Abstract
This paper estimates the yields of DNA double-strand breaks (DSBs) induced by ultrasoft X-rays and uses the DSB yields and the repair outcomes to evaluate the relative biological effectiveness (RBE) of ultrasoft X-rays. We simulated the yields of DSB induction and predicted them in the presence and absence of oxygen, using a Monte Carlo damage simulation (MCDS) software, to calculate the RBE. Monte Carlo excision repair (MCER) simulations were also performed to calculate the repair outcomes (correct repairs, mutations, and DSB conversions). Compared to 60Co γ-rays, the RBE values for ultrasoft X-rays (titanium K-shell, aluminum K-shell, copper L-shell, and carbon K-shell) for DSB induction were respectively 1.3, 1.9, 2.3, and 2.6 under aerobic conditions and 1.3, 2.1, 2.5, and 2.9 under a hypoxic condition (2% O2). The RBE values for enzymatic DSBs were 1.6, 2.1, 2.3, and 2.4, respectively, indicating that the enzymatic DSB yields are comparable to the yields of DSB induction. The synergistic effects of DSB induction and enzymatic DSB formation further facilitate cell killing and the advantage in cancer treatment.
1. Introduction
The relative biological effectiveness (RBE) depends on photon energy. In particular, RBE of ultrasoft X-rays (<20 keV) increases with decreasing energy [1,2,3]. Ultrasoft X-rays are more effective up to a factor of 4 than 250 keV X-rays with regard to double-strand break (DSB) yield, cell death, and mutations [4,5,6]. The higher RBE is on account of the short traveling distances (~7 nm) of the secondary electrons [7], which is similar to the width of DNA helix. These very-low-energy electrons which produce through photoelectric interactions or secondary electrons scattering [8] cause highly clustered energy depositions and are the predominant contributor to complex DNA damage, which leads to an increment in genetic mutation, DNA damage yield, and apoptosis.
DSBs can be directly induced by energy deposition or from the outcome of a mis-repairing process dealing with non-DSB clustered lesions via base excision repair (BER) pathways [9,10] and nucleotide excision repair (NER) pathways [11,12,13], known as enzymatic DSBs. Monte Carlo simulations and experiments performed on cell lines demonstrate that the yield of enzymatic DSB is comparable to the yield of direct DSB induction, which is also known as prompt DSB yield [14,15]. These extra DSBs that are formed after the mis-repair process contribute further to the DSB yield and result in a higher RBE value [16]. In these cases, the greater numbers of DSBs and damage clusters increase the DNA damage complexity [17].
Most clustered damage on DNA involves the presence of O2 [17]. When clustered DNA damage occurs under aerobic conditions, it is firstly induced by free radicals and then modified by dissolved O2. Thus, if O2 is present when the DNA damage occurs, the damage is fixed and cannot be reversible repaired by chemical repair by thiols, which is widely regarded as the oxygen fixation hypothesis [18]. However, the O2 does not fix the damage directly; rather, it modifies the pathway and final chemical products [19].
In general, ultrasoft X-rays are not used for conventional X-ray therapy (XT) because they penetrate to a shallower depth [7]. However, they are used for dermatological radiotherapy, known as Grenz therapy (GT) [20]. GT is used to reduce allergic skin conditions such as eczema, contact dermatitis and psoriasis, etc. [20,21]. It can also be used to treat malignant cells such as lentigo maligna (LM) and LM melanoma [22]. In contrast to XT, GT has very few side effects, and its most adverse effects are erythema and pigmentation [23]. Since 1959, however, GT has been reported to induce carcinoma, and the pattern of tumor development is similar to that resulting from exposure to ultraviolet light [23].
For cells that are irradiated by GT, the processes of DSB yield are almost exclusively due to the low-energy secondary electrons [24], and the data obtained from simulations of GT or low-energy secondary electrons [25,26] show that these electrons could produce higher yields of complex DSB damage than those induced by XT, leading to other varieties of DSB [27,28]. Here, we present a study covering DSB induction, DNA repair, and the effects of O2 on damage yields for cells irradiated by GT and further investigate the RBE of DSB induction and DNA repair for cells irradiated by GT.
2. Materials and Methods
2.1. Irradiation Geometries and Materials
A cylindrical geometry has been used to mimic the typical cell culture experiments [29,30,31]. The cylinder was designed with a radius of 26 mm and a height of 10 μm (the thickness of a mammalian cell) to mimic a monolayer cell culture geometry. The photon sources were put 5 μm above the cells and were set like a pencil beam incident on the cylinder. The cell was approximated by water at a density of 1.0 g cm−3, as used in a previous study [29].
2.2. Determination of Secondary-Electron Fluence
The PENCYL program in the version 2011 penetration and energy loss of positrons and electron (PENELOPE) Monte Carlo code [32,33,34] was used to calculate the fluence of secondary electrons passing over single cells. All simulations were performed with at least 2 × 108 source particle histories, using the 2011 version of PENELOPE. All primary and secondary electrons were followed down to 50 eV [35]. PENELOPE uses a scattering model [32] that merges analytical cross-sectional models for different interaction mechanisms with numerical databases. The modified version of PENELOPE 2011 uses a more realistic continuous energy-loss distribution to replace the previous discrete optical oscillator-strength model, which increases both the reliability and the generality of the code.
The simulation of electron transport processes in PENELOPE was controlled by the parameters C1, C2, WCC, and WCR whose definitions and roles were explained previously [36]. Briefly, C1 controls the average angular deflection that is produced by multiple elastic scattering of electrons along the step between hard events and C2 controls the maximum average fractional energy loss in the step. WCC and WCR respectively represent the cut-off energy loss for hard inelastic collisions and for hard Bremsstrahlung emission. The values of parameters in this study were set as C1 = 0.00, C2 = 0.01, WCC = 10 eV, and WCR = 10 eV, as suggested for the simulation of very low energy (0.1–10 keV) electrons in microdosimetry applications [37].
2.3. Monte Carlo Damage Simulation (MCDS)
The MCDS is a fast algorithm that calculates the yield of clustered damage within a cell that is irradiated with photons, mono-energetic electrons, protons, or heavy ions, such as 56Fe [27,38,39]. The MCDS software is free and available online for commercial, educational, or research purposes [39] and simulates the induction and clustering of DNA lesions in both hypoxic and normoxic cells (>21% O2) [27,38,39].
The MCDS is not a track structure simulation code and its algorithm generates DNA damage data similar to those using computationally-expensive but detailed event-by-event simulations. The types of DNA damage can be grouped into base damage (BD), single strand breaks (SSBs), and DSB. Various complex SSB or DSB, i.e., SSB+, 2SSB, DSB+ and DSB++, and total SSB and DSB were defined previously [40]. Briefly, the types and classifications of DNA damage and their abbreviations are: two or more strand breaks on the same strand (SSB+), two or more strand breaks on opposite strands that do not constitute DSB (2SSB), DSBs with additional break(s) on a strand within 10 base pairs (DSB+), and more than one DSB within 10 base pairs (DSB++).
