Take Me Home, Protein Roads: Structural Insights into Signal Peptide Interactions during ER Translocation
Abstract
:1. Introduction
2. SP Recognition in the Cytosol and ER Targeting
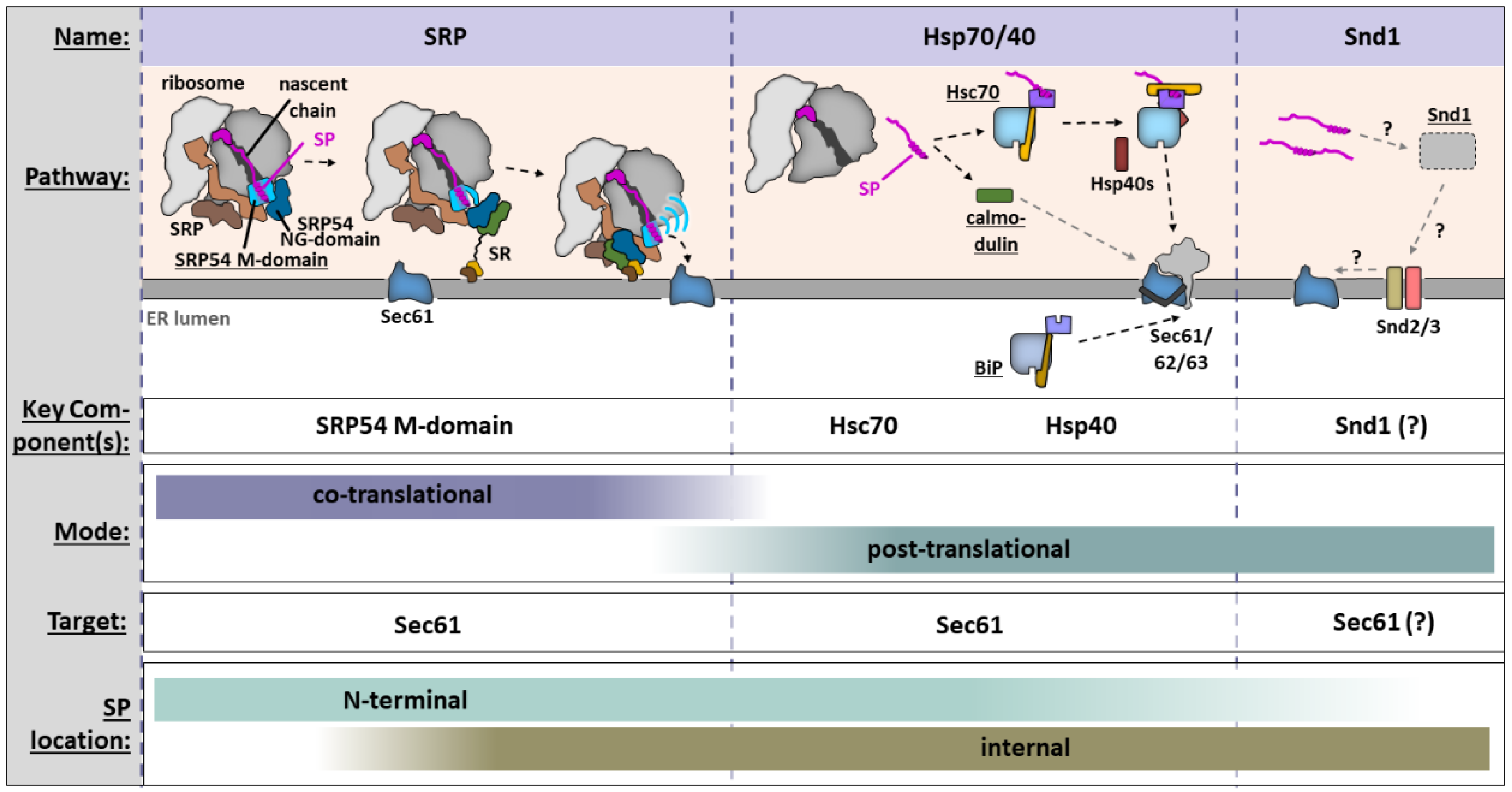
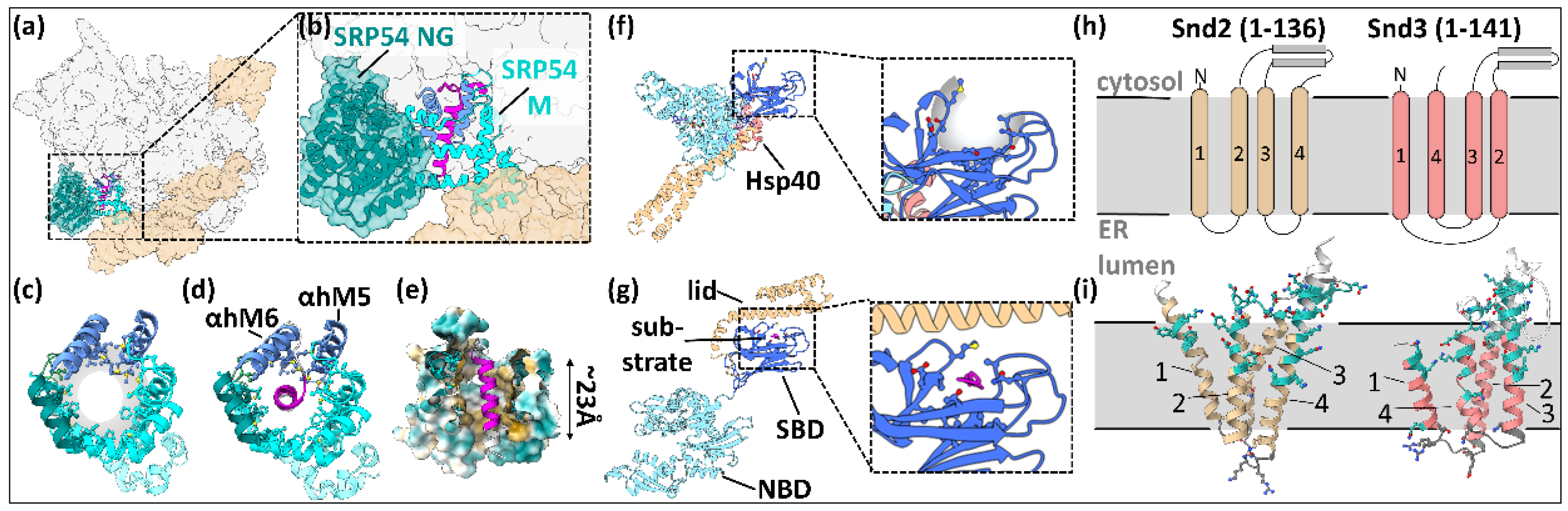
2.1. Life in the Fast Lane: Co-Translational Recognition and ER Targeting by the SRP
2.2. With a Little Help from My Friends: Post-Translational Recognition and ER Targeting
2.2.1. Stairway to Lumen: Hsp70/40-Mediated Chaperoning in the Cytosol and ER Delivery
2.2.2. The Joker: The Snd Pathway
3. Protein Translocation and Insertion at the ER Membrane
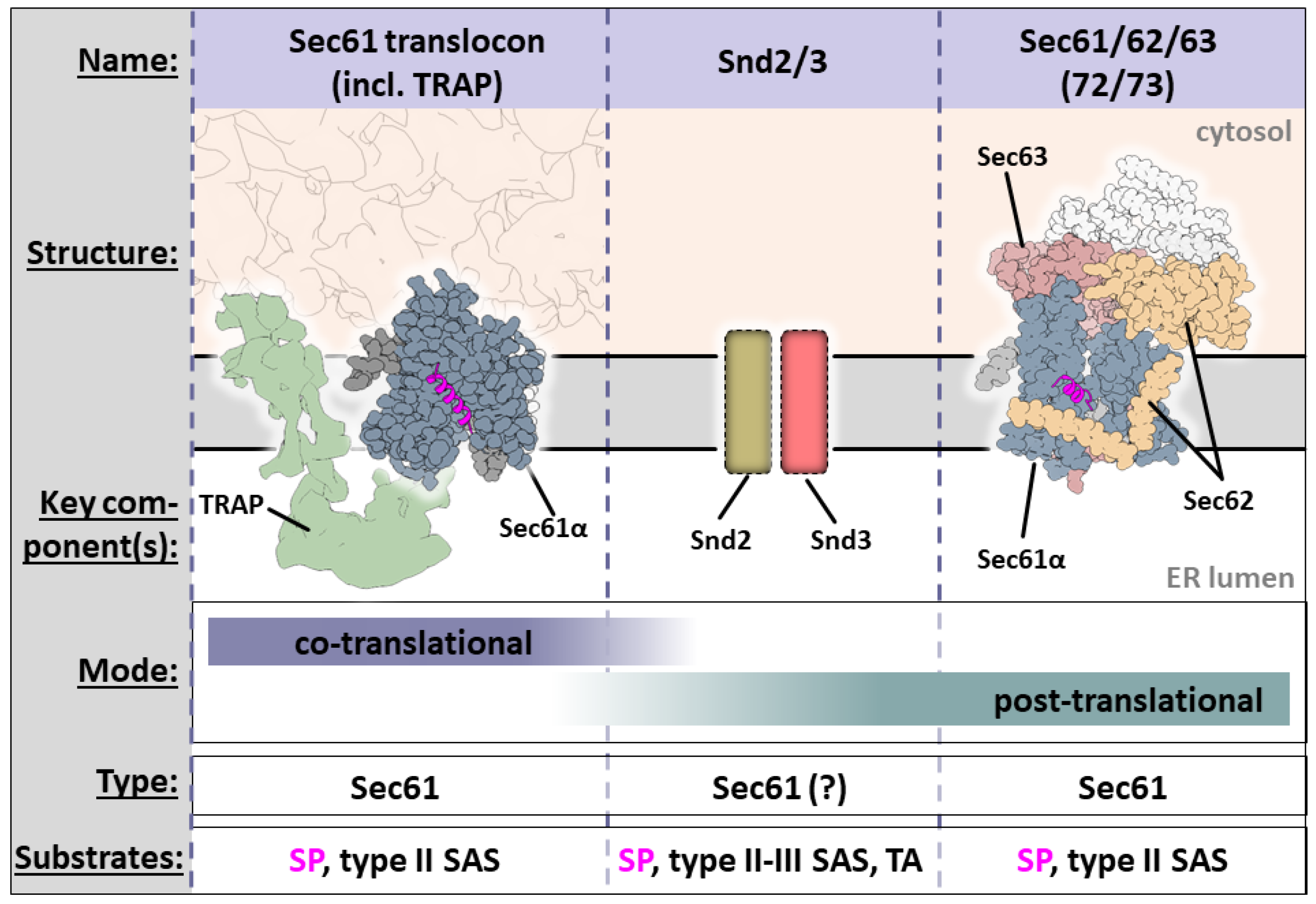
3.1. Through the Barricades: Co-Translational Translocation/Insertion
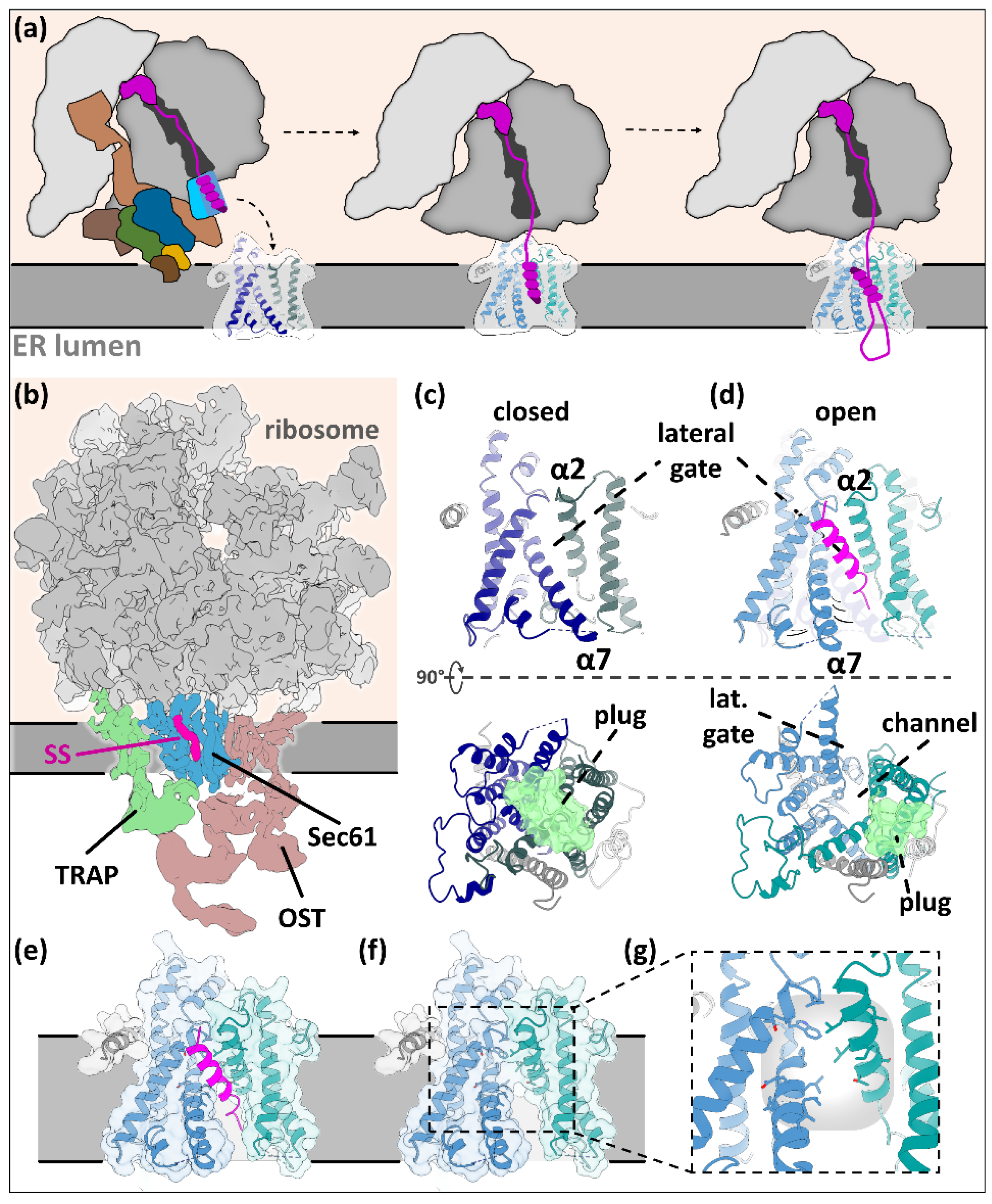
3.1.1. Tunnel of Love: Protein Translocation by Sec61
3.1.2. Smooth Operator: TRAP Is a Sec61 Assistant
3.2. Insane in the Membrane: Post-Translational Translocation/Insertion
With Arms Wide Open: Post-Translational Insertion and Translocation Assisted by Sec62/63
4. SP Removal and Post-Targeting Functions
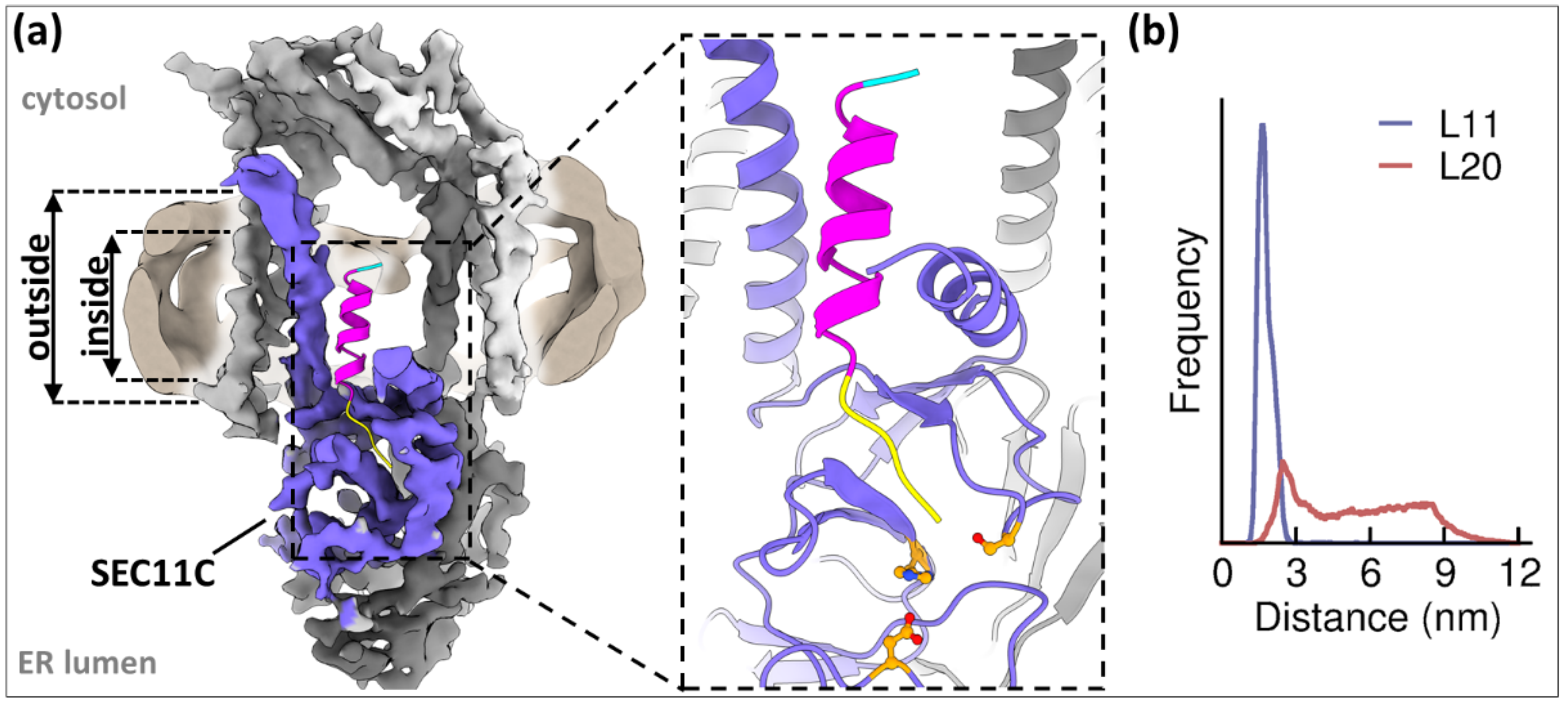
4.1. Time to Say Goodbye: SP Removal by the Signal Peptidase Complex
4.2. The Show Must Go On: Post-Targeting Functions of SPs
5. Summary and Outlook
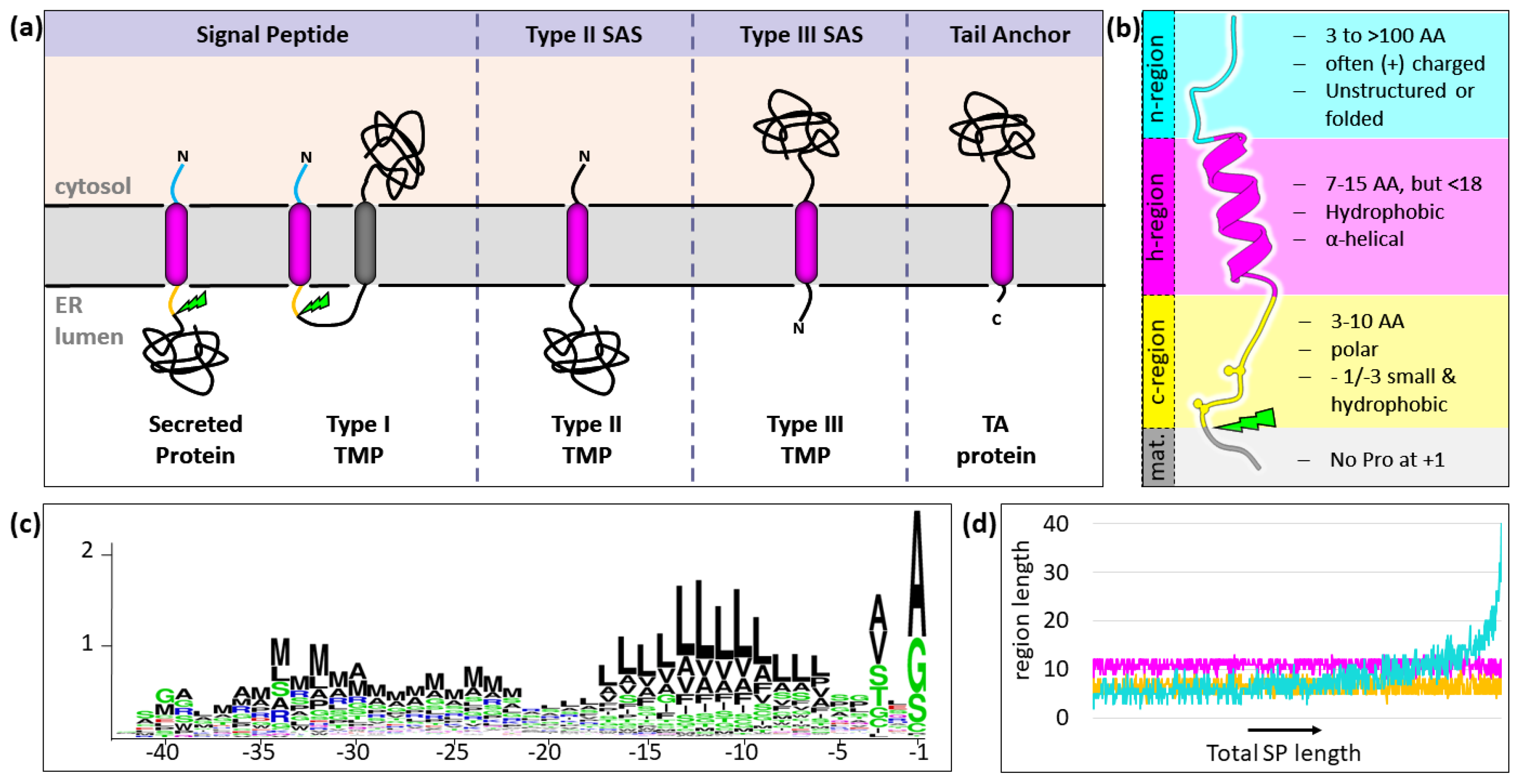
5.1. The Long and Winding Road: Principles of SP Recognition
5.2. Imagine: Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Palade, G.; Arch, A.; Locke, M.; Locke, E.; Palade, G. Intracellular aspects of the process of protein synthesis. Science 1975, 189, 347–358. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Aviram, N.; Schuldiner, M. Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J. Cell Sci. 2017, 130, 4079–4085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapoport, T.A.; Li, L.; Park, E. Structural and mechanistic insights into protein translocation. Annu. Rev. Cell Dev. Biol. 2017, 33, 369–390. [Google Scholar] [CrossRef]
- Liaci, A.M.; Steigenberger, B.; Tamara, S.; Telles de Souza, P.C.; Gröllers-Mulderij, M.; Ogrissek, P.; Marrink, S.J.; Scheltema, R.; Förster, F. Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage. Mol. Cell 2021, 81, 3934–3948. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, C.A.; Preuss, D.; Grisafi, P.; Botstein, D. Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science 1987, 235, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Von Heijne, G. The signal peptide. J. Membr. Biol. 1990, 115, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Liaci, A.M.; Förster, F. Empirical Analysis of Eukaryotic ER Signal Peptides; Mendeley Data; Data Archiving and Networked Services (DANS): The Hague, The Netherlands, 2021. [Google Scholar] [CrossRef]
- McDowell, M.A.; Heimes, M.; Sinning, I. Structural and molecular mechanisms for membrane protein biogenesis by the Oxa1 superfamily. Nat. Struct. Mol. Biol. 2021, 28, 234–239. [Google Scholar] [CrossRef]
- Hegde, R.S.; Keenan, R.J. The mechanisms of integral membrane protein biogenesis. Nat. Rev. Mol. Cell Biol. 2021, 1–18. [Google Scholar] [CrossRef]
- O’Keefe, S.; Pool, M.R.; High, S. Membrane protein biogenesis at the ER: The highways and byways. FEBS J. 2021, 1–28. [Google Scholar] [CrossRef]
- Hegde, R.S.; Bernstein, H.D. The surprising complexity of signal sequences. Trends Biochem. Sci. 2006, 31, 563–571. [Google Scholar] [CrossRef]
- Borgese, N.; Coy-Vergara, J.; Colombo, S.F.; Schwappach, B. The ways of tails: The GET pathway and more. Protein J. 2019, 38, 289–305. [Google Scholar] [CrossRef]
- Jomaa, A.; Eitzinger, S.; Zhu, Z.; Chandrasekar, S.; Kobayashi, K.; Shan, S.; Ban, N. Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle. Cell Rep. 2021, 36, 109350. [Google Scholar] [CrossRef] [PubMed]
