Djnedd4L Is Required for Head Regeneration by Regulating Stem Cell Maintenance in Planarians
Abstract
:1. Background
2. Results
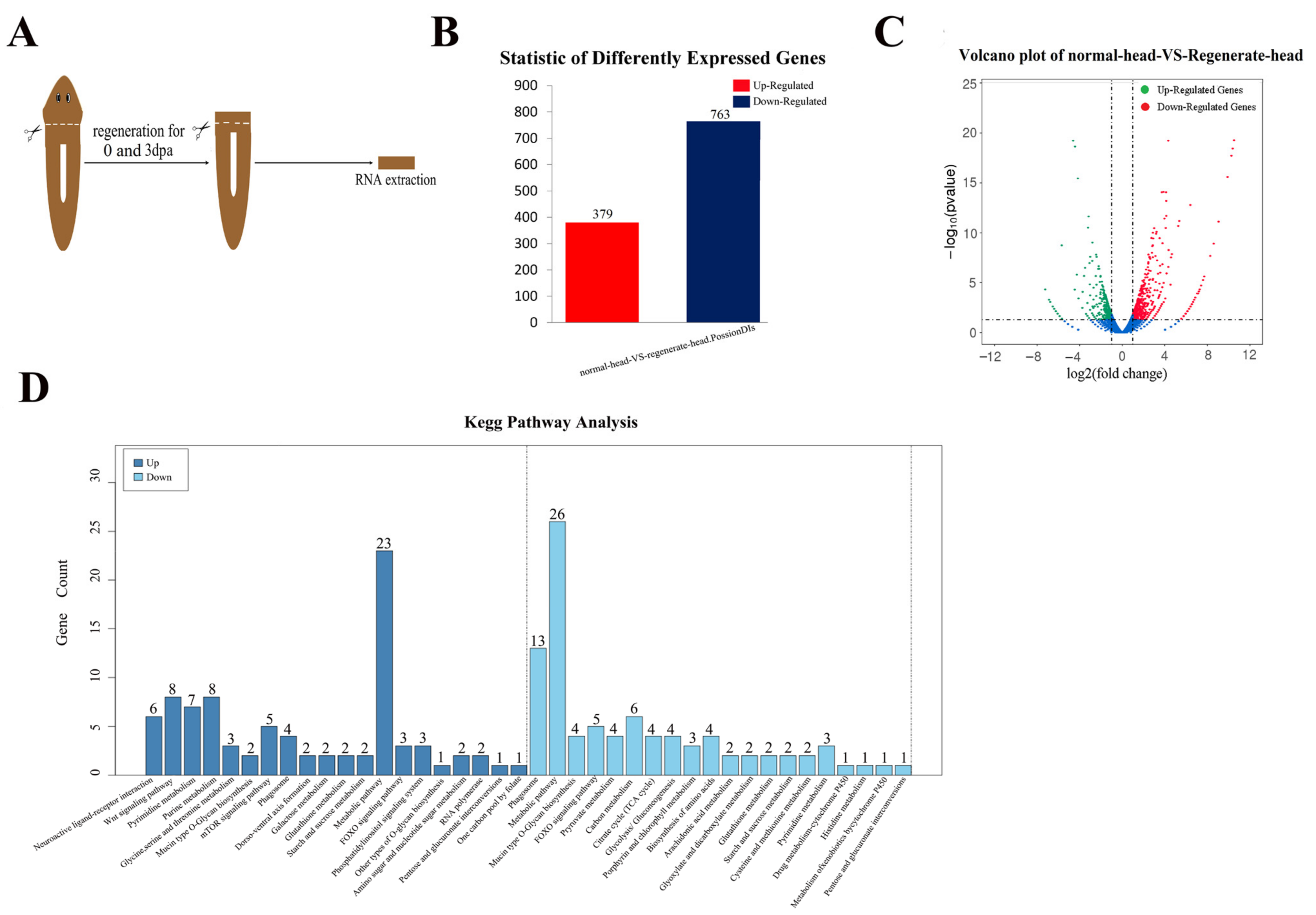
2.1. Transcriptome Analysis Clarified the Genes Involved in Head Regeneration in the Planarian Dugesia japonica
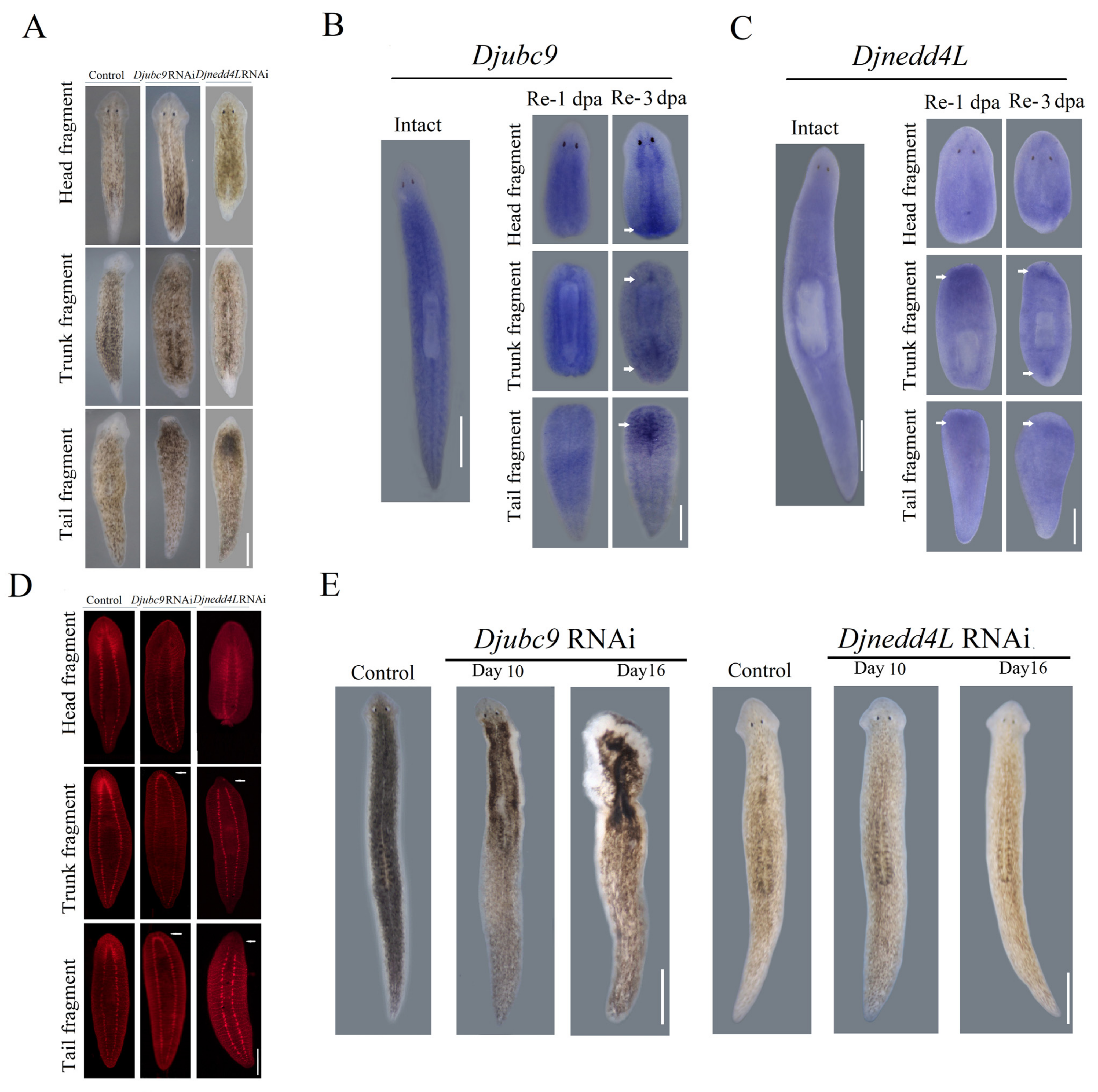
2.2. Djnedd4L and Djubc9 RNAi Cause Head Regeneration Defects
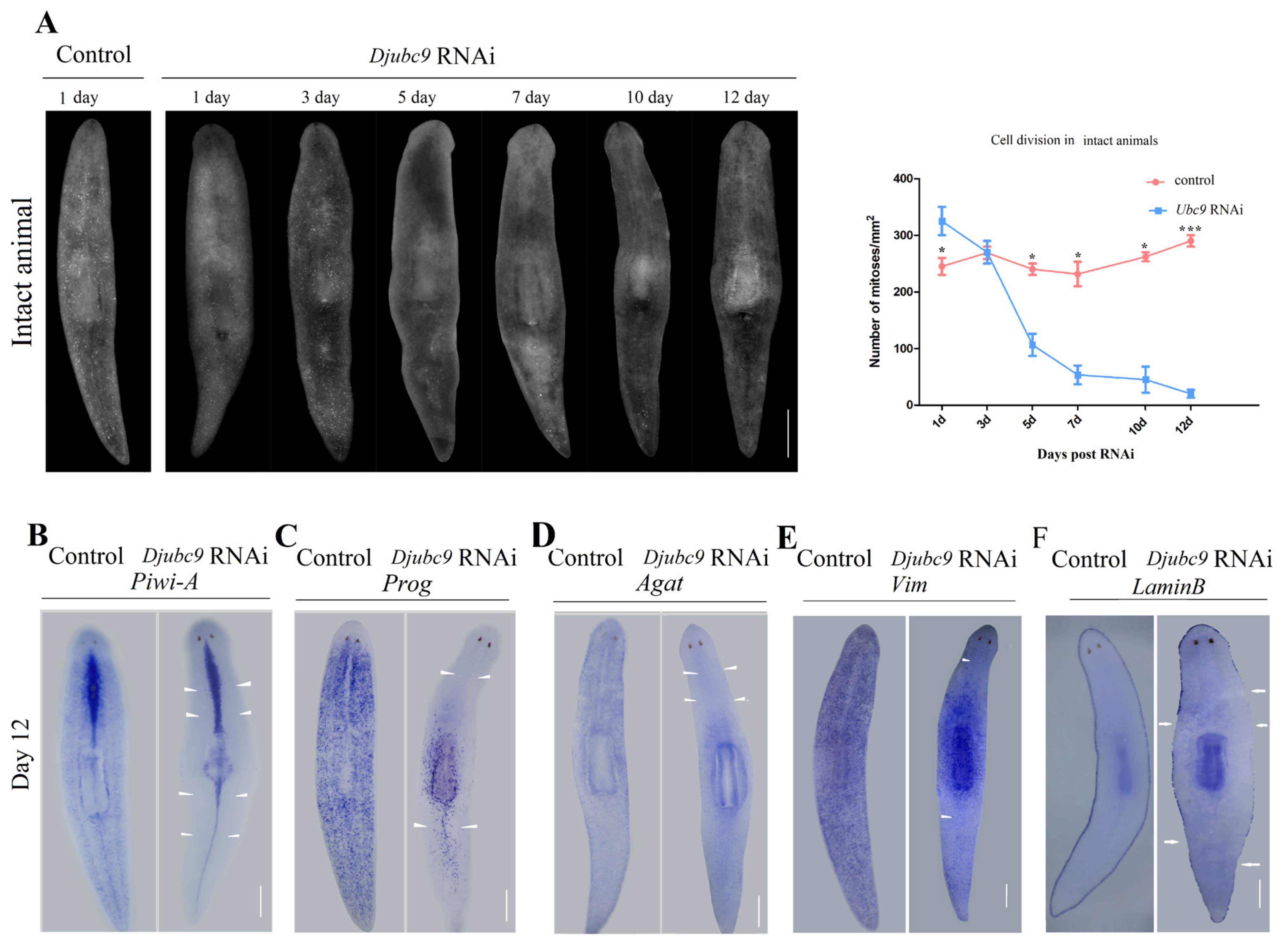
2.3. Djubc9 (RNAi) Phenotype Defects Indicate That Djubc9 Is Required for the Maintenance of Stem Cells
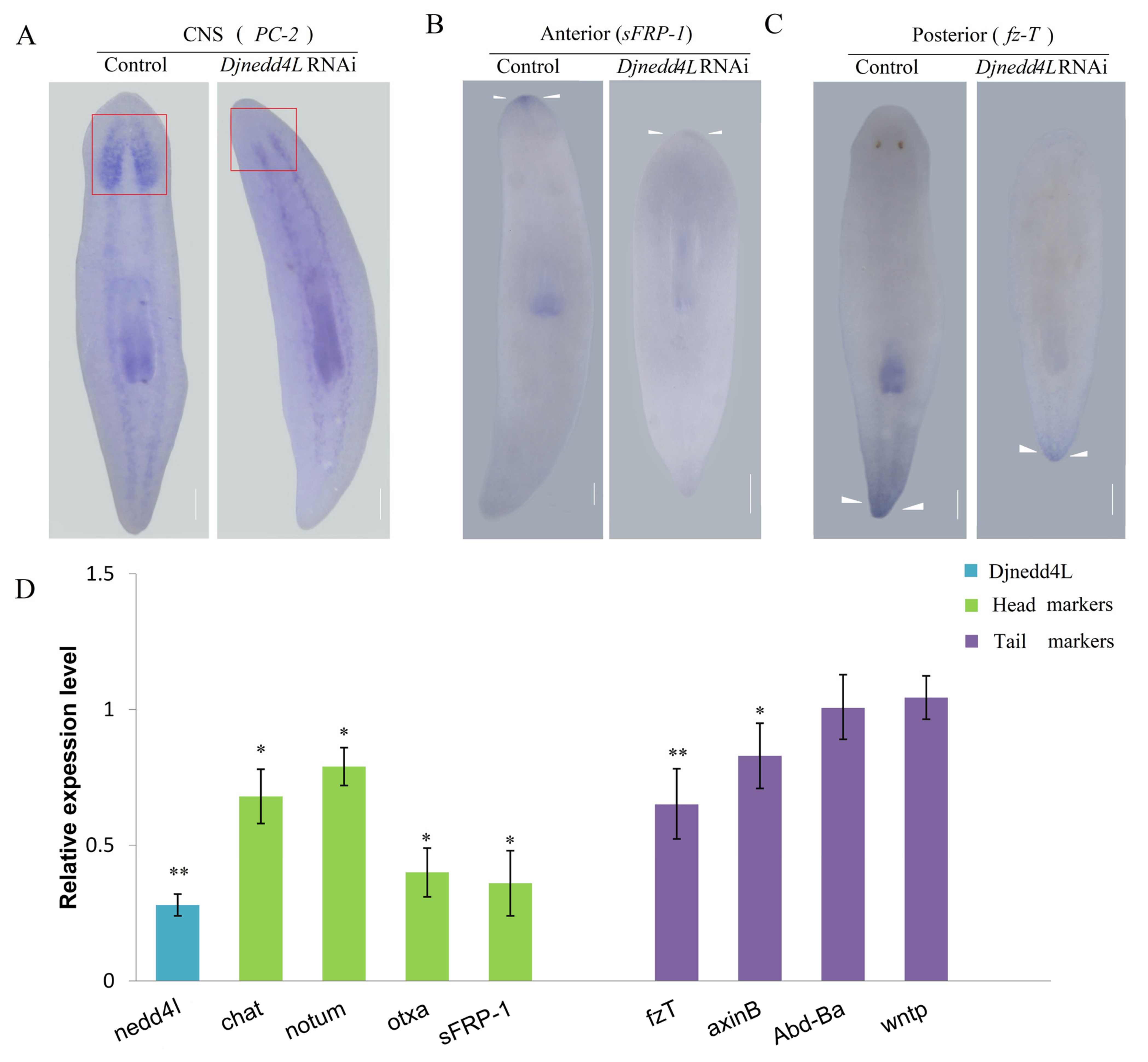
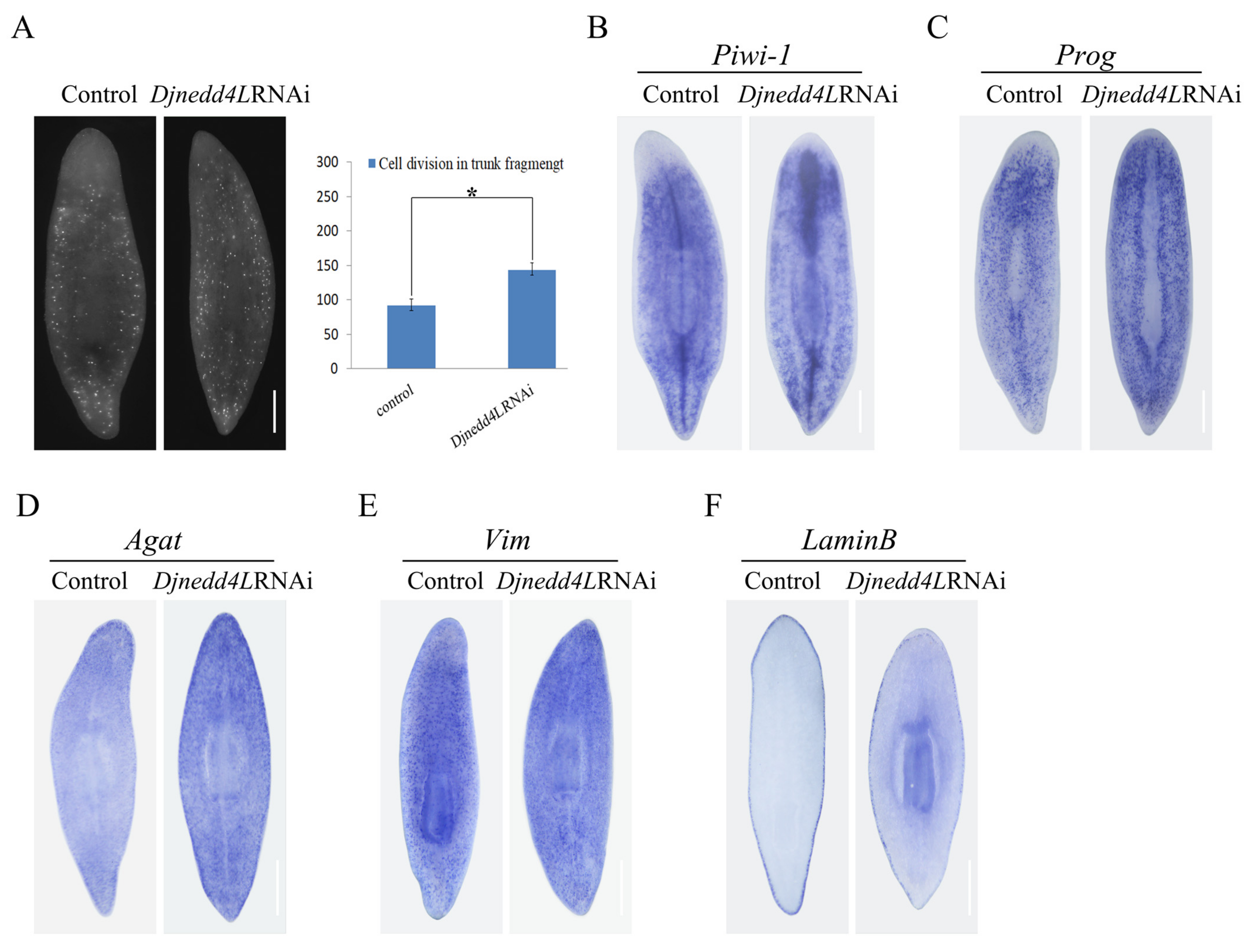
2.4. The Effect Djnedd4L (RNAi) on Cell Division and Differentiation during Regeneration
3. Discussion
4. Materials and Methods
4.1. Planarian Culture
4.2. cDNA Clones and RNAi Experiments
4.3. In Situ Hybridization
4.4. Whole-Mount Immunostaining
4.5. Quantitative Real-Time PCR Analysis of Gene Expression
4.6. Sample Collection and Head Regeneration RNA-Seq
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, C.; Matesic, L.E. The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 2007, 26, 587–604. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [Green Version]
- Tsuchida, S.; Satoh, M.; Takiwaki, M.; Nomura, F. Ubiquitination in Periodontal Disease: A Review. Int. J. Mol. Sci. 2017, 18, 1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strand, N.S.; Allen, J.M.; Zayas, R.M. Post-translational regulation of planarian regeneration. Semin. Cell Dev. Biol. 2018, 87, 58–68. [Google Scholar] [CrossRef]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010, 11, 861–871. [Google Scholar] [CrossRef] [Green Version]