2.4. Monte Carlo Excision Repair (MCER) Simulation
The MCER simulation was used to estimate the probabilities of the repair outcomes in the BER and BER/NER pathways for DNA damage in cells that are irradiated with electrons, protons, and helium ions [41,42] and has been used for several studies [43,44]. The MCER simulation simulated the key steps of repair pathways, i.e., short-patch BER (SP BER), long-patch BER (LP BER), SP BER/NER, and LP BER/NER [41,42]. A single nucleotide replacement generally occurs in the SP BER pathway and the LP BER pathway removes a fragment of 2–13 nucleotides. MCER output the probabilities of the repair outcomes that a DNA clustered damage repair correctly, repair with a mutation and converted into a DSB.
The following input parameters were used and are the same as those for a previous study [42]: inhibition distance = 8 base pairs, probability of selecting a lesion from the first strand break = 0.5, polymerase error for SP BER = 1.0−4, polymerase error for LP BER and for NER = 1.0−6, probability of incorrect insertion opposite a damaged base = 0.75, and probability of incorrect insertion opposite a lost base = 0.75. All four pathways (SP BER, LP BER, NER/SP BER, and NER/LP BER) were simulated. The results show that the NER/LP BER pathway has the lowest probability of correct repair and highest probability for mutation and DSB formation, which is similar to the results for 60Co γ-rays [44], protons [40], and helium ions [44]. These results are ascribed to the fact that the repair of non-DSB clusters occurs mainly through the BER pathway, but other bulk damage is mended by the NER pathway. Therefore, the LP BER pathway was selected to represent enzymatic DSB.
2.5. Calculation of DSB Conversion from DNA Damage
The enzymatic DSBs for ultrasoft X-rays and 60Co γ-rays were calculated with the dose-weighted formula [29]:
where Φ(E) is the energy fluence of secondary electrons of energy E that are generated in the cell medium from ultrasoft X-rays or 60Co photon interactions. The value for unrestricted LET (stopping power) from the National Institute of Standards and Technology is used for electrons with energy that is greater than 1 keV [45]. For energies below 1 keV, the LET was calculated by fitting to the data of Emfietzoglou and Nikjoo (2007) [46], i.e.,
where T is the electron energy in eV.
2.6. Hypoxia Reduction Factor (HRF) and RBE
The oxygen enhancement ratio is generally used to quantify the effects of oxygen concentration on DNA damage and cell killing and defined as the ratio of the hypoxic dose to the aerated dose that is required to achieve the same biological outcome [18]. Some studies also used it as a scaling factor to quantify the biological outcomes or radiation sensitivity of cells under hypoxia conditions [47,48]. To avoid inconsistency, we used another term, HRF, previously defined as the ratio of DSB yield under aerobic conditions (21% O2) to that under hypoxic conditions [47] to quantify the effects of oxygen concentration on DNA damage induction, i.e.,
where the DSB yield with the subscript a and h, respectively denote aerobic and hypoxia conditions.
The RBE is originally defined as the ratio of the dose of low LET reference radiation to the dose of any other radiation that is required to achieve an equal biological effect [18]. The RBE can also be interpreted as the ratio of the DSB yields of cells irradiated with different radiation sources because DSB induction is linearly proportional to the absorbed dose, D, up to a hundred Gy under aerobic condition (21% O2) and severe hypoxia (0.1% O2) [49,50], as shown below:
The subscripts, U and γ, respectively denote ultrasoft X-rays and γ-rays. The DSB yield for 60Co γ-rays is the reference for all reported RBE values.
3. Results
The RBE calculated from the measured DSB induction relative to 60Co γ-rays increased from 1.4 to 2.7 as the photon energy decreased from 4.55 keV to 280 eV [3]. The MCDS-derived RBE for DSB induction agreed well (within ±5.3%) with the RBE values using the measured DSB induction for photon energies of 280 eV to 1.49 keV (Table 1). The yield of more complicated types of DNA damage, such as DSB+ and DSB++, increased as photon energy decreased (Table 2), resulting in an increasing yield of total DSBs as photon energy decreased. In contrast, the yield for simpler types of damage, such as BD and SSBs, decreased as photon energy decreased, thus total SSBs and total damage decreased as photon energy decreased. The increase in the yields for more complicated types of DNA damage resulted in an increase in the DNA damage complexity as photon energy decreased.

Table 1.
Absolute yields of DSBs (per Gy per Gbp) induced by ultrasoft X-rays and 60Co γ-rays.

Table 2.
Absolute yields of DNA damage (per Gy per Gbp) induced by ultrasoft X-rays and 60Co γ-rays at a normal oxygen concentration (21%).
The complexity of the DNA damage was significantly affected by indirect action and oxygen. The yield of DNA damage was simulated at an oxygen concentration of 2% (Table 3) and lower (0.1% O2, Table 4). The limits of the standard deviations for all subtypes of DNA damage (i.e., BD, SSB, SSB+, 2SSB, DSB, DSB+, and DSB++) listed in Table 2, Table 3 and Table 4 are within 0.2%. The resulting yields of SSBs, DSBs, DSBs+, and DSBs++ of Table 3 and Table 4 were lower than those at an oxygen concentration of 21%, regardless of the level of photon energy (Table 2). There was no clear relationship between the yield of BD and oxygen concentration but the portion of BD of all energy levels are decreasing as oxygen concentration decreases. The yield of BD from copper K-shell and carbon K-shell photons at lower oxygen concentrations (≤2%) (Table 3 and Table 4) was slightly higher than that at an oxygen concentration of 21% (Table 2). As the oxygen concentration decreased to 2% or 0.1%, the RBE values estimated based on DSB induction relative to 60Co γ-rays were increasing to the range 1.3–2.9 and 1.4–4.4, respectively (Table 3 and Table 4).

Table 3.
Absolute yields of DNA damage (per Gy per Gbp) induced by ultrasoft X-rays and 60Co γ-rays under moderate hypoxia (2%).

Table 4.
Absolute yields of DNA damage (per Gy per Gbp) induced by ultrasoft X-rays and 60Co γ-rays under severe hypoxia (0.1%).
The RBE for DSB induction for cells irradiated with copper L-shell photons (0.96 keV) increased from 2.3 (Table 1) to 2.5 (8.6% increase; Table 3) and the RBE for cells irradiated with carbon K-shell photons (280 eV) increased from 2.7 to 2.9 (7% increase) as oxygen concentration decreases from 21% to 2%. Under severe hypoxia (0.1% O2), the RBE values for cells irradiated with copper L-shell and carbon K-shell photons further increase to 3.3 and 4.4, respectively. The RBE values for lower energy photons such as carbon K-shell photons (280 eV) under hypoxic conditions were greater, because there was a greater decrease in the yield of DSBs for 60Co γ-rays. The change in DSB yield for different oxygen concentrations is quantified using HRF, considering that carbon K-shell photons (280 eV) had a lower HRF than 60Co γ-rays (see Figure 1).
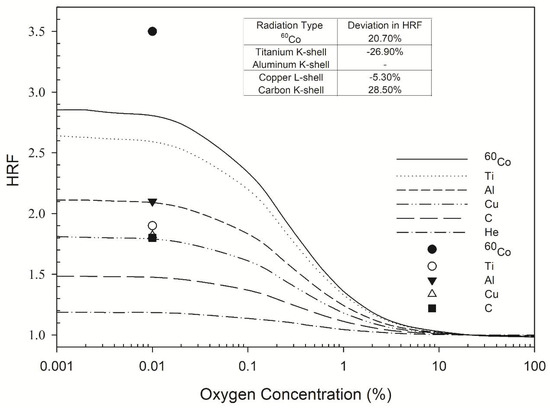
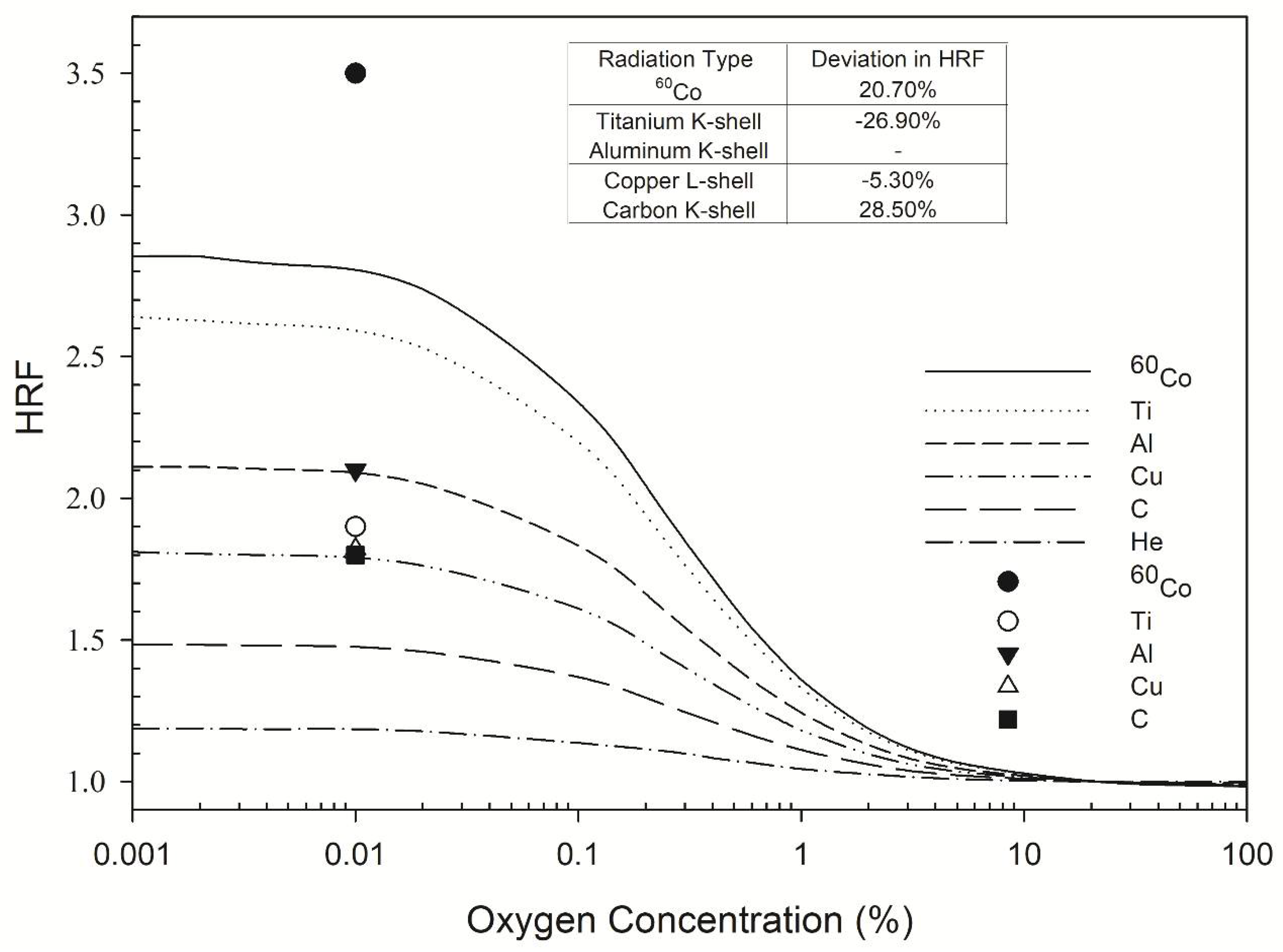
Figure 1.
HRF for DSB induction as a function of oxygen concentration. The symbols Ti, Al, Cu, and C represent titanium K-shell, aluminum K-shell, copper L-shell, and carbon K-shell radiation, respectively. The lines indicate the Monte Carlo damage simulation results (this work) and the individual symbols represent the experimental results of de Lara et al. (2001) [3].
The highest HRF values calculated by the MCDS for 60Co γ-rays, titanium K-shell, aluminum K-shell, copper L-shell, and carbon K-shell X-rays were 2.9, 2.6, 2.1, 1.9, and 1.4, respectively, for an oxygen concentration of less than 0.01% (Figure 1). All of these HRF values were constant within 0–0.1% O2 and gradually decreased as the oxygen concentration increased to 10% O2. The respective measured values of the HRF for 60Co γ-rays, titanium K-shell, aluminum K-shell, copper L-shell, and carbon K-shell were 3.5, 1.9, 2.1, 1.8, and 1.8 [3] for an oxygen concentration of less than 0.01% (Figure 1). The measured values of the HRF for ultrasoft X-rays with different energies differed from those calculated by the MCDS by up to 28.5% (Figure 1, inset table).
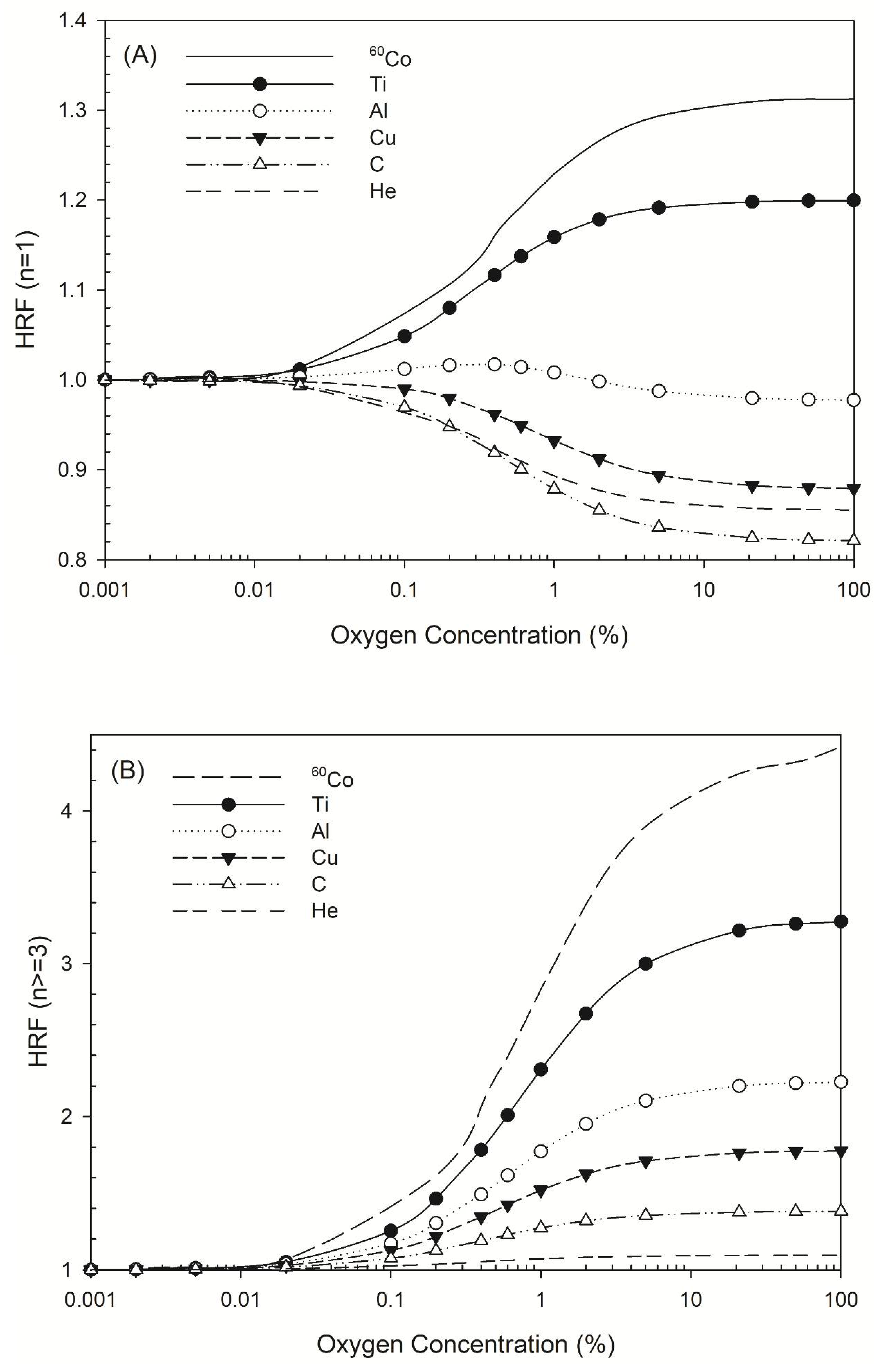
The HRF for DSB induction by all radiation types considered here increased as oxygen concentration decreased, but the HRF for non-DSB clusters varied in different ways. Figure 2A shows the HRF for a non-DSB cluster with just one lesion (n = 1), for which the HRF for 60Co γ-rays and titanium K-shell X-rays increased as oxygen concentration increased, while that for aluminum K-shell, copper L-shell, carbon K-shell, and 3.31 MeV helium ions (linear energy transfer (LET) =120 keV/μm) [44] decreased. There was a greater decrease in the HRF for carbon K-shell X-rays than for 3.31 MeV helium ions (Figure 2A). For complex DNA damage, such as DNA with three lesions or more, the HRF for all types of radiation increased as oxygen concentration increased (Figure 2B).
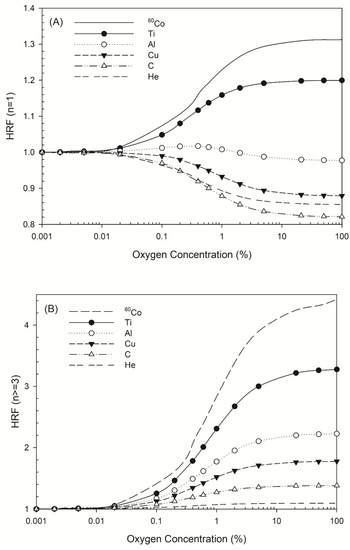
Figure 2.
HRF for different types of radiation at various oxygen concentrations. The symbols Ti, Al, Cu, C, and He represent titanium K-shell, aluminum K-shell, copper L-shell, and carbon K-shell X-rays, and helium ions. (A) HRF for DNA damage with n = 1 lesion as a function of oxygen concentration and (B) HRF for DNA damage with n ≥ 3 lesions as a function of oxygen concentration.
In terms of DNA repair via the LP BER pathway for cells irradiated by 60Co γ-rays, titanium K-shell, aluminum K-shell, copper L-shell, and carbon K-shell ultrasoft X-rays, the probability of correct repair, mutation, and DSB formation were calculated (Table 5). The probability of correct repair decreased as photon energy decreased and the probability of mutation and DSB conversion increased. In regards to pathways, the combined NER/LP BER pathway had the lowest probability of correct repair and the highest probability of mutation and DSB conversion. The probability of repair outcomes for carbon K-shell X-rays was comparable to that reported for helium ions (LET = 120 keV/μm) [44]. The probability of correct repair, mutation, and DSB conversion for carbon K-shell X-rays was 0.822, 0.128, and 0.05, and for helium ions it was 0.78, 0.16, and 0.06, suggesting a possible application for carbon K-shell X-rays in tumor killing.

Table 5.
Average repair outcome probabilities for all types of DNA damage for cells irradiated with ultrasoft X-rays or 60Co γ-rays.
The yield for DSB conversion was less than that for DSB induction for all energy levels, because the lost proportion of DSB conversion increased as photon energy decreased (from 88% to 69% for 4.55 keV to 0.28 keV). The range of RBE values (1.3–2.6) for DSB induction was greater than that calculated from DSB conversion (1.6–2.4; Table 6).

Table 6.
The relative biological effectiveness (RBE) of double-strand break (DSB) induction and enzymatic DSBs for long-patch base excision repair (LP BER) for cells irradiated with ultrasoft X-rays or 60Co γ-rays.
4. Discussion
In this study, the yields of induction and conversion of DSB for cells irradiated by ultrasoft X-rays were calculated. RBE values were also computed under various oxygen concentrations, using 60Co γ-rays as the reference radiation. In the following sections, we compare our results with experimental results and discuss the effects of oxygen.
4.1. DNA Damage Profile
Relative to 60Co γ-rays, the measured RBE for DSB induction in cells irradiated by titanium K-shell, aluminum K-shell, copper L-shell, and carbon K-shell X-rays increased from 1.4 to 2.7 [3] as the photon energy decreased from 4.55 keV to 280 eV (Table 1), which was within a 7% difference to the simulated values. Other studies have shown that the RBE of MCDS-derived DSB yields were comparable (within 7.1%) to the experimentally measured yield of ultrasoft X-rays (4.55 keV to 280 eV) [29,31]. The MCDS-derived DSB yield (21.6 per Gy per Gbp) for cells irradiated with 280 eV photons in the study by Streitmatter et al. (2017) [31] gave an RBE of 2.7 relative to 60Co γ-rays (8.6 per Gy per Gbp), which was the same value as that experimentally obtained by de Lara et al. (2001) [3].
Data summarized in Table 2, Table 3 and Table 4 showed that the DSB yields for all energy levels are decreasing as oxygen concentration decreases from 21% to 0.1% O2. The RBE for DSB induction increases from 1.3–2.6 under aerobic condition (21% O2) to 1.3–2.9 under moderate hypoxia (2% O2) and further 1.4–4.4 under severe hypoxia (0.1% O2). These results demonstrated that oxygen plays a significant role in DSB induction via indirect action, especially for photon energy above 4.55 keV. The DSB induction for the reference radiation, 60Co γ-rays, reduces 53% as oxygen concentration reduces from 21% to 0.1% while that for carbon K shell (280 eV) reduces 27% only.
In addition to DSB yield, the MCDS provided specified complexity of the DNA damage, and these results were approximated to those of track structure simulations [27]. The portion of simple DNA damage, i.e., BD, declined as photon energy decreased, while that of other types of DNA damage, such as SSB+, 2SSB, DSBs, DSBs+, and DSBs++, increased (Table 2, Table 3 and Table 4). It has been shown that the increase in the ionization density as photon energy decreases results in an increment in the DNA damage complexity, possibly due to the increase in the LET of secondary low energy electrons. However, the absence of oxygen could reduce the yields of DNA damage. If oxygen is present, the DNA damage would be fixed by oxygen; otherwise thiols could repair DNA damage, such as glutathione, which has been referred as chemical repair [17]. Values in Table 2, Table 3 and Table 4 show that only BD yields of carbon K-shell photons increase as oxygen concentration decreases from 21% to 0.1% while the yields of other types of DNA damage decrease. Because higher-LET radiations such as carbon K-shell photons may produce more complex damage, it is possible that the chemical repair process reduces the complex DNA damage such as SSB+, 2SSB, or DSB to BD under hypoxia.
4.2. HRF Comparison
The presence of oxygen contributed to the DNA damage complexity. The yield of total DSBs for carbon K-shell X-rays (280 eV) at an oxygen concentration of 21% (Table 2) was 21.2 per Gy per Gbp, and was 20.0 per Gy per Gbp (4.5% decrease) at a 2% oxygen concentration (Table 3) and reduced to 15.5 per Gy per Gbp (27% decrease) at 0.1% oxygen concentration (Table 4). The MCDS-derived HRF values (Figure 1) were in the range of 1.4–2.6, which agreed well with the range (1.8–2.1) for V79 cells [51] for all four energy levels of the ultrasoft X-rays.
However, the HRF values obtained from a Monte Carlo simulation using track structures, which simulate the effect of oxygen, were about 3.0 for X-rays or particles with an LET of < 10 keV/μm [52]. Experimental data from another study also showed that the HRF value for murine squamous cell carcinoma cells irradiated with 200 kVp X-rays (LET ~1 keV/μm) was 1.68 [53]. The differences in HRF values between experimental data and simulations suggest that the HRF value may vary depending on cell type [52].
Our simulation data (Figure 2A) revealed the HRF values for non-DSB clustered damage with one lesion caused by 60Co γ-rays or titanium K-shell X-rays, aluminum K-shell, copper L-shell, and carbon K-shell X-rays. The HRF for 60Co γ-rays and titanium K-shell X-rays increased as oxygen concentration increased, which shows that the presence of oxygen increased the number of instances of single-lesion DNA damage due to oxygen fixation [17]. The HRF for titanium K-shell X-rays and 60Co γ-rays was similar, possibly due to the energy of the secondary electrons induced by titanium K-shell X-rays, which is about 4.55 keV. The DNA damage profiles are similar when the electron energy is >2 keV [54], resulting in a near-identical DSB yield for titanium K-shell X-rays and 60Co γ-rays [24]. In contrast, the HRF for damage that produced one lesion decreased as oxygen concentration increased for the other three types of ultrasoft X-rays, which is in accordance with helium ions, a type of high LET radiation (LET = 120 keV/μm). That may be explained that the sparse DNA damage pattern is present for low-LET radiations (the secondary electrons produced through the irradiation of 60Co γ-rays and titanium K-shell X-rays on cells), while the denser pattern is present for higher-LET radiations. The presence of oxygen intensifies the effects of indirect actions and it turns out that the number of DNA damage with one lesion increases as the oxygen concentration increases (Figure 2A) for low-LET radiations (sparse pattern). In terms of complex DNA damage, the HRF values for ultrasoft X-rays such as aluminum K-shell, copper L-shell, and carbon K-shell X-rays were 1.4–2.0, which are comparable to that for helium ions (1.1; Figure 2B). The values for titanium K-shell X-rays and 60Co γ-rays were 3.3 and 4.5, respectively. The HRF values decrease as photon energy decreases (Figure 2B).
4.3. Comparison of Repair Outcomes
The respective RBE values obtained from the MCER-derived probability of mutation frequency for cells that are irradiated with aluminum K-shell and carbon K-shell X-rays were 2.9 and 4.4, using 60Co γ-rays as the reference radiation (Table 5). The RBE values obtained from the measured mutation frequencies of HF19 human diploid fibroblast cells irradiated with aluminum K-shell and carbon K-shell X-rays were 2.6 and 2.2, respectively, and the RBE values for V79 cells were 2.1 and 3.0, respectively, using 250 keV X-rays as the reference radiation [4,5]. Accordingly, the RBE values for the mutation frequency of cells irradiated with aluminum K-shell X-rays were in the range of 2.1–2.6 [4,5], while that generated by the MCER simulation was 2.9. For carbon K-shell X-rays, the RBE values were 2.2–3.0 [4,5] and the MCER-derived RBE was 4.4. The disagreement between the MCER-derived results and the experimental data could be due to several factors. Cell type, cell cycle length, and assay type all affect the measured RBE values, which are determined using genetic effects such as mutations and aberrations [5,55]. The RBE for the mutation frequency of Chinese hamster V79 cells (6.1 ± 1.0 μm cell thickness) [56] irradiated with carbon K shell X-rays, relative to 60Co γ-rays, was 3.0 [5], but it has also been measured as 2.0–2.6 [7] for mouse C3H10T1/2 cells (2.9 ± 0.6 μm cell thickness) [56]. If chromosomal aberrations are taken into account, the RBE values for Chinese hamster V79 cells, baby hamster kidney (BHK) fibroblasts, and human lymphocyte cells irradiated with carbon K-shell X-rays are 2.4, 2.6, and 2.1, respectively, relative to 60Co γ-rays [5]. The variation in RBE for genetic effects could be partly attributed to the highly localized energy deposition of secondary electrons on a micrometer or nanometer scale [57,58]. Intracellular factors such as cell thickness and type also play a role [59,60]. The MCER-derived results provide useful information about repair outcomes, but may be overestimated (Table 5).
Table 6 summarizes the RBE values for DSB induction and enzymatic DSB yields. The RBE values for DSB induction and enzymatic DSBs were in the range 1.3–2.6 and 1.6–2.4, respectively, suggesting that the enzymatic DSB yields are comparable to the yields of DSB induction. Under hypoxia conditions, the enzymatic DSB yields may be reduced due to the reduction of non-DSB DNA damage (Table 2, Table 3 and Table 4), but the RBE values for enzymatic DSB yields may also be higher, as the RBE values for DSB induction, due to a higher reduction in the yields of 60Co γ-rays. It has been reported that the oxygen level is around 4–7.5% in most normal tissues, but reduces to 0.3–4.2% in tumors [61]. That suggests that GT has a higher RBE than XT under hypoxia conditions and has an advantage in cancer treatment.
5. Conclusions
The MCDS- and MCER-derived RBE values for ultrasoft X-rays estimated based on DSB induction and enzymatic DSBs under an aerobic condition are in the range of 1.3–2.6 and 1.6–2.4, respectively. The RBE values for DSB induction under moderate hypoxia (2% O2) and severe hypoxia (0.1% O2) increase to the range of 1.3–2.9 and 1.4–4.4, respectively, indicating the advantage of GT for hypoxic tumor cells.
Author Contributions
Conceptualization, Y.-Y.H.; methodology, Y.-Y.H. and C.-C.C.; software, C.-C.C.; validation, Y.-Y.H., F.-H.C. and C.-C.T.; formal analysis, C.-C.C.; investigation, C.-C.C.; data curation, C.-C.C.; writing—original draft preparation, Y.-Y.H. and C.-C.C.; writing—review and editing, Y.-Y.H., F.-H.C. and C.-C.T.; supervision, C.-C.T.; project administration, C.-C.T.; funding acquisition, Y.-Y.H., F.-H.C. and C.-C.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants CSMU-INT-109-08 supported by Chung Shan Medical University, Taichung, Taiwan awarded to Ya-Yun Hsiao; CMRPD1H0473, CMRPD1J0322 supported by Chang Gung Memorial Hospital at Linkou, Taiwan; and MOST 109-2314-B-182-078-MY3, 110-2628-B-182-008 supported by the Ministry of Sciences and Technology, Taiwan awarded to Fang-Hsin Chen; and by grant MOST 108-2638-E-005-001-MY2 supported by the Ministry of Sciences and Technology, Taiwan awarded to Ching-Chih Tsai.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This research was supported by the Chung Shan Medical University, Taichung, Taiwan, Linkou-Chang Gung Memorial Hospital and the Ministry of Sciences and Technology, Taiwan.
Conflicts of Interest
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- Goodhead, D.T.; Thacker, J. Inactivation and mutation of cultured mammalian cells by aluminium characteristic ultrasoft x-rays. I. Properties of aluminium x-rays and preliminary experiments with chinese hamster cells. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1977, 31, 541–559. [Google Scholar] [CrossRef]
- Raju, M.R.; Carpenter, S.G.; Chmielewski, J.J.; Schillaci, M.E.; Wilder, M.E.; Freyer, J.P.; Johnson, N.F.; Schor, P.L.; Sebring, R.J.; Goodhead, D.T. Radiobiology of ultrasoft x rays. I. Cultured hamster cells (v79). Radiat. Res. 1987, 110, 396–412. [Google Scholar] [CrossRef] [PubMed]
- De Lara, C.M.; Hill, M.A.; Jenner, T.J.; Papworth, D.; O’Neill, P. Dependence of the yield of DNA double-strand breaks in chinese hamster v79-4 cells on the photon energy of ultrasoft X-rays. Radiat. Res. 2001, 155, 440–448. [Google Scholar] [CrossRef]
- Cox, R.; Thacker, J.; Goodhead, D.T. Inactivation and mutation of cultured mammalian cells by aluminium characteristic ultrasoft X-rays. II. Dose-responses of chinese hamster and human diploid cells to aluminium x-rays and radiations of different let. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1977, 31, 561–576. [Google Scholar] [CrossRef] [PubMed]
- Goodhead, D.T.; Nikjoo, H. Current status of ultrasoft X-rays and track structure analysis as tools for testing and developing biophysical models of radiation action. Radiat. Prot. Dosim. 1990, 31, 343–350. [Google Scholar] [CrossRef]
- Hannam, S.; Webster, M.R.; Nixon, R.L. Radiotherapy in the treatment of hand eczema. In Textbook of Hand Eczema; Alikhan, A., Lachapelle, J., Maibach, H., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 353–359. [Google Scholar]
- Frankenberg, D.; Kuhn, H.; Frankenberg-Schwager, M.; Lenhard, W.; Beckonert, S. 0.3 kev carbon k ultrasoft X-rays are four times more effective than gamma-rays when inducing oncogenic cell transformation at low doses. Int. J. Radiat. Biol. 1995, 68, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.M.; Gibbons, J.P. Khan’s the Physics of Radiation Therapy, 5th ed.; Lippincott Williams & Wilkins/Wolters Kluwer Health: Philadelphia, PA, USA, 2014; pp. 1–12. [Google Scholar]
- Shikazono, N.; Noguchi, M.; Fujii, K.; Urushibara, A.; Yokoya, A. The yield, processing, and biological consequences of clustered DNA damage induced by ionizing radiation. J. Radiat. Res. 2009, 50, 27–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nickoloff, J.A.; Sharma, N.; Taylor, L. Clustered DNA double-strand breaks: Biological effects and relevance to cancer radiotherapy. Genes 2020, 11, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.C.; Hsu, D.S.; Kazantsev, A.; Sancar, A. Substrate spectrum of human excinuclease: Repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc. Natl. Acad. Sci. USA 1994, 91, 12213–12217. [Google Scholar] [CrossRef] [Green Version]
- Reardon, J.T.; Bessho, T.; Kung, H.C.; Bolton, P.H.; Sancar, A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: Possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc. Natl. Acad. Sci. USA 1997, 94, 9463–9468. [Google Scholar] [CrossRef] [Green Version]
- Blaisdell, J.O.; Harrison, L.; Wallace, S.S. Base excision repair processing of radiation-induced clustered DNA lesions. Radiat. Prot. Dosim. 2001, 97, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Lomax, M.E.; Gulston, M.K.; O’Neill, P. Chemical aspects of clustered DNA damage induction by ionising radiation. Radiat. Prot. Dosim. 2002, 99, 63–68. [Google Scholar] [CrossRef]
- Eccles, L.J.; O’Neill, P.; Lomax, M.E. Delayed repair of radiation induced clustered DNA damage: Friend or foe? Mutat. Res. 2011, 711, 134–141. [Google Scholar] [CrossRef]
- Blocher, D. DNA double-strand break repair determines the rbe of alpha-particles. Int. J. Radiat. Biol. 1988, 54, 761–771. [Google Scholar] [CrossRef]
- Von Sonntag, C. Free-Radical-Induced DNA Damage and Its Repair, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 357–482. [Google Scholar]
- Hall, E.J.; Giaccia, A.J. Radiobiology for the Radiologist, 7th ed.; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2012; pp. 16–29. [Google Scholar]
- Lomax, M.E.; Folkes, L.K.; O’Neill, P. Biological consequences of radiation-induced DNA damage: Relevance to radiotherapy. Clin. Oncol. 2013, 25, 578–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panizzon, R.G.; Seegenschmiedt, M.H. Radiation Treatment and Radiation Reactions in Dermatology; Springer: Berlin/Heidelberg, Germany, 2015; pp. 73–87. [Google Scholar]
- Hanlon, A. A Practical Guide to Skin Cancer; Springer: Cham, Switzerland, 2018; pp. 249–267. [Google Scholar]
- Drakensjö, I.R.T.; Rosen, E.; Frohm Nilsson, M.; Girnita, A. Ten-year follow-up study of grenz ray treatment for lentigo maligna and early lentigo maligna melanoma. Acta Derm. Venereol. 2020, 100, adv00282. [Google Scholar]
- Breneman, J.C.; Goldschmidt, H.; Gorson, R.O.; Panizzon, R.G.; Lindelöf, B. Modern Dermatologic Radiation Therapy; Springer: New York, NY, USA, 2012; pp. 155–159. [Google Scholar]
- Kochan, J.A.; van den Belt, M.; von der Lippe, J.; Desclos, E.C.B.; Steurer, B.; Hoebe, R.A.; Scutigliani, E.M.; Verhoeven, J.; Stap, J.; Bosch, R.; et al. Ultra-soft X-ray system for imaging the early cellular responses to x-ray induced DNA damage. Nucleic Acids Res. 2019, 47, e100. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.F. The complexity of DNA damage: Relevance to biological consequences. Int. J. Radiat. Biol. 1994, 66, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Nikjoo, H.; O’Neill, P.; Goodhead, D.T.; Terrissol, M. Computational modelling of low-energy electron-induced DNA damage by early physical and chemical events. Int. J. Radiat. Biol. 1997, 71, 467–483. [Google Scholar] [CrossRef]
- Semenenko, V.A.; Stewart, R.D. A fast monte carlo algorithm to simulate the spectrum of DNA damages formed by ionizing radiation. Radiat. Res. 2004, 161, 451–457. [Google Scholar] [CrossRef]
- Liang, Y.; Fu, Q.; Wang, X.; Liu, F.; Yang, G.; Luo, C.; Ouyang, Q.; Wang, Y. Relative biological effectiveness for photons: Implication of complex DNA double-strand breaks as critical lesions. Phys. Med. Biol. 2017, 62, 2153–2175. [Google Scholar] [CrossRef]
- Hsiao, Y.; Stewart, R.D. Monte carlo simulation of DNA damage induction by x-rays and selected radioisotopes. Phys. Med. Biol. 2008, 53, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Kirkby, C.; Ghasroddashti, E.; Poirier, Y.; Tambasco, M.; Stewart, R.D. Rbe of kv cbct radiation determined by monte carlo DNA damage simulations. Phys. Med. Biol. 2013, 58, 5693–5704. [Google Scholar] [CrossRef]
- Streitmatter, S.W.; Stewart, R.D.; Jenkins, P.A.; Jevremovic, T. DNA double strand break (dsb) induction and cell survival in iodine-enhanced computed tomography (ct). Phys. Med. Biol. 2017, 62, 6164–6184. [Google Scholar] [CrossRef] [PubMed]
- Baró, J.; Sempau, J.; Fernández-Varea, J.M.; Salvat, F. Penelope: An algorithm for monte carlo simulation of the penetration and energy loss of electrons and positrons in matter. Nucl. Inst. Methods Phys. Res. B 1995, 100, 31–46. [Google Scholar] [CrossRef]
- Sempau, J.; Acosta, E.; Baró, J.; Fernández-Varea, J.M.; Salvat, F. An algorithm for monte carlo simulation of coupled electron-photon transport. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 1997, 132, 377–390. [Google Scholar] [CrossRef]
- Salvat, F.; Fernandez-Varea, J.M.; Acosta, E.; Sempau, J. Penelope—A Code System for Monte Carlo Simulation of Electron and Photon Transport; Nuclear Energy Agency of the OECD (NEA): Organisation for Economic Co-Operation and Development—Nuclear Energy Agency: Paris, France, 2001. [Google Scholar]
- Salvat, F.; Fernández-Varea, J.; Sempau, J. Penelope 2011: A Code System for Monte Carlo Simulation of Electron and Photon Transport; OECD Nuclear Energy Agency: Issy-les-Moulineaux, France, 2011. [Google Scholar]
- Salvat, F.; Fernandez-Varea, J.M.; Baro, J.; Sempau, J. Penelope, an Algorithm and Computer Code for Monte Carlo Simulation of Electron-Photon Showers; CIEMAT: Madrid, Spain, 1996. [Google Scholar]
- Stewart, R.D.; Wilson, W.E.; McDonald, J.C.; Strom, D.J. Microdosimetric properties of ionizing electrons in water: A test of the penelope code system. Phys. Med. Biol. 2002, 47, 79–88. [Google Scholar] [CrossRef]
- Semenenko, V.A.; Stewart, R.D. Fast monte carlo simulation of DNA damage formed by electrons and light ions. Phys. Med. Biol. 2006, 51, 1693–1706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, R.D.; Yu, V.K.; Georgakilas, A.G.; Koumenis, C.; Park, J.H.; Carlson, D.J. Effects of radiation quality and oxygen on clustered DNA lesions and cell death. Radiat. Res. 2011, 176, 587–602. [Google Scholar] [CrossRef]
- Luo, W.R.; Chen, F.H.; Huang, R.J.; Chen, Y.P.; Hsiao, Y.Y. Effects of indirect actions and oxygen on relative biological effectiveness: Estimate of dsb inductions and conversions induced by therapeutic proton beams. Int. J. Radiat. Biol. 2020, 96, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Semenenko, V.A.; Stewart, R.D.; Ackerman, E.J. Monte carlo simulation of base and nucleotide excision repair of clustered DNA damage sites. I. Model properties and predicted trends. Radiat. Res. 2005, 164, 180–193. [Google Scholar] [CrossRef]
- Semenenko, V.A.; Stewart, R.D. Monte carlo simulation of base and nucleotide excision repair of clustered DNA damage sites. Ii. Comparisons of model predictions to measured data. Radiat. Res. 2005, 164, 194–201. [Google Scholar] [CrossRef]
- Tavares, A.A.S.; Tavares, J.M.R.S. Evaluating 99mtc auger electrons for targeted tumor radiotherapy by computational methods. Med. Phys. 2010, 37, 3551–3559. [Google Scholar] [CrossRef]
- Tsai, J.Y.; Chen, F.H.; Hsieh, T.Y.; Hsiao, Y.Y. Effects of indirect actions and oxygen on relative biological effectiveness: Estimate of dsb induction and conversion induced by gamma rays and helium ions. J. Radiat. Res. 2015, 56, 691–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, M.J.; Coursey, J.S.; Zucker, M.A.; Chang, J. Estar, Pstar, and Astar: Computer Programs for Calculating Stopping-Power and Range Tables for Electrons, Protons, and Helium Ions. Available online: http://physics.nist.gov/Star (accessed on 12 December 2020).
- Emfietzoglou, D.; Nikjoo, H. Accurate electron inelastic cross sections and stopping powers for liquid water over the 0.1-10 kev range based on an improved dielectric description of the bethe surface. Radiat. Res. 2007, 167, 110–120. [Google Scholar] [CrossRef]
- Carlson, D.J.; Stewart, R.D.; Semenenko, V.A. Effects of oxygen on intrinsic radiation sensitivity: A test of the relationship between aerobic and hypoxic linear-quadratic (lq) model parameters. Med. Phys. 2006, 33, 3105. [Google Scholar] [CrossRef] [PubMed]
- Nahum, A.E.; Movsas, B.; Horwitz, E.M.; Stobbe, C.C.; Chapman, J.D. Incorporating clinical measurements of hypoxia into tumor local control modeling of prostate cancer: Implications for the α/β ratio. Int. J. Radiat. Oncol. Biol. Phys. 2003, 57, 391–401. [Google Scholar] [CrossRef]
- Rothkamm, K.; Lobrich, M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc. Natl. Acad. Sci. USA 2003, 100, 5057–5062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sapora, O.; Barone, F.; Belli, M.; Maggi, A.; Quintiliani, M.; Tabocchini, M.A. Relationships between cell killing, mutation induction and DNA damage in x-irradiated v79 cells: The influence of oxygen and dmso. Int. J. Radiat. Biol. 1991, 60, 467–482. [Google Scholar] [CrossRef]
- Botchway, S.W.; Stevens, D.L.; Hill, M.A.; Jenner, T.J.; O’Neill, P. Induction and rejoining of DNA double-strand breaks in chinese hamster v79-4 cells irradiated with characteristic aluminum k and copper l ultrasoft x rays. Radiat. Res. 1997, 148, 317–324. [Google Scholar] [CrossRef]
- Forster, J.C.; Douglass, M.J.J.; Phillips, W.M.; Bezak, E. Monte carlo simulation of the oxygen effect in DNA damage induction by ionizing radiation. Radiat. Res. 2018, 190, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, R.; Uzawa, A.; Matsumoto, Y.; Noguchi, M.; Kase, Y.; Takase, N.; Ito, A.; Koike, S.; Ando, K.; Okayasu, R.; et al. Induction of DNA dsb and its rejoining in clamped and non-clamped tumours after exposure to carbon ion beams in comparison to x rays. Radiat. Prot. Dosim. 2011, 143, 508–512. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, Y.Y.; Hung, T.H.; Tu, S.J.; Tung, C.J. Fast monte carlo simulation of DNA damage induction by auger-electron emission. Int. J. Radiat. Biol. 2014, 90, 392–400. [Google Scholar] [CrossRef]
- Barendsen, G.W. Rbe-let relationships for different types of lethal radiation damage in mammalian cells: Comparison with DNA dsb and an interpretation of differences in radiosensitivity. Int. J. Radiat. Biol. 1994, 66, 433–436. [Google Scholar] [CrossRef]
- Bettega, D.; Calzolari, P.; Doglia, S.M.; Dulio, B.; Tallone, L.; Villa, A.M. Technical report: Cell thickness measurements by confocal fluorescence microscopy on c3h10t1/2 and v79 cells. Int. J. Radiat. Biol. 1998, 74, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Famulari, G.; Pater, P.; Enger, S.A. Microdosimetry calculations for monoenergetic electrons using geant4-DNA combined with a weighted track sampling algorithm. Phys. Med. Biol. 2017, 62, 5495–5508. [Google Scholar] [CrossRef] [PubMed]
- Yachi, Y.; Yoshii, Y.; Matsuya, Y.; Mori, R.; Oikawa, J.; Date, H. Track structure study for energy dependency of electrons and x-rays on DNA double-strand break induction. Sci. Rep. 2019, 9, 17649. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, S.; Cornforth, M.N.; Harvey, W.F.; Raju, M.R.; Schillaci, M.E.; Wilder, M.E.; Goodhead, D.T. Radiobiology of ultrasoft x rays. Iv. Flat and round-shaped hamster cells (cho-10b, hs-23). Radiat. Res. 1989, 119, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Nikjoo, H.; Lindborg, L. Rbe of low energy electrons and photons. Phys. Med. Biol. 2010, 55, R65–R109. [Google Scholar] [CrossRef] [Green Version]
- McKeown, S.R. Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br. J. Radiol. 2014, 87, 20130676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).