, 476, 88–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Temple, B.R.; Jones, A.M. The plant heterotrimeric G-protein complex. Annu. Rev. Plant Biol. 2007, 58, 249–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullah, H.; Chen, J.G.; Young, J.C.; Im, K.H.; Sussman, M.R.; Jones, A.M. Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 2001, 292, 2066–2069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urano, D.; Jones, A.M. Heterotrimeric G protein-coupled signaling in plants. Annu. Rev. Plant Biol. 2014, 65, 365–384. [Google Scholar] [CrossRef] [Green Version]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef]
- Marquez, Y.; Brown, J.W.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef] [Green Version]
- Reddy, A.S.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 3657–3683. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.B.; Brendel, V. Genome wide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA 2006, 103, 7175–7180. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Regulski, M.; Tseng, E.; Olson, A.; Goodwin, S.; McCombie, W.R.; Ware, D. A comparative transcriptional landscape of maize and sorghum obtained by single-molecule sequencing. Genome Res. 2018, 28, 921–932. [Google Scholar] [CrossRef] [Green Version]
- Wendel, J.F. Genome evolution in polyploids. Plant Mol. Biol. 2000, 42, 225–249. [Google Scholar] [CrossRef]
- Hu, Z.; Han, Z.; Song, N.; Chai, L.; Yao, Y.; Peng, H.; Ni, Z.; Sun, Q. Epigenetic modification contributes to the expression divergence of three TaEXPA1 homoeologs in hexaploid wheat (Triticum aestivum). New Phytol. 2013, 197, 1344–1352. [Google Scholar] [CrossRef]
- Shitsukawa, N.; Tahira, C.; Kassai, K.; Hirabayashi, C.; Shimizu, T.; Takumi, S.; Mochida, K.; Kawaura, K.; Ogihara, Y.; Murai, K. Genetic and epigenetic alteration among three homoeologous genes of a class E MADS box gene in hexaploid wheat. Plant Cell 2007, 19, 1723–1737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Fan, X.; Gao, Y.; Liu, L.; Sun, L.; Su, Q.; Han, J.; Zhang, N.; Cui, F.; Ji, J.; et al. Chromatin modification contributes to the expression divergence of three TaGS2 homoeologs in hexaploid wheat. Sci. Rep. 2017, 7, 44677. [Google Scholar] [CrossRef]
- Wang, Z.; Ji, H.; Yuan, B.; Wang, S.; Su, C.; Yao, B.; Zhao, H.; Li, X. ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat. Commun. 2015, 6, 8138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, S.; Wang, L.; Mao, H.; Shao, L.; Li, X.; Xiao, J.; Ouyang, Y.; Zhang, Q. A G-protein pathway determines grain size in rice. Nat. Commun. 2018, 9, 851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Liu, J.; Zhang, Y.; Yang, Z.; Gao, C. Biolistic genetic transformation of a wide range of Chinese elite wheat (Triticum aestivum L.) varieties. J. Genet. Genomics 2015, 42, 39–42. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, X.; Zhi, L.; Liu, L.; Meng, D.; Su, Q.; Batool, A.; Ji, J.; Song, L.; Zhang, N.; Guo, L.; et al. Alternative Splicing of TaGS3 Differentially Regulates Grain Weight and Size in Bread Wheat. Int. J. Mol. Sci. 2021, 22, 11692. https://doi.org/10.3390/ijms222111692
Ren X, Zhi L, Liu L, Meng D, Su Q, Batool A, Ji J, Song L, Zhang N, Guo L, et al. Alternative Splicing of TaGS3 Differentially Regulates Grain Weight and Size in Bread Wheat. International Journal of Molecular Sciences. 2021; 22(21):11692. https://doi.org/10.3390/ijms222111692
Chicago/Turabian StyleRen, Xiaoli, Liya Zhi, Lei Liu, Deyuan Meng, Qiannan Su, Aamana Batool, Jun Ji, Liqiang Song, Na Zhang, Lin Guo, and et al. 2021. "Alternative Splicing of TaGS3 Differentially Regulates Grain Weight and Size in Bread Wheat" International Journal of Molecular Sciences 22, no. 21: 11692. https://doi.org/10.3390/ijms222111692
APA StyleRen, X., Zhi, L., Liu, L., Meng, D., Su, Q., Batool, A., Ji, J., Song, L., Zhang, N., Guo, L., Liu, X., Li, J., & Zhang, W. (2021). Alternative Splicing of TaGS3 Differentially Regulates Grain Weight and Size in Bread Wheat. International Journal of Molecular Sciences, 22(21), 11692. https://doi.org/10.3390/ijms222111692