- Geiss-Friedlander, R.; Melchior, F. Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell Biol. 2007, 8, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Hay, R.T. SUMO-specific proteases: A twist in the tail. Trends Cell Biol. 2007, 17, 370–376. [Google Scholar] [CrossRef]
- Kim, K.I.; Baek, S.H. Small ubiquitin-like modifiers in cellular malignancy and metastasis. Int. Rev. Cell Mol. Biol. 2009, 273, 265–311. [Google Scholar] [PubMed]
- Newmark, P.A.; Sanchez Alvarado, A. Not your father’s planarian: A classic model enters the era of functional genomics. Nat. Rev. Genet. 2002, 3, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Reddien, P.W.; Sanchez Alvarado, A. Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 2004, 20, 725–757. [Google Scholar] [CrossRef] [Green Version]
- Salo, E. The power of regeneration and the stem-cell kingdom: Freshwater planarians (Platyhelminthes). Bioessays 2006, 28, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Rossi, L.; Salvetti, A. Planarian stem cell niche, the challenge for understanding tissue regeneration. Semin Cell Dev. Biol. 2018, 87, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, T.; Vogg, M.C.; Owlarn, S.; Boutros, M.; Bartscherer, K. The head-regeneration transcriptome of the planarian Schmidtea mediterranea. Genome Biol. 2011, 12, R76. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, E.M.; Reddien, P.W. The cellular basis for animal regeneration. Dev. Cell 2011, 21, 172–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, D.E.; Wang, I.E.; Reddien, P.W. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 2011, 332, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.E.; Ho, J.J.; Reddien, P.W. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem. Cell 2012, 10, 299–311. [Google Scholar] [CrossRef] [Green Version]
- Wenemoser, D.; Lapan, S.W.; Wilkinson, A.W.; Bell, G.W.; Reddien, P.W. A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 2012, 26, 988–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malinowski, P.T.; Cochet-Escartin, O.; Kaj, K.J.; Ronan, E.; Groisman, A.; Diamond, P.H.; Collins, E.S. Mechanics dictate where and how freshwater planarians fission. Proc. Natl. Acad. Sci. USA 2017, 114, 10888–10893. [Google Scholar] [CrossRef] [Green Version]
- Scimone, M.L.; Cote, L.E.; Reddien, P.W. Orthogonal muscle fibres have different instructive roles in planarian regeneration. Nature 2017, 551, 623–628. [Google Scholar] [CrossRef]
- Cebria, F.; Kobayashi, C.; Umesono, Y.; Nakazawa, M.; Mineta, K.; Ikeo, K.; Gojobori, T.; Itoh, M.; Taira, M.; Sanchez Alvarado, A.; et al. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature 2002, 419, 620–624. [Google Scholar] [CrossRef]
- Cebria, F.; Newmark, P.A. Morphogenesis defects are associated with abnormal nervous system regeneration following roboA RNAi in planarians. Development 2007, 134, 833–837. [Google Scholar] [CrossRef] [Green Version]
- Pearson, B.J.; Sanchez Alvarado, A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development 2010, 137, 213–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenhoffer, G.T.; Kang, H.; Sanchez Alvarado, A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell 2008, 3, 327–339. [Google Scholar] [CrossRef] [Green Version]