- Aviram, N.; Ast, T.; Costa, E.A.; Arakel, E.C.; Chartzman, S.G.; Jan, C.H.; Haßdenteufel, S.; Dudek, J.; Jung, M.; Schorr, S.; et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 2016, 540, 134–138. [Google Scholar] [CrossRef]
- Haßdenteufel, S.; Sicking, M.; Schorr, S.; Aviram, N.; Fecher-Trost, C.; Schuldiner, M.; Jung, M.; Zimmermann, R.; Lang, S. hSnd2 protein represents an alternative targeting factor to the endoplasmic reticulum in human cells. FEBS Lett. 2017, 591, 3211–3224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamerdinger, M.; Hanebuth, M.A.; Frickey, T.; Deuerling, E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science 2015, 348, 201–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakkaraju, A.K.K.; Thankappan, R.; Mary, C.; Garrison, J.L.; Taunton, J.; Strub, K. Efficient secretion of small proteins in mammalian cells relies on Sec62-dependent posttranslational translocation. Mol. Biol. Cell 2012, 23, 2712–2722. [Google Scholar] [CrossRef]
- Shao, S.; Hegde, R.S. Membrane protein insertion at the endoplasmic reticulum. Annu. Rev. Cell Dev. Biol. 2011, 27, 25–56. [Google Scholar] [CrossRef] [Green Version]
- Johnson, N.; Vilardi, F.; Lang, S.; Leznicki, P.; Zimmermann, R.; High, S. TRC40 can deliver short secretory proteins to the Sec61 translocon. J. Cell Sci. 2012, 125, 4414. [Google Scholar] [CrossRef] [Green Version]
- Costa, E.A.; Subramanian, K.; Nunnari, J.; Weissman, J.S. Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 2018, 359, 689–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chirico, W.J.; Waters, M.G.; Blobel, G. 70K heat shock related proteins stimulate protein translocation into microsomes. Nature 1988, 332, 805–810. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Koch, B.D.; Werner-Washburne, M.; Craig, E.A.; Schekman, R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 1988, 332, 800–805. [Google Scholar] [CrossRef]
- Ng, D.T.W.; Brown, J.D.; Walter, P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 1996, 134, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Plath, K.; Mothes, W.; Wilkinson, B.M.; Stirling, C.J.; Rapoport, T.A. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell 1998, 94, 795–807. [Google Scholar] [CrossRef] [Green Version]
- Garcia, P.D.; Walter, P. Full-length prepro-α-factor can be translocated across the mammalian microsomal membrane only if translation has not terminated. J. Cell Biol. 1988, 106, 1043–1048. [Google Scholar] [CrossRef] [Green Version]
- Von Heijne, G. Signal sequences. The limits of variation. J. Mol. Biol. 1985, 184, 99–105. [Google Scholar] [CrossRef]
- Schibich, D.; Gloge, F.; Pöhner, I.; Björkholm, P.; Wade, R.C.; Von Heijne, G.; Bukau, B.; Kramer, G. Global profiling of SRP interaction with nascent polypeptides. Nature 2016, 536, 219–223. [Google Scholar] [CrossRef]
- Chartron, J.W.; Hunt, K.C.L.; Frydman, J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature 2016, 536, 224–228. [Google Scholar] [CrossRef]
- O’Keefe, S.; Zong, G.; Duah, K.B.; Andrews, L.E.; Shi, W.Q.; High, S. An alternative pathway for membrane protein biogenesis at the endoplasmic reticulum. Commun. Biol. 2021, 4, 828. [Google Scholar] [CrossRef]
- Raue, U.; Oellerer, S.; Rospert, S. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J. Biol. Chem. 2007, 282, 7809–7816. [Google Scholar] [CrossRef] [Green Version]
- Noriega, T.R.; Tsai, A.; Elvekrog, M.M.; Petrov, A.; Neher, S.B.; Chen, J.; Bradshaw, N.; Puglisi, J.D.; Walter, P. Signal recognition particle-ribosome binding is sensitive to nascent chain length. J. Biol. Chem. 2014, 289, 19294–19305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, N.; Haßdenteufel, S.; Theis, M.; Paton, A.W.; Paton, J.C.; Zimmermann, R.; High, S. The signal sequence influences post-translational ER translocation at distinct stages. PLoS ONE 2013, 8, e75394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goder, V.; Crottet, P.; Spiess, M. In vivo kinetics of protein targeting to the endoplasmic reticulum determined by site-specific phosphorylation. EMBO J. 2000, 19, 6704–6712. [Google Scholar] [CrossRef] [Green Version]
- Voorhees, R.M.; Hegde, R.S. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. eLife 2015, 4, e07975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bornemann, T.; Jöckel, J.; Rodnina, M.V.; Wintermeyer, W. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 2008, 15, 494–499. [Google Scholar] [CrossRef]
- Denks, K.; Sliwinski, N.; Erichsen, V.; Borodkina, B.; Origi, A.; Koch, H.G. The signal recognition particle contacts uL23 and scans substrate translation inside the ribosomal tunnel. Nat. Microbiol. 2017, 2, 16265. [Google Scholar] [CrossRef]
- Jomaa, A.; Boehringer, D.; Leibundgut, M.; Ban, N. Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat. Commun. 2016, 7, 10471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berndt, U.; Oellerer, S.; Zhang, Y.; Johnson, A.E.; Rospert, S. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc. Natl. Acad. Sci. USA 2009, 106, 1398–1403. [Google Scholar] [CrossRef] [Green Version]
- Flanagan, J.J.; Chen, J.C.; Miao, Y.; Shao, Y.; Lin, J.; Bock, P.E.; Johnson, A.E. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J. Biol. Chem. 2003, 278, 18628–18637. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Chandrasekar, S.; Chung, S.Y.; Fu, Y.H.H.; Liu, D.; Weiss, S.; Shan, S.O. Sequential activation of human signal recognition particle by the ribosome and signal sequence drives efficient protein targeting. Proc. Natl. Acad. Sci. USA 2018, 115, E5487–E5496. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, I.; Lara, P.; Hessa, T.; Johnson, A.E.; von Heijne, G.; Karamyshev, A.L. The code for directing proteins for translocation across ER membrane: SRP cotranslationally recognizes specific features of a signal sequence. J. Mol. 2015, 427, 1191–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janda, C.Y.; Li, J.; Oubridge, C.; Hernández, H.; Robinson, C.V.; Nagai, K. Recognition of a signal peptide by the signal recognition particle. Nature 2010, 465, 507–510. [Google Scholar] [CrossRef]
- Hainzl, T.; Huang, S.; Meriläinen, G.; Brännström, K.; Sauer-Eriksson, A.E. Structural basis of signal-sequence recognition by the signal recognition particle. Nat. Struct. Mol. Biol. 2011, 18, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Jomaa, A.; Fu, Y.H.H.; Boehringer, D.; Leibundgut, M.; Shan, S.O.; Ban, N. Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome. Nat. Commun. 2017, 8, 15470. [Google Scholar] [CrossRef]
- Wang, S.; Jomaa, A.; Jaskolowski, M.; Yang, C.I.; Ban, N.; Shan, S. The molecular mechanism of cotranslational membrane protein recognition and targeting by SecA. Nat. Struct. Mol. Biol. 2019, 26, 919–929. [Google Scholar] [CrossRef]
- Powers, T.; Walter, P. The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr. Biol. 1996, 6, 331–338. [Google Scholar] [CrossRef] [Green Version]
- Walter, P.; Blobel, G. Translocation of proteins across the endoplasmic reticulum. III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 1981, 91, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Mason, N.; Ciufo, L.F.; Brown, J.D. Elongation arrest is a physiologically important function of signal recognition particle. EMBO J. 2000, 19, 4164–4174. [Google Scholar] [CrossRef] [Green Version]
- Lakkaraju, A.K.K.; Mary, C.; Scherrer, A.; Johnson, A.E.; Strub, K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell 2008, 133, 440–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariosa, A.R.; Duncan, S.S.; Saraogi, I.; Lu, X.; Brown, A.; Phillips, G.J.; Shan, S.O. Fingerloop activates cargo delivery and unloading during cotranslational protein targeting. Mol. Biol. Cell 2013, 24, 63–73. [Google Scholar] [CrossRef]
- Egea, P.F.; Shan, S.O.; Napetschnig, J.; Savage, D.F.; Walter, P.; Stroud, R.M. Substrate twinning activates the signal recognition particle and its receptor. Nature 2004, 427, 215–221. [Google Scholar] [CrossRef]
- Freymann, D.M.; Keenan, R.J.; Stroud, R.M.; Walter, P. Structure of the conserved GTPase domain of the signal recognition particle. Nature 1997, 385, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, S.; Freymann, D.M. The conformation of bound GMPPNP suggests a mechanism for gating the active site of the SRP GTPase. Structure 2001, 9, 859–867. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, K.; Jomaa, A.; Lee, J.H.; Chandrasekar, S.; Boehringer, D.; Shan, S.; Ban, N. Structure of a prehandover mammalian ribosomal SRP · SRP receptor targeting complex. Science 2018, 327, 323–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, K.; Arslan, S.; Akopian, D.; Ha, T.; Shan, S. Activated GTPase movement on an RNA scaffold drives co-translational protein targeting. Nature 2012, 492, 271–275. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, H.-H.; Lee, J.H.; Chandrasekar, S.; Shan, S. A ribosome-associated chaperone enables substrate triage in a cotranslational protein targeting complex. Nat. Commun. 2020, 11, 5840. [Google Scholar] [CrossRef] [PubMed]
- Karamyshev, A.L.; Patrick, A.E.; Karamysheva, Z.N.; Griesemer, D.S.; Hudson, H.; Tjon-Kon-Sang, S.; Nilsson, I.; Otto, H.; Liu, Q.; Rospert, S.; et al. Inefficient SRP interaction with a nascent chain triggers a MRNA quality control pathway. Cell 2014, 156, 146–157. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, R.; Sagstetter, M.; Lewis, M.J.; Pelham, H.R. Seventy-kilodalton heat shock proteins and an additional component from reticulocyte lysate stimulate import of M13 procoat protein into microsomes. EMBO J. 1988, 7, 2875–2880. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Hegde, R.S. A calmodulin-dependent translocation pathway for small secretory proteins. Cell 2011, 147, 1576–1588. [Google Scholar] [CrossRef] [Green Version]
- Johnson, N.; Powis, K.; High, S. Post-translational translocation into the endoplasmic reticulum. Biochim. Biophys. Acta—Mol. Cell Res. 2013, 1833, 2403–2409. [Google Scholar] [CrossRef] [PubMed]
- Schorr, S.; Nguyen, D.; Haßdenteufel, S.; Nagaraj, N.; Cavalié, A.; Greiner, M.; Weissgerber, P.; Loi, M.; Paton, A.W.; Paton, J.C.; et al. Identification of signal peptide features for substrate specificity in human Sec62/Sec63-dependent ER protein import. FEBS J. 2020, 287, 4612–4640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, D.; Stutz, R.; Schorr, S.; Lang, S.; Pfeffer, S.; Freeze, H.H.; Förster, F.; Helms, V.; Dudek, J.; Zimmermann, R. Proteomics reveals signal peptide features determining the client specificity in human TRAP-dependent ER protein import. Nat. Commun. 2018, 9, 3765. [Google Scholar] [CrossRef] [Green Version]
- Rüdiger, S.; Germeroth, L.; Schneider-Mergener, J.; Bukau, B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997, 16, 1501–1507. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, A.; Mandon, E.C.; Gilmore, R.; Rapoport, T.A. Two alternative binding mechanisms connect the protein translocation Sec71-Sec72 complex with heat shock proteins. J. Biol. Chem. 2017, 292, 8007–8018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngosuwan, J.; Wang, N.M.; Fung, K.L.; Chirico, W.J. Roles of cytosolic Hsp70 and Hsp40 molecular chaperones in post-translational translocation of presecretory proteins into the endoplasmic reticulum. J. Biol. Chem. 2003, 278, 7034–7042. [Google Scholar] [CrossRef] [Green Version]
- Ast, T.; Cohen, G.; Schuldiner, M. A network of cytosolic factors targets SRP-independent proteins to the endoplasmic reticulum. Cell 2013, 152, 1134–1145. [Google Scholar] [CrossRef] [Green Version]
- Becker, T.; Song, J.; Pfanner, N. Versatility of preprotein transfer from the cytosol to mitochondria. Trends Cell Biol. 2019, 29, 534–548. [Google Scholar] [CrossRef]
- Caplan, A.J.; Cyr, D.M.; Douglas, M.G. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell 1992, 71, 1143–1155. [Google Scholar] [CrossRef]
- Craig, E.A. Hsp70 at the membrane: Driving protein translocation. BMC Biol. 2018, 16, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kityk, R.; Kopp, J.; Mayer, M.P. Molecular mechanism of J-domain-triggered ATP hydrolysis by Hsp70 chaperones. Mol. Cell 2018, 69, 227–237. [Google Scholar] [CrossRef]
- Craig, E.A.; Marszalek, J. How do J-proteins get Hsp70 to do so many different things? Trends Biochem. Sci. 2017, 42, 355–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabu, C.; Wipf, P.; Brodsky, J.L.; High, S. A precursor-specific role for Hsp40/Hsc70 during tail-anchored protein integration at the endoplasmic reticulum. J. Biol. Chem. 2008, 283, 27504–27513. [Google Scholar] [CrossRef] [Green Version]
- Müller, G.; Zimmermann, R. Import of honeybee prepromelittin into the endoplasmic reticulum: Structural basis for independence of SRP and docking protein. EMBO J. 1987, 6, 2099–2107. [Google Scholar] [CrossRef] [PubMed]
- Guna, A.; Volkmar, N.; Christianson, J.C.; Hegde, R.S. The ER membrane protein complex is a transmembrane domain insertase. Science 2018, 359, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Haßdenteufel, S.; Johnson, N.; Paton, A.W.; Paton, J.C.; High, S.; Zimmermann, R. Chaperone-mediated Sec61 channel gating during ER import of small precursor proteins overcomes Sec61 inhibitor-reinforced energy barrier. Cell Rep. 2018, 23, 1373–1386. [Google Scholar] [CrossRef]
- Casson, J.; McKenna, M.; Haßdenteufel, S.; Aviram, N.; Zimmerman, R.; High, S. Multiple pathways facilitate the biogenesis of mammalian tail-anchored proteins. J. Cell Sci. 2017, 130, 3851–3861. [Google Scholar] [CrossRef] [Green Version]
- Fleischer, T.C.; Weaver, C.M.; McAfee, K.J.; Jennings, J.L.; Link, A.J. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006, 20, 1294–1307. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Xiao, Y.; Li, P.; Xie, P.; Wang, H.; Huang, S.; Song, P.; Zhao, Y. hSnd2/TMEM208 is an HIF-1α-targeted gene and contains a WH2 motif. Acta Biochim. Biophys. Sin. 2020, 52, 328–331. [Google Scholar] [CrossRef]
- Gemmer, M.; Förster, F. A clearer picture of the ER translocon complex. J. Cell Sci. 2020, 133, jcs231340. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Dudek, J.; Schaffer, M.; Ng, B.G.; Albert, S.; Plitzko, J.M.; Baumeister, W.; Zimmermann, R.; Freeze, H.H.; Engel, B.D.; et al. Dissecting the molecular organization of the translocon-associated protein complex. Nat. Commun. 2017, 8, 14516. [Google Scholar] [CrossRef]
- McGilvray, P.T.; Anghel, S.A.; Sundaram, A.; Zhong, F.; Trnka, M.J.; Fuller, J.R.; Hu, H.; Burlingame, A.L.; Keenan, R.J. An ER translocon for multi-pass membrane protein biogenesis. eLife 2020, 9, e56889. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Siggel, M.; Ovchinnikov, S.; Mi, W.; Svetlov, V.; Nudler, E.; Liao, M.; Hummer, G.; Rapoport, T.A. Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex. Science 2020, 368, eaaz2449. [Google Scholar] [CrossRef]
- Itskanov, S.; Park, E. Structure of the posttranslational Sec protein-translocation channel complex from yeast. Science 2019, 363, 84–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pleiner, T.; Tomaleri, G.P.; Januszyk, K.; Inglis, A.J.; Hazu, M.; Voorhees, R.M. Structural basis for membrane insertion by the human ER membrane protein complex. Science 2020, 369, 433–436. [Google Scholar] [CrossRef]
- Bai, L.; You, Q.; Feng, X.; Kovach, A.; Li, H. Structure of the ER membrane complex, a transmembrane-domain insertase. Nature 2020, 584, 475–478. [Google Scholar] [CrossRef]
- Miller-Vedam, L.E.; Bräuning, B.; Popova, K.D.; Oakdale, N.T.S.; Bonnar, J.L.; Prabu, J.R.; Boydston, E.A.; Sevillano, N.; Shurtleff, M.J.; Stroud, R.M.; et al. Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients. eLife 2020, 9, e62611. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, J.P.; Phillips, B.P.; Yagita, Y.; Juszkiewicz, S.; Wagner, A.; Malinverni, D.; Keenan, R.J.; Miller, E.A.; Hegde, R.S. The architecture of EMC reveals a path for membrane protein insertion. eLife 2020, 9, e57887. [Google Scholar] [CrossRef]
- Wild, R.; Kowal, J.; Eyring, J.; Ngwa, E.M.; Aebi, M.; Locher, K.P. Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science 2018, 5140, 545–550. [Google Scholar] [CrossRef] [Green Version]
- Braunger, K.; Pfeffer, S.; Shrimal, S.; Gilmore, R.; Berninghausen, O.; Mandon, E.C.; Becker, T.; Förster, F.; Beckmann, R. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science 2018, 360, 215–219. [Google Scholar] [CrossRef] [Green Version]
- Chitwood, P.J.; Juszkiewicz, S.; Guna, A.; Shao, S.; Hegde, R.S. EMC is required to initiate accurate membrane protein topogenesis. Cell 2018, 175, 1507–1519. [Google Scholar] [CrossRef]
- Voorhees, R.M.; Hegde, R.S. Structure of the Sec61 channel opened by a signal sequence. Science 2016, 351, 88–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, T.; Steinchen, W.; Beatrix, B.; Berninghausen, O.; Becker, T.; Bange, G.; Cheng, J.; Beckmann, R. Architecture of the active post-translational Sec translocon. EMBO J. 2021, 40, e105643. [Google Scholar] [CrossRef]
- Van Den Berg, B.; Clemons, W.M.; Collinson, I.; Modis, Y.; Hartmann, E.; Harrison, S.C.; Rapoport, T.A. X-ray structure of a protein-conducting channel. Nature 2004, 427, 36–44. [Google Scholar] [CrossRef]
- Voorhees, R.M.; Fernández, I.S.; Scheres, S.H.W.; Hegde, R.S. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 2014, 157, 1632–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gogala, M.; Becker, T.; Beatrix, B.; Armache, J.P.; Barrio-Garcia, C.; Berninghausen, O.; Beckmann, R. Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 2014, 506, 107–110. [Google Scholar] [CrossRef]
- Park, E.; Rapoport, T.A. Mechanisms of Sec61SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 2012, 41, 21–40. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Park, E.; Ling, J.J.; Ingram, J.; Ploegh, H.; Rapoport, T.A. Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature 2016, 531, 395–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamayun, I.; O’Keefe, S.; Pick, T.; Klein, M.C.; Nguyen, D.; McKibbin, C.; Piacenti, M.; Williams, H.M.; Flitsch, S.L.; Whitehead, R.C.; et al. Eeyarestatin compounds selectively enhance Sec61-mediated Ca2+ leakage from the endoplasmic reticulum. Cell Chem. Biol. 2019, 26, 571–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kriegler, T.; Magoulopoulou, A.; Amate Marchal, R.; Hessa, T. Measuring endoplasmic reticulum signal sequences translocation efficiency using the Xbp1 arrest peptide. Cell Chem. Biol. 2018, 25, 880–890. [Google Scholar] [CrossRef] [PubMed]
- Cymer, F.; Von Heijne, G. Cotranslational folding of membrane proteins probed by arrest-peptide- mediated force measurements. Proc. Natl. Acad. Sci. USA 2013, 110, 14640–14645. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.J.; Mitra, D.; Salerno, J.R.; Hegde, R.S. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev. Cell 2002, 2, 207–217. [Google Scholar] [CrossRef] [Green Version]
- Shaffer, K.L.; Sharma, A.; Snapp, E.L.; Hegde, R.S. Regulation of protein compartmentalization expands the diversity of protein function. Dev. Cell 2005, 9, 545–554. [Google Scholar] [CrossRef] [Green Version]
- Fons, R.D.; Bogert, B.A.; Hegde, R.S. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003, 160, 529–539. [Google Scholar] [CrossRef] [Green Version]
- Spiess, M.; Junne, T.; Janoschke, M. Membrane Protein Integration and Topogenesis at the ER. Protein J. 2019, 38, 306–316. [Google Scholar] [CrossRef]
- Pfeffer, S.; Burbaum, L.; Unverdorben, P.; Pech, M.; Chen, Y.; Zimmermann, R.; Beckmann, R.; Forster, F. Structure of the native Sec61 protein-conducting channel. Nat. Commun. 2015, 6, 8403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niesen, M.J.M.; Müller-Lucks, A.; Hedman, R.; von Heijne, G.; Miller, T.F. Forces on nascent polypeptides during membrane insertion and translocation via the Sec translocon. Biophys. J. 2018, 115, 1885–1894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kriegler, T.; Lang, S.; Notari, L.; Hessa, T. Prion protein translocation mechanism revealed by pulling force studies. J. Mol. Biol. 2020, 432, 4447–4465. [Google Scholar] [CrossRef]
- Kriegler, T.; Kiburg, G.; Hessa, T. Translocon-associated protein complex (TRAP) is crucial for co-translational translocation of pre-proinsulin. J. Mol. Biol. 2020, 432, 166694. [Google Scholar] [CrossRef]
- Pfeffer, S.; Dudek, J.; Zimmermann, R.; Forster, F. Organization of the native ribosome-translocon complex at the mammalian endoplasmic reticulum membrane. Biochim. Biophys. Acta 2016, 1860, 2122–2129. [Google Scholar] [CrossRef]
- Sommer, N.; Junne, T.; Kalies, K.U.; Spiess, M.; Hartmann, E. TRAP assists membrane protein topogenesis at the mammalian ER membrane. Biochim. Biophys. Acta—Mol. Cell Res. 2013, 1833, 3104–3111. [Google Scholar] [CrossRef] [Green Version]
- Cabelli, R.J.; Chen, L.; Tai, P.C.; Oliver, D.B. SecA protein is required for secretory protein translocation into E. coli membrane vesicles. Cell 1988, 55, 683–692. [Google Scholar] [CrossRef]
- Brundage, L.; Hendrick, J.P.; Schiebel, E.; Driessen, A.J.M.; Wickner, W. The purified E. coli integral membrane protein SecY E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell 1990, 62, 649–657. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Rhoads, D.; Tai, P.C. Alkaline phosphatase and OmpA protein can be translocated posttranslationally into membrane vesicles of Escherichia coli. J. Bacteriol. 1985, 161, 973–980. [Google Scholar] [CrossRef] [Green Version]
- Rothblatt, J.A.; Deshaies, R.J.; Sanders, S.L.; Daum, G.; Schekman, R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J. Cell Biol. 1989, 109, 2641–2652. [Google Scholar] [CrossRef] [Green Version]
- Meyer, H.A.; Grau, H.; Kraft, R.; Kostka, S.; Prehn, S.; Kalies, K.U.; Hartmann, E. Mammalian Sec61 is associated with Sec62 and Sec63. J. Biol. Chem. 2000, 275, 14550–14557. [Google Scholar] [CrossRef] [Green Version]
- Tyedmers, J.; Lerner, M.; Bies, C.; Dudek, J.; Skowronek, M.H.; Haas, I.G.; Heim, N.; Nastainczyk, W.; Volkmer, J.; Zimmermann, R. Homologs of the yeast Sec complex subunits Sec62p and Sec63p are abundant proteins in dog pancreas microsomes. Proc. Natl. Acad. Sci. USA 2000, 97, 7214–7219. [Google Scholar] [CrossRef] [Green Version]
- Deshaies, R.J.; Sanders, S.L.; Feldheim, D.A.; Schekman, R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 1991, 349, 806–808. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cabanos, C.; Rapoport, T.A. Structure of the post-translational protein translocation machinery of the ER membrane. Nature 2019, 566, 136–139. [Google Scholar] [CrossRef]
- Itskanov, S.; Kuo, K.M.; Gumbart, J.C.; Park, E. Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62. Nat. Struct. Mol. Biol. 2021, 28, 162–172. [Google Scholar] [CrossRef]
- Okun, M.M.; Eskridge, E.M.; Shields, D. Truncations of a secretory protein define minimum lengths required for binding to signal recognition particle and translation across the endoplasmic reticulum membrane. J. Biol. Chem. 1990, 265, 7478–7484. [Google Scholar] [CrossRef]
- Feldheim, D.; Rothblatt, J.; Schekman, R. Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol. Cell. Biol. 1992, 12, 3288–3296. [Google Scholar] [CrossRef] [Green Version]
- Matlack, K.E.S.; Misselwitz, B.; Plath, K.; Rapoport, T.A. BIP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell 1999, 97, 553–564. [Google Scholar] [CrossRef] [Green Version]
- Sanders, S.L.; Whitfield, K.M.; Vogel, J.P.; Rose, M.D.; Schekman, R.W. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell 1992, 69, 353–365. [Google Scholar] [CrossRef]
- Schäuble, N.; Lang, S.; Jung, M.; Cappel, S.; Schorr, S.; Ulucan, Ö.; Linxweiler, J.; Dudek, J.; Blum, R.; Helms, V.; et al. BiP-mediated closing of the Sec61 channel limits Ca2+ leakage from the ER. EMBO J. 2012, 31, 3282–3296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blau, M.; Mullapudi, S.; Becker, T.; Dudek, J.; Zimmermann, R.; Penczek, P.A.; Beckmann, R. ERj1p uses a universal ribosomal adaptor site to coordinate the 80S ribosome at the membrane. Nat. Struct. Mol. Biol. 2005, 12, 1015–1016. [Google Scholar] [CrossRef]
- Melnyk, A.; Rieger, H.; Zimmermann, R. The Networking of Chaperones by Co-Chaperones: Control of Cellular Protein Homeostasis; Blatch, G.L., Edkins, A.L., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 179–200. [Google Scholar]
- Collins, P.G.; Gilmore, R. Ribosome binding to the endoplasmic reticulum: A 180-kD protein identified by crosslinking to membrane-bound ribosomes is not required for ribosome binding activity. J. Cell Biol. 1991, 114, 639–649. [Google Scholar] [CrossRef]
- Conti, B.J.; Devaraneni, P.K.; Yang, Z.; David, L.L.; Skach, W.R. Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate-driven translocation events. Mol. Cell 2015, 58, 269–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, S.; Benedix, J.; Fedeles, S.V.; Schorr, S.; Schirra, C.; Schäuble, N.; Jalal, C.; Greiner, M.; Haßdenteufel, S.; Tatzelt, J.; et al. Different effects of Sec61α, Sec62 and Sec63 depletion on transport of polypeptides into the endoplasmic reticulum of mammalian cells. J. Cell Sci. 2012, 125, 1958–1969. [Google Scholar]
- Jadhav, B.; McKenna, M.; Johnson, N.; High, S.; Sinning, I.; Pool, M.R. Mammalian SRP receptor switches the Sec61 translocase from Sec62 to SRP-dependent translocation. Nat. Commun. 2015, 6, 10133. [Google Scholar] [CrossRef] [Green Version]
- Mothes, W.; Prehn, S.; Rapoport, T.A. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO J. 1994, 13, 3973–3982. [Google Scholar] [CrossRef]
- Nilsson, I.M.; Johnson, A.E.; Von Heijne, G. Cleavage of a tail-anchored protein by signal peptidase. FEBS Lett. 2002, 516, 106–108. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, I.M.; Whitley, P.; Von Heijne, G. The COOH-terminal ends of internal signal and signal-anchor sequences are positioned differently in the ER translocase. J. Cell Biol. 1994, 126, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, N.; Wu, M.J.; Shanmugam, S.; Yi, M.K. Delayed by design: Role of suboptimal signal peptidase processing of viral structural protein precursors in flaviviridae virus assembly. Viruses 2020, 12, 1090. [Google Scholar] [CrossRef]
- Pfeffer, S.; Dudek, J.; Gogala, M.; Schorr, S.; Linxweiler, J.; Lang, S.; Becker, T.; Beckmann, R.; Zimmermann, R.; Förster, F. Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nat. Commun. 2014, 5, 3072. [Google Scholar] [CrossRef] [Green Version]
- Paetzel, M.; Karla, A.; Strynadka, N.C.J.; Dalbey, R.E. Signal peptidases. Chem. Rev. 2002, 102, 4549–4579. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Chen, W.; Sun, J.; Guo, H.; Madley, R.; Xiong, Y.; Pan, X.; Wang, H.; Tai, A.W.; Weiss, M.A.; et al. Competitive inhibition of the endoplasmic reticulum signal peptidase by non-cleavable mutant preprotein cargos. J. Biol. Chem. 2015, 290, 28131–28140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lobