Panx1b Modulates the Luminance Response and Direction of Locomotion in the Zebrafish
Abstract
:1. Introduction
2. Results
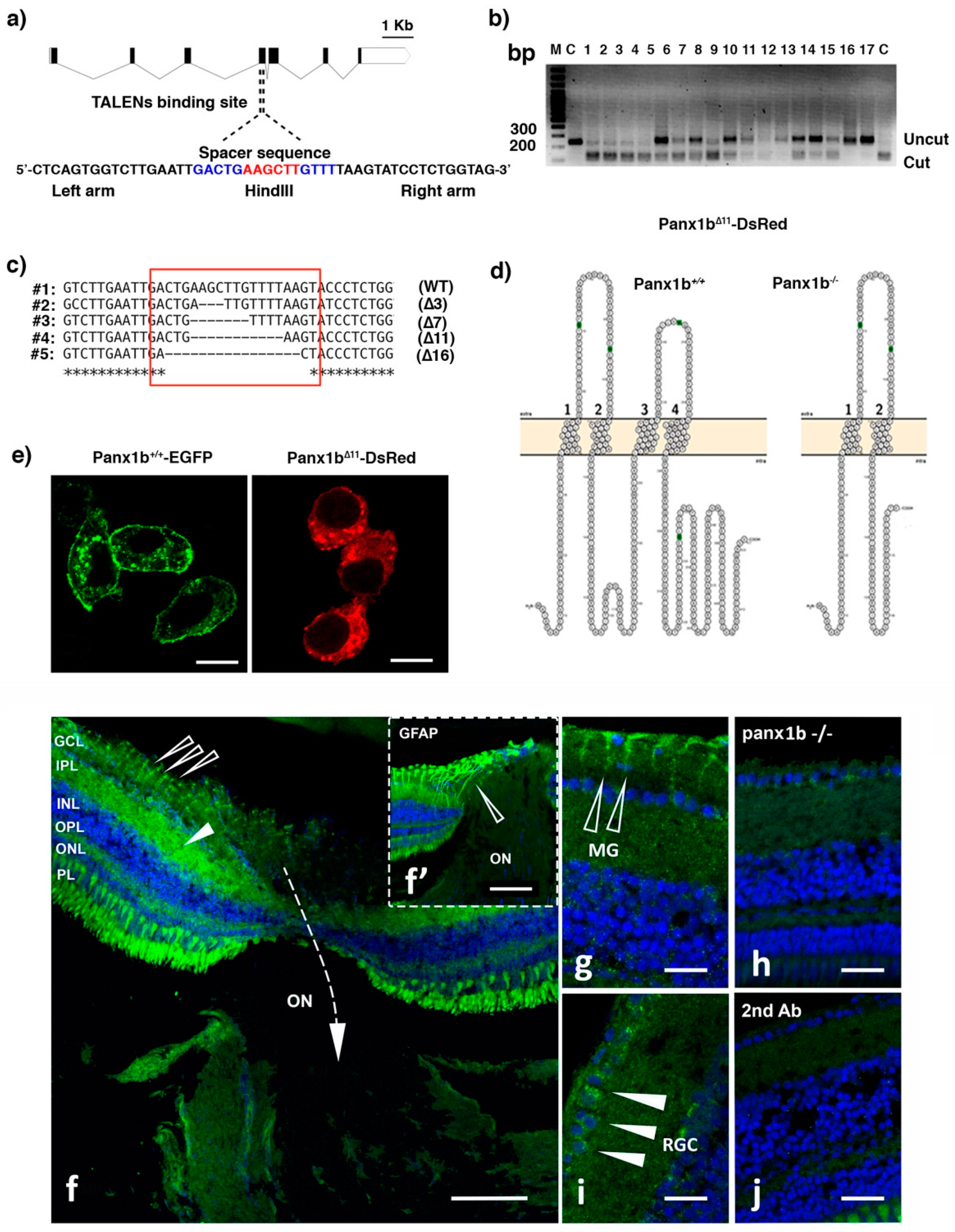
2.1. Targeted Disruption of the panx1b Gene
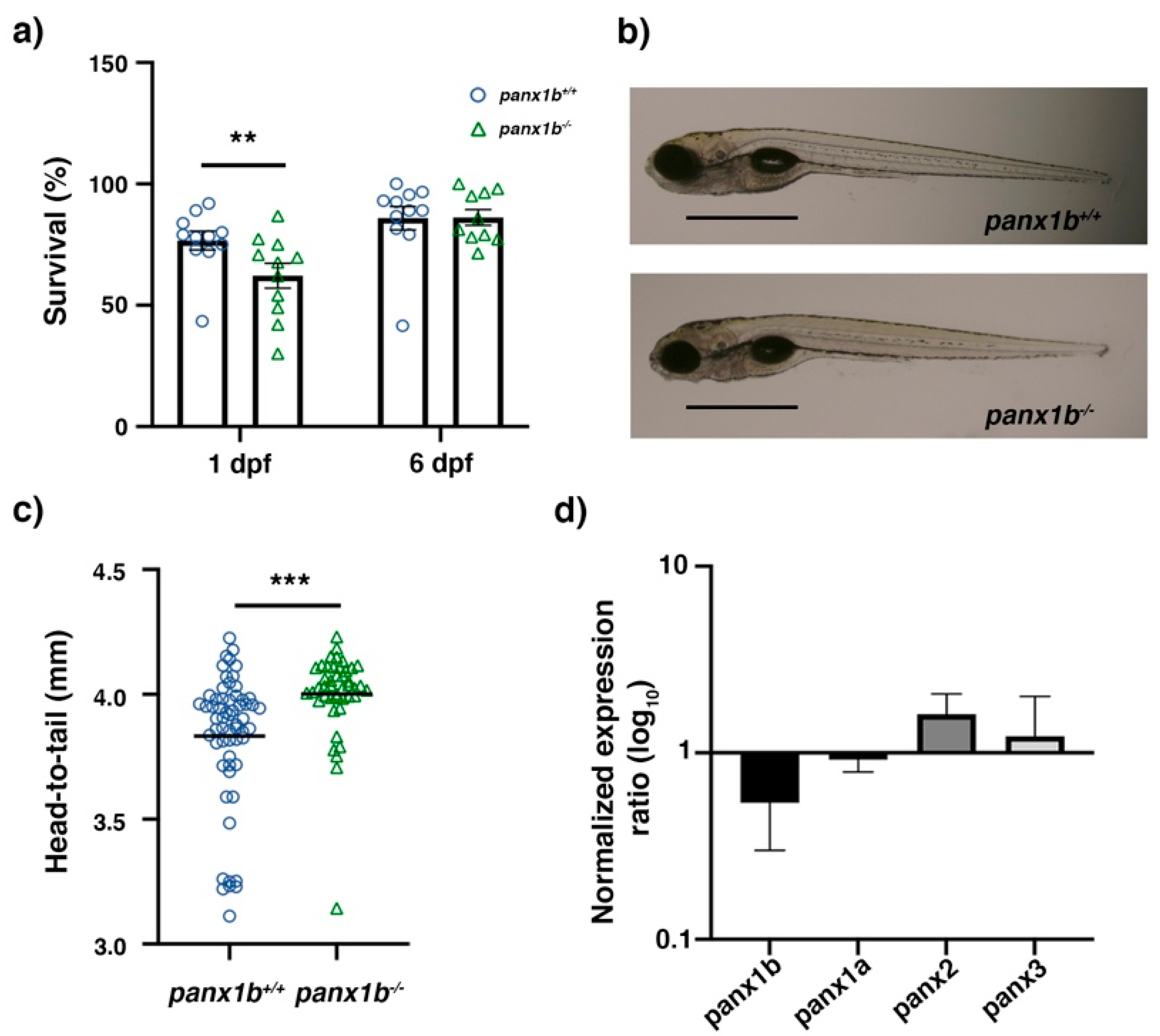
2.2. Characterization of panx1b−/− Larvae
2.3. Transcriptome Profiling of 6 dpf panx1b−/− Larvae
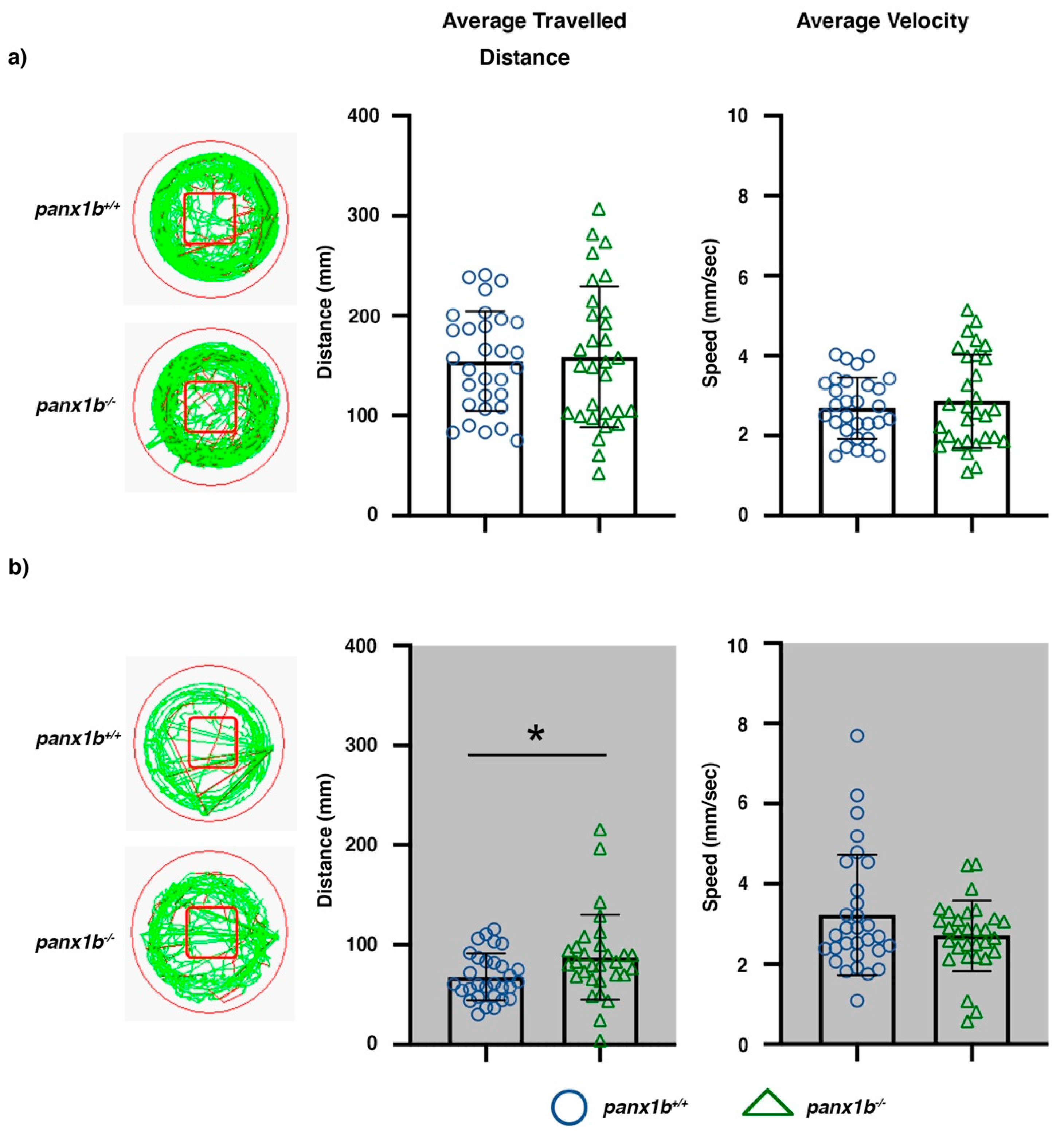
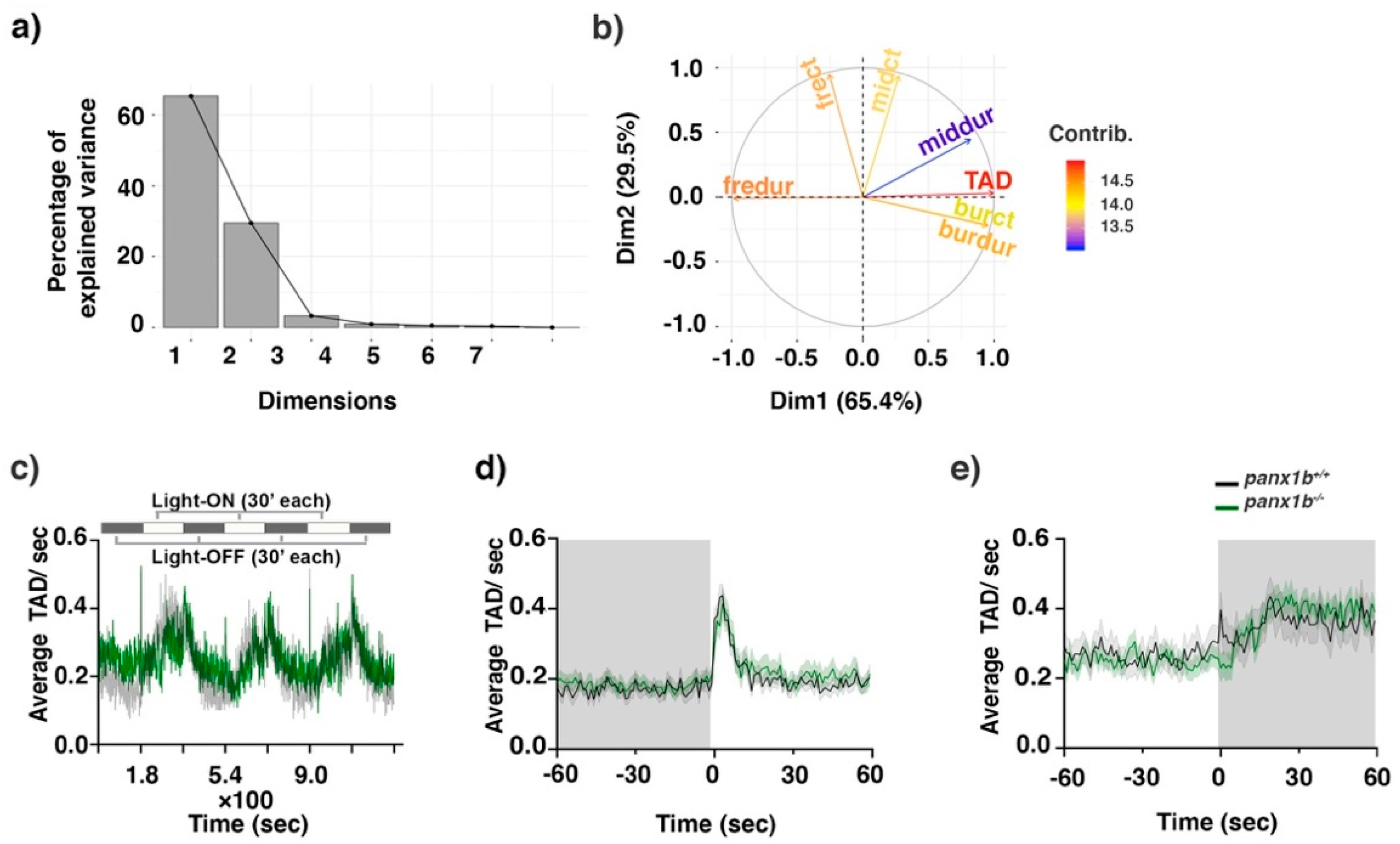
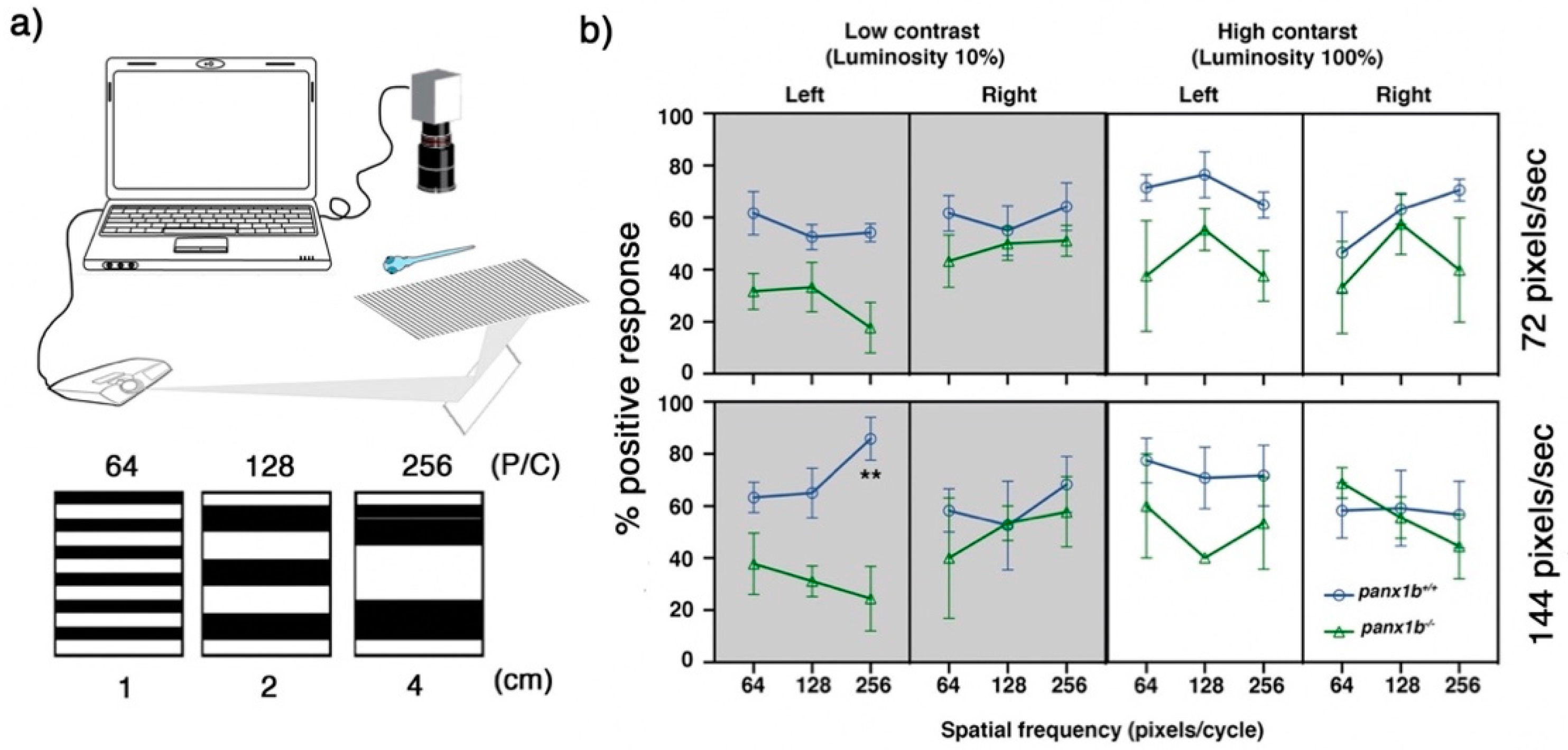
2.4. Loss of Panx1b Affected Visually Guided Locomotor Activities of Larvae
3. Discussion
4. Materials and Methods
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dahl, G. ATP release through pannexon channels. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140191. [Google Scholar] [CrossRef]
- Mim, C.; Perkins, G.; Dahl, G. Structure versus function: Are new conformations of pannexin 1 yet to be resolved? J. Gen. Physiol. 2021, 153, e202012754. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; He, Z.; Maksaev, G.; Bitter, R.M.; Rau, M.; Fitzpatrick, J.A.J.; Yuan, P. Cryo-EM structures of the ATP release channel pannexin 1. Nat. Struct. Mol. Biol. 2020, 27, 373–381. [Google Scholar] [CrossRef]
- Jin, Q.; Zhang, B.; Zheng, X.; Li, N.; Xu, L.; Xie, Y.; Song, F.; Bhat, E.A.; Chen, Y.; Gao, N.; et al. Cryo-EM structures of human pannexin 1 channel. Cell Res. 2020, 30, 449–451. [Google Scholar] [CrossRef] [PubMed]
- Michalski, K.; Syrjanen, J.L.; Henze, E.; Kumpf, J.; Furukawa, H.; Kawate, T. The Cryo-EM structure of pannexin 1 reveals unique motifs for ion selection and inhibition. eLife 2020, 9, e54670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mou, L.; Ke, M.; Song, M.; Shan, Y.; Xiao, Q.; Liu, Q.; Li, J.; Sun, K.; Pu, L.; Guo, L.; et al. Structural basis for gating mechanism of Pannexin 1 channel. Cell Res. 2020, 30, 452–454. [Google Scholar] [CrossRef] [Green Version]
- Qu, R.; Dong, L.; Zhang, J.; Yu, X.; Wang, L.; Zhu, S. Cryo-EM structure of human heptameric Pannexin 1 channel. Cell Res. 2020, 30, 446–448. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Z.; Orozco, I.J.; Du, J.; Lü, W. Structures of human pannexin 1 reveal ion pathways and mechanism of gating. Nature 2020, 584, 646–651. [Google Scholar] [CrossRef]