- Tu, K.C.; Cheng, L.C.; Vu, H.T.; Lange, J.J.; McKinney, S.A.; Seidel, C.W.; Sanchez Alvarado, A. Egr-5 is a post-mitotic regulator of planarian epidermal differentiation. eLife 2015, 4, e10501. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.J.; Hallows, S.E.; Currie, K.W.; Xu, C.; Pearson, B.J. A mex3 homolog is required for differentiation during planarian stem cell lineage development. eLife 2015, 4, e07025. [Google Scholar] [CrossRef] [PubMed]
- Van Wolfswinkel, J.C.; Wagner, D.E.; Reddien, P.W. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell 2014, 15, 326–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wurtzel, O.; Oderberg, I.M.; Reddien, P.W. Planarian Epidermal Stem Cells Respond to Positional Cues to Promote Cell-Type Diversity. Dev. Cell 2017, 40, 491–504.e5. [Google Scholar] [CrossRef] [Green Version]
- Rossi, L.; Bonuccelli, L.; Iacopetti, P.; Evangelista, M.; Ghezzani, C.; Tana, L.; Salvetti, A. Prohibitin 2 regulates cell proliferation and mitochondrial cristae morphogenesis in planarian stem cells. Stem Cell Rev. 2014, 10, 871–887. [Google Scholar] [CrossRef] [PubMed]
- Rossi, L.; Salvetti, A.; Lena, A.; Batistoni, R.; Deri, P.; Pugliesi, C.; Loreti, E.; Gremigni, V. DjPiwi-1, a member of the PAZ-Piwi gene family, defines a subpopulation of planarian stem cells. Dev. Genes Evol. 2006, 216, 335–346. [Google Scholar] [CrossRef]
- Tian, Q.; Zhao, G.; Sun, Y.; Yuan, D.; Guo, Q.; Zhang, Y.; Liu, J.; Zhang, S. Exportin-1 is required for the maintenance of the planarian epidermal lineage. Int. J. Biol. Macromol. 2019, 126, 1050–1055. [Google Scholar] [CrossRef]
- Shibata, N.; Hayashi, T.; Fukumura, R.; Fujii, J.; Kudome-Takamatsu, T.; Nishimura, O.; Sano, S.; Son, F.; Suzuki, N.; Araki, R.; et al. Comprehensive gene expression analyses in pluripotent stem cells of a planarian, Dugesia japonica. Int. J. Dev. Biol. 2012, 56, 93–102. [Google Scholar] [CrossRef] [Green Version]
- Durant, F.; Morokuma, J.; Fields, C.; Williams, K.; Adams, D.S.; Levin, M. Long-Term, Stochastic Editing of Regenerative Anatomy via Targeting Endogenous Bioelectric Gradients. Biophys. J. 2017, 112, 2231–2243. [Google Scholar] [CrossRef] [Green Version]
- Gurley, K.A.; Rink, J.C.; Sanchez Alvarado, A. Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 2008, 319, 323–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reuter, H.; Marz, M.; Vogg, M.C.; Eccles, D.; Grifol-Boldu, L.; Wehner, D.; Owlarn, S.; Adell, T.; Weidinger, G.; Bartscherer, K. Beta-catenin-dependent control of positional information along the AP body axis in planarians involves a teashirt family member. Cell Rep. 2015, 10, 253–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owen, J.H.; Wagner, D.E.; Chen, C.C.; Petersen, C.P.; Reddien, P.W. teashirt is required for head-versus-tail regeneration polarity in planarians. Development 2015, 142, 1062–1072. [Google Scholar] [CrossRef] [Green Version]
- Petersen, C.P.; Reddien, P.W. Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 2011, 332, 852–855. [Google Scholar] [CrossRef]
- Iglesias, M.; Almuedo-Castillo, M.; Aboobaker, A.A.; Salo, E. Early planarian brain regeneration is independent of blastema polarity mediated by the Wnt/beta-catenin pathway. Dev. Biol. 2011, 358, 68–78. [Google Scholar] [CrossRef] [Green Version]
- Yazawa, S.; Umesono, Y.; Hayashi, T.; Tarui, H.; Agata, K. Planarian Hedgehog/Patched establishes anterior-posterior polarity by regulating Wnt signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 22329–22334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adell, T.; Salo, E.; Boutros, M.; Bartscherer, K. Smed-Evi/Wntless is required for beta-catenin-dependent and -independent processes during planarian regeneration. Development 2009, 136, 905–910. [Google Scholar] [CrossRef] [Green Version]
- Nogi, T.; Watanabe, K. Position-specific and non-colinear expression of the planarian posterior (Abdominal-B-like) gene. Dev. Growth Differ. 2001, 43, 177–184. [Google Scholar] [CrossRef]
- Thiruvalluvan, M.; Barghouth, P.G.; Tsur, A.; Broday, L.; Oviedo, N.J. SUMOylation controls stem cell proliferation and regional cell death through Hedgehog signaling in planarians. Cell Mol. Life Sci. 2018, 75, 1285–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, J.M.; Nisperos, S.V.; Weeks, J.; Ghulam, M.; Marin, I.; Zayas, R.M. Identification of HECT E3 ubiquitin ligase family genes involved in stem cell regulation and regeneration in planarians. Dev. Biol. 2015, 404, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Strand, N.S.; Allen, J.M.; Ghulam, M.; Taylor, M.R.; Munday, R.K.; Carrillo, M.; Movsesyan, A.; Zayas, R.M. Dissecting the function of Cullin-RING ubiquitin ligase complex genes in planarian regeneration. Dev. Biol. 2018, 433, 210–217. [Google Scholar] [CrossRef]
- Novellasdemunt, L.; Kucharska, A.; Jamieson, C.; Prange-Barczynska, M.; Baulies, A.; Antas, P.; van der Vaart, J.; Gehart, H.; Maurice, M.M.; Li, V.S. NEDD4 and NEDD4L regulate Wnt signalling and intestinal stem cell priming by degrading LGR5 receptor. EMBO J. 2020, 39, e102771. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhang, Y.; Xu, C.; Tao, Q.H.; Chen, Y.G. HECT domain-containing E3 ubiquitin ligase NEDD4L negatively regulates Wnt signaling by targeting dishevelled for proteasomal degradation. J. Biol. Chem. 2013, 288, 8289–8298. [Google Scholar] [CrossRef] [Green Version]
- Reddien, P.W. Positional Information and Stem Cells Combine to Result in Planarian Regeneration. Cold Spring Harb. Perspect Biol. 2021, a040717. [Google Scholar] [CrossRef]
- Tian, Q.N.; Bao, Z.X.; Lu, P.; Qin, Y.F.; Chen, S.J.; Liang, F.; Mai, J.; Zhao, J.M.; Zhu, Z.Y.; Zhang, Y.Z.; et al. Differential expression of microRNA patterns in planarian normal and regenerative tissues. Mol. Biol. Rep. 2012, 39, 2653–2658. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Zhao, G.; Ni, J.; Guo, Y.; Zhang, Y.; Tian, Q.; Zhang, S. Down-regulate of Djrfc2 causes tissues hypertrophy during planarian regeneration. Biochem. Biophys. Res. Commun. 2017, 493, 1224–1229. [Google Scholar] [CrossRef]
- Rouhana, L.; Weiss, J.A.; Forsthoefel, D.J.; Lee, H.; King, R.S.; Inoue, T.; Shibata, N.; Agata, K.; Newmark, P.A. RNA interference by feeding in vitro-synthesized double-stranded RNA to planarians: Methodology and dynamics. Dev. Dyn 2013, 242, 718–730. [Google Scholar] [CrossRef] [Green Version]
- Pearson, B.J.; Eisenhoffer, G.T.; Gurley, K.A.; Rink, J.C.; Miller, D.E.; Sanchez Alvarado, A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn 2009, 238, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Cebria, F.; Newmark, P.A. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development 2005, 132, 3691–3703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Y.F.; Fang, H.M.; Tian, Q.N.; Bao, Z.X.; Lu, P.; Zhao, J.M.; Mai, J.; Zhu, Z.Y.; Shu, L.L.; Zhao, L.; et al. Transcriptome profiling and digital gene expression by deep-sequencing in normal/regenerative tissues of planarian Dugesia japonica. Genomics 2011, 97, 364–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Q.; Sun, Y.; Gao, T.; Li, J.; Fang, H.; Zhang, S. Djnedd4L Is Required for Head Regeneration by Regulating Stem Cell Maintenance in Planarians. Int. J. Mol. Sci. 2021, 22, 11707. https://doi.org/10.3390/ijms222111707
Tian Q, Sun Y, Gao T, Li J, Fang H, Zhang S. Djnedd4L Is Required for Head Regeneration by Regulating Stem Cell Maintenance in Planarians. International Journal of Molecular Sciences. 2021; 22(21):11707. https://doi.org/10.3390/ijms222111707
Chicago/Turabian StyleTian, Qingnan, Yujia Sun, Tingting Gao, Jiaxin Li, Huimin Fang, and Shoutao Zhang. 2021. "Djnedd4L Is Required for Head Regeneration by Regulating Stem Cell Maintenance in Planarians" International Journal of Molecular Sciences 22, no. 21: 11707. https://doi.org/10.3390/ijms222111707
APA StyleTian, Q., Sun, Y., Gao, T., Li, J., Fang, H., & Zhang, S. (2021). Djnedd4L Is Required for Head Regeneration by Regulating Stem Cell Maintenance in Planarians. International Journal of Molecular Sciences, 22(21), 11707. https://doi.org/10.3390/ijms222111707