- Ray, A.; Zoidl, G.; Weickert, S.; Wahle, P.; Dermietzel, R. Site-Specific and developmental expression of pannexin1 in the mouse nervous system. Eur. J. Neurosci. 2005, 21, 3277–3290. [Google Scholar] [CrossRef]
- Dvoriantchikova, G.; Ivanov, D.; Barakat, D.; Grinberg, A.; Wen, R.; Slepak, V.Z.; Shestopalov, V.I. Genetic Ablation of Pannexin1 Protects Retinal Neurons from Ischemic Injury. PLoS ONE 2012, 7, e31991. [Google Scholar] [CrossRef]
- Dvoriantchikova, G.; Ivanov, D.; Panchin, Y.; Shestopalov, V.I. Expression of pannexin family of proteins in the retina. FEBS Lett. 2006, 580, 2178–2182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kranz, K.; Dorgau, B.; Pottek, M.; Herrling, R.; Schultz, K.; Bolte, P.; Monyer, H.; Penuela, S.; Laird, D.W.; Dedek, K.; et al. Expression of Pannexin1 in the outer plexiform layer of the mouse retina and physiological impact of its knockout. J. Comp. Neurol. 2012, 521, 1119–1135. [Google Scholar] [CrossRef] [PubMed]
- Pronin, A.; Pham, D.; An, W.; Dvoriantchikova, G.; Reshetnikova, G.; Qiao, J.; Kozhekbaeva, Z.; Reiser, A.E.; Slepak, V.Z.; Shestopalov, V.I. Inflammasome Activation Induces Pyroptosis in the Retina Exposed to Ocular Hypertension Injury. Front. Mol. Neurosci. 2019, 12, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravi, V.; Venkatesh, B. The Divergent Genomes of Teleosts. Annu. Rev. Anim. Biosci. 2018, 6, 47–68. [Google Scholar] [CrossRef]
- Bond, S.R.; Wang, N.; Leybaert, L.; Naus, C.C. Pannexin 1 Ohnologs in the Teleost Lineage. J. Membr. Biol. 2012, 245, 483–493. [Google Scholar] [CrossRef]
- Kurtenbach, S.; Prochnow, N.; Kurtenbach, S.; Klooster, J.; Zoidl, C.; Dermietzel, R.; Kamermans, M.; Zoidl, G. Pannexin1 Channel Proteins in the Zebrafish Retina Have Shared and Unique Properties. PLoS ONE 2013, 8, e77722. [Google Scholar] [CrossRef] [Green Version]
- Prochnow, N.; Hoffmann, S.; Vroman, R.; Klooster, J.; Bunse, S.; Kamermans, M.; Dermietzel, R.; Zoidl, G. Pannexin1 in the outer retina of the zebrafish, Danio rerio. Neuroscience 2009, 162, 1039–1054. [Google Scholar] [CrossRef]
- Cenedese, V.; de Graaff, W.; Csikós, T.; Poovayya, M.; Zoidl, G.; Kamermans, M. Pannexin 1 Is Critically Involved in Feedback from Horizontal Cells to Cones. Front. Mol. Neurosci. 2017, 10, 403. [Google Scholar] [CrossRef] [Green Version]
- Safarian, N.; Whyte-Fagundes, P.; Zoidl, C.; Grigull, J.; Zoidl, G. Visuomotor deficiency in panx1a knockout zebrafish is linked to dopaminergic signaling. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Bedell, V.; Wang, Y.; Campbell, J.M.; Poshusta, T.L.; Starker, C.; Ii, R.G.K.; Tan, W.; Penheiter, S.G.; Ma, A.C.; Leung, A.Y.H.; et al. In Vivo genome editing using a high-efficiency TALEN system. Nature 2012, 491, 114–118. [Google Scholar] [CrossRef] [Green Version]
- Prochnow, N.; Hoffmann, S.; Dermietzel, R.; Zoidl, G. Replacement of a single cysteine in the fourth transmembrane region of zebrafish pannexin1 alters hemichannel gating behavior. Exp. Brain Res. 2009, 199, 255–264. [Google Scholar] [CrossRef]
- Hirayama, J.; Kaneko, M.; Cardone, L.; Cahill, G.; Sassone-Corsi, P. Analysis of Circadian Rhythms in Zebrafish. Methods Enzymol. 2005, 393, 186–204. [Google Scholar] [CrossRef]
- Emran, F.; Rihel, J.; Dowling, J.E. A Behavioral Assay to Measure Responsiveness of Zebrafish to Changes in Light Intensities. J. Vis. Exp. 2008, e923. [Google Scholar] [CrossRef]
- Bilotta, J. Effects of abnormal lighting on the development of zebrafish visual behavior. Behav. Brain Res. 2000, 116, 81–87. [Google Scholar] [CrossRef]
- Neuhauss, S.C. Behavioral genetic approaches to visual system development and function in zebrafish. J. Neurobiol. 2002, 54, 148–160. [Google Scholar] [CrossRef] [Green Version]
- Portugues, R.; Engert, F. The neural basis of visual behaviors in the larval zebrafish. Curr. Opin. Neurobiol. 2009, 19, 644–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiebel-Kalish, H.; Reich, E.; Rainy, N.; Vatine, G.; Nisgav, Y.; Tovar, A.; Gothilf, Y.; Bach, M. Gucy2f zebrafish knockdown—A model for Gucy2d-related leber congenital amaurosis. Eur. J. Hum. Genet. 2012, 20, 884–889. [Google Scholar] [CrossRef] [Green Version]
- Prochnow, N.; Abdulazim, A.; Kurtenbach, S.; Wildförster, V.; Dvoriantchikova, G.; Hanske, J.; Petrasch-Parwez, E.; Shestopalov, V.I.; Dermietzel, R.; Manahan-Vaughan, D.; et al. Pannexin1 Stabilizes Synaptic Plasticity and Is Needed for Learning. PLoS ONE 2012, 7, e51767. [Google Scholar] [CrossRef] [Green Version]
- Dvoriantchikova, G.; Pronin, A.; Kurtenbach, S.; Toychiev, A.; Chou, T.-H.; Yee, C.W.; Prindeville, B.; Tayou, J.; Porciatti, V.; Sagdullaev, B.; et al. Pannexin 1 sustains the electrophysiological responsiveness of retinal ganglion cells. Sci. Rep. 2018, 8, 5797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kist, A.; Portugues, R. Optomotor Swimming in Larval Zebrafish Is Driven by Global Whole-Field Visual Motion and Local Light-Dark Transitions. Cell Rep. 2019, 29, 659–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, A.; Wu, Y.; Baier, H.; Kubo, F. Neuronal Architecture of a Visual Center that Processes Optic Flow. Neuron 2019, 103, 118–132. [Google Scholar] [CrossRef]
- Naumann, E.A.; Fitzgerald, J.E.; Dunn, T.W.; Rihel, J.; Sompolinsky, H.; Engert, F. From Whole-Brain Data to Functional Circuit Models: The Zebrafish Optomotor Response. Cell 2016, 167, 947–960. [Google Scholar] [CrossRef] [Green Version]
- Portugues, R.; Engert, F. Adaptive Locomotor Behavior in Larval Zebrafish. Front. Syst. Neurosci. 2011, 5, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orger, M.; Baier, H. Channeling of red and green cone inputs to the zebrafish optomotor response. Vis. Neurosci. 2005, 22, 275–281. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Bear, J.; Roberts, P.A.; Janiak, F.K.; Semmelhack, J.; Yoshimatsu, T.; Baden, T. Zebrafish Retinal Ganglion Cells Asymmetrically Encode Spectral and Temporal Information across Visual Space. Curr. Biol. 2020, 30, 2927–2942. [Google Scholar] [CrossRef] [PubMed]
- Abbas, F.; Triplett, M.A.; Goodhill, G.; Meyer, M.P. A Three-Layer Network Model of Direction Selective Circuits in the Optic Tectum. Front. Neural Circuits 2017, 11, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tikidji-Hamburyan, A.; Reinhard, K.; Seitter, H.; Hovhannisyan, A.; Procyk, C.; Allen, A.E.; Schenk, M.; Lucas, R.; Münch, T.A. Retinal output changes qualitatively with every change in ambient illuminance. Nat. Neurosci. 2014, 18, 66–74. [Google Scholar] [CrossRef]
- Trenholm, S.; McLaughlin, A.J.; Schwab, D.J.; Awatramani, G.B. Dynamic Tuning of Electrical and Chemical Synaptic Transmission in a Network of Motion Coding Retinal Neurons. J. Neurosci. 2013, 33, 14927–14938. [Google Scholar] [CrossRef] [Green Version]
- Trenholm, S.; McLaughlin, A.J.; Schwab, D.J.; Turner, M.H.; Smith, R.G.; Rieke, F.; Awatramani, G.B. Nonlinear dendritic integration of electrical and chemical synaptic inputs drives fine-scale correlations. Nat. Neurosci. 2014, 17, 1759–1766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaney, D.I. Territorial organization of direction-selective ganglion cells in rabbit retina. J. Neurosci. 1994, 14, 6301–6316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, X.; Cafaro, J.; McLaughlin, A.J.; Postma, F.R.; Paul, D.L.; Awatramani, G.; Field, G.D. Gap Junctions Contribute to Differential Light Adaptation across Direction-Selective Retinal Ganglion Cells. Neuron 2018, 100, 216–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, Y.; Bennett, D.; Rubinov, M.; Narayan, S.; Yang, C.-T.; Tanimoto, M.; Mensh, B.D.; Looger, L.L.; Ahrens, M.B. Glia Accumulate Evidence that Actions Are Futile and Suppress Unsuccessful Behavior. Cell 2019, 178, 27–43. [Google Scholar] [CrossRef] [Green Version]
- Halassa, M.M.; Fellin, T.; Haydon, P.G. Tripartite synapses: Roles for astrocytic purines in the control of synaptic physiology and behavior. Neuropharmacology 2009, 57, 343–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazargani, N.; Attwell, D. Astrocyte calcium signaling: The third wave. Nat. Neurosci. 2016, 19, 182–189. [Google Scholar] [CrossRef]
- Suadicani, S.O.; Iglesias, R.; Wang, J.; Dahl, G.; Spray, D.C.; Scemes, E. ATP signaling is deficient in cultured pannexin1-null mouse astrocytes. Glia 2012, 60, 1106–1116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckel, J.M.; Argall, A.J.; Lim, J.C.; Xia, J.; Lu, W.; Coffey, E.E.; Macarak, E.J.; Shahidullah, M.; Delamere, N.; Zode, G.S.; et al. Mechanosensitive release of adenosine 5′-triphosphate through pannexin channels and mechanosensitive upregulation of pannexin channels in optic nerve head astrocytes: A mechanism for purinergic involvement in chronic strain. Glia 2014, 62, 1486–1501. [Google Scholar] [CrossRef]
- Marpegan, L.; Swanstrom, A.E.; Chung, K.; Simon, T.; Haydon, P.G.; Khan, S.K.; Liu, A.C.; Herzog, E.; Beaulé, C. Circadian Regulation of ATP Release in Astrocytes. J. Neurosci. 2011, 31, 8342–8350. [Google Scholar] [CrossRef] [Green Version]
- Kovalzon, V.; Moiseenko, L.; Ambaryan, A.; Kurtenbach, S.; Shestopalov, V.; Panchin, Y. Sleep-Wakefulness cycle and behavior in pannexin1 knockout mice. Behav. Brain Res. 2017, 318, 24–27. [Google Scholar] [CrossRef]
- Shestopalov, V.I.; Panchin, Y.; Tarasova, O.S.; Gaynullina, D.; Kovalzon, V.M. Pannexins Are Potential New Players in the Regulation of Cerebral Homeostasis during Sleep-Wake Cycle. Front. Cell. Neurosci. 2017, 11. [Google Scholar] [CrossRef] [Green Version]
- Neff, K.L.; Argue, D.P.; Ma, A.C.; Lee, H.B.; Clark, K.J.; Ekker, S.C. Mojo Hand, a TALEN design tool for genome editing applications. BMC Bioinform. 2013, 14, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedell, V.; Ekker, S.C. Using Engineered Endonucleases to Create Knockout and Knockin Zebrafish Models. In Chromosomal Mutagenesis; Humana Press: New York, NY, USA, 2015; pp. 291–305. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Štih, V.; Petrucco, L.; Kist, A.M.; Portugues, R. Stytra: An open-source, integrated system for stimulation, tracking and closed-loop behavioral experiments. PLoS Comput. Biol. 2019, 15, e1006699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
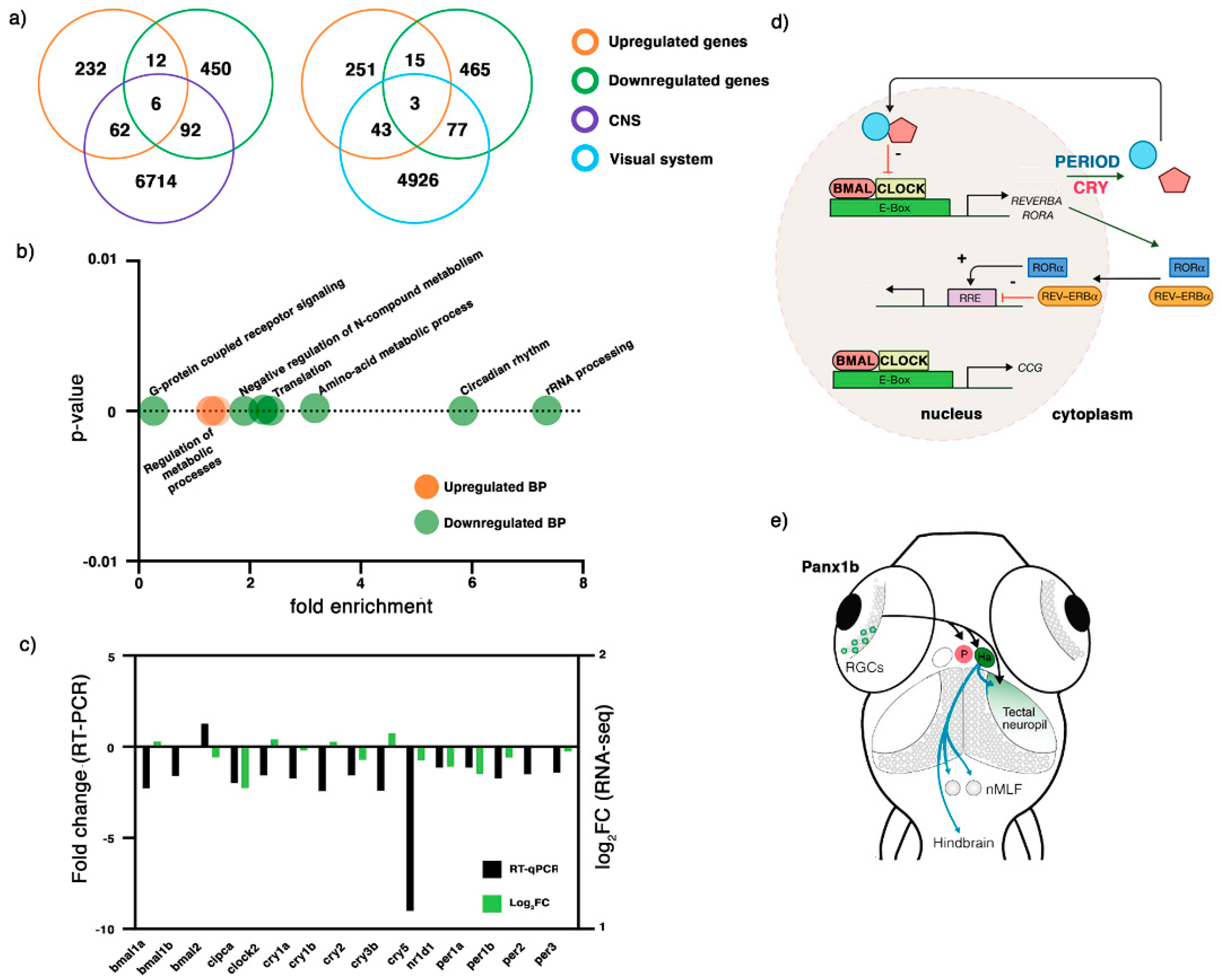
| Biological Process (Up) | Danio Rerio (REF) # | Upload # | Fold Enrichment | p-Value |
| Regulation of primary metabolic process (GO:0080090) | 4132 | 170 | 1.4 | 0.0000062 |
| Regulation of nitrogen compound metabolic process (GO:0051171) | 4076 | 167 | 1.39 | 0.000009 |
| Regulation of macromolecule metabolic process (GO:0060255) | 4368 | 178 | 1.39 | 0.00000623 |
| Biological Process (Down) | Danio Rerio (REF) # | Upload # | Fold Enrichment | p-Value |
| cleavage involved in rRNA processing (GO:0000469) | 26 | 8 | 7.35 | 0.0000474 |
| circadian regulation of gene expression (GO:0032922) | 45 | 11 | 5.84 | 0.0000119 |
| cellular modified amino acid metabolic process (GO:0006575) | 113 | 15 | 3.17 | 0.000199 |
| mRNA processing (GO:0006397) | 332 | 33 | 2.37 | 0.0000185 |
| translation (GO:0006412) | 320 | 30 | 2.24 | 0.0000975 |
| negative regulation of nitrogen compound metabolic process (GO:0051172) | 756 | 60 | 1.9 | 0.00000729 |
| G protein-coupled receptor signaling pathway (GO:0007186) | 1134 | 13 | 0.27 | 0.00000000543 |
| Gene | RT-qPCR Expression (REST) | SEM (REST) | p-Value (REST) | Result |
|---|---|---|---|---|
| bmal1a | −2.277 | 0.050 | 0.005 | Down |
| bmal1b | −1.609 | 0.053 | 0.019 | Down |
| bmal2 | 1.273 | 0.135 | 0.246 | |
| cipca | −1.988 | 0.060 | 0.055 | |
| clock2 | −1.568 | 0.063 | 0.017 | Down |
| cry1a | −1.749 | 0.109 | 0.004 | Down |
| cry1b | −2.43 | 0.086 | 0.002 | Down |
| cry2 | −1.569 | 0.073 | 0.058 | |
| cry3b | −2.42 | 0.052 | 0.11 | |
| cry5 | −9.002 | 0.033 | 0.0012 | Down |
| nr1d1 | −1.142 | 0.065 | 0.279 | |
| per1a | −1.142 | 0.062 | 0.451 | |
| per1b | −1.732 | 0.159 | 0.245 | |
| per2 | −1.503 | 0.082 | 0.154 | |
| per3 | −1.422 | 0.184 | 0.289 |
| TL | Panx1b−/− | ||||||
|---|---|---|---|---|---|---|---|
| Stimulus | Time Bin | Average TAD/s | S.E.M | Average TAD/s | S.E.M | Effect Estimator EE | Adjusted p-Value |
| Light-ON | Before (60–0 s) | 0.17 | 0.13 | 0.18 | 0.14 | 0.48 | 0.00 |
| AT (0–1 s) | 0.38 | 0.19 | 0.33 | 0.22 | 0.57 | 0.08 | |
| After (1–60 s) | 0.21 | 0.15 | 0.23 | 0.20 | 0.50 | 0.70 | |
| Light-OFF | Before (60–0 s) | 0..27 | 0.27 | 0.25 | 0.19 | 0.53 | 0.00 |
| AT (0–1 s) | 0.36 | 0.31 | 0.24 | 0.24 | 0.67 | 0.00 | |
| After (1–60 s) | 0.36 | 0.33 | 0.37 | 0.23 | 0.49 | 0.06 |
| Contrast Levels | Low (10%) | High (100%) | |||||
|---|---|---|---|---|---|---|---|
| Speed (pixels/s) | Direction | Spatial Frequencies (pixels/cycles) | |||||
| 64 | 128 | 256 | 64 | 128 | 256 | ||
| 72 | Leftward | 0.061 | 0.51 | 0.16 | 0.23 | 0.22 | 0.22 |
| Rightward | 0.52 | 0.77 | 0.44 | 0.89 | 0.8 | 0.33 | |
| 144 | Leftward | 0.22 | 0.13 | 0.0012 | 0.63 | 0.26 | 0.66 |
| Rightward | 0.97 | 0.97 | 0.97 | 0.65 | 0.99 | 0.77 | |
| Gene | ID | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Amplicon (bp) |
|---|---|---|---|---|
| per1a | NM_001030183 | GGAAAAGGCTCAGCCACAGA | TGAACTTCCGCTCAAAAGAC | 105 |
| per1b | NM_212439 | GAGGGAGACAGTCAAACAGAAG | AGAAGGGCTGTGAGAGTTTG | 116 |
| per2 | NM_182857 | CGTGCCCATTACAGTGAAGAG | ATGTTGACAGACCTGACTGC | 149 |
| per3 | NM_131584 | CAAGTACAAGCAAACAGCGAG | ACTACCACAAAAGAGTCCGTG | 146 |
| arntl1a/bmal1a | NM_131577 | TCAGATTGTTCCTGGTATTCCTG | GCGTGGCAGTGATGTTTAAAG | 149 |
| arntl1b/bmal1b | NM_178300 | GAGTTCATGTCTCCCAGTTCC | GTCCTTGTCTGTGTCCATACTG | 147 |
| arntl2/bmal2 | NM_131578 | ATGAACTCAAGCACCTCGTC | CTCTGTCCAATTAACTCCGTCC | 139 |
| nr1d1 | NM_205729 | AGCGACCTCCAAACAGTAAC | GGGTCTGAGTAAGGATGCATG | 142 |
| clocka | NM_130957 | CATCTCTTGTGTCTCAGCCTC | GTCTCTGGATGTTTGCCTCA | 150 |
| clockb | NM_178295 | GGTGGATCATGGAGAAGTGT | ATATACAGGCGGTTTCTTGGG | 148 |
| clock2/npas2 | NM_178299 | AACATCCTGAACTTCCTGCC | GTTTCCCCTCGCTATATGACAG | 150 |
| cipca | NM_001327903 | TTGTCCTCAAGCAGCCGAAC | GTACTGTCACCGCTGGGTAA | 136 |
| cry1a | NM_001077297 | CTCCACGACAATCCTTCACTC | CGTCCAAGTCTTCCAAACAC | 148 |
| cry1b | NM_131786 | GGTTGGACAAGCTCACAAAC | TTCCTTGGTTAGATGCCGTC | 126 |
| cry2 | NM_131786 | CTGTCATGTCGCGTGTTTTAC | CAAAGTTAGGGTTATTGGTGCC | 142 |
| cry3b | NM_131792 | CAGCAAATGTAAACCGTGGAG | CCTGACCCTATAAGACGATGC | 80 |
| cry5 | NM_131788 | CATGACTCAGTTACGACAGGAG | TCAGCGACCAATCAGAATCC | 149 |
| 18s rRNA | NM_001098396 | TGCATGGCCGTTCTTAGTTG | AGTCTCGTTCGTTATCGGAATGA | 86 |
| panx1a | NM_200916 | TGGATACAAACAGCTCTGTGATAG | ATCCTTTAGAGTAGCGCTTGG | 96 |
| panx1b | NM_001100030 | AAGTATAAAGGCGTGCGGCTGG | TGATCTGAGTACCCACAGAGAC | 129 |
| panx2 | NM_001256641 | TCGACTGAGAGGACAAACCC | TCGCCACCAACTTCACAACTC | 99 |
| panx3 | XM_001919826 | CATACAACCGCTCTGTCCGT | GCTTGCGTTTGGCTCTCTC | 105 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Safarian, N.; Houshangi-Tabrizi, S.; Zoidl, C.; Zoidl, G.R. Panx1b Modulates the Luminance Response and Direction of Locomotion in the Zebrafish. Int. J. Mol. Sci. 2021, 22, 11750. https://doi.org/10.3390/ijms222111750
Safarian N, Houshangi-Tabrizi S, Zoidl C, Zoidl GR. Panx1b Modulates the Luminance Response and Direction of Locomotion in the Zebrafish. International Journal of Molecular Sciences. 2021; 22(21):11750. https://doi.org/10.3390/ijms222111750
Chicago/Turabian StyleSafarian, Nickie, Sarah Houshangi-Tabrizi, Christiane Zoidl, and Georg R. Zoidl. 2021. "Panx1b Modulates the Luminance Response and Direction of Locomotion in the Zebrafish" International Journal of Molecular Sciences 22, no. 21: 11750. https://doi.org/10.3390/ijms222111750
APA StyleSafarian, N., Houshangi-Tabrizi, S., Zoidl, C., & Zoidl, G. R. (2021). Panx1b Modulates the Luminance Response and Direction of Locomotion in the Zebrafish. International Journal of Molecular Sciences, 22(21), 11750. https://doi.org/10.3390/ijms222111750






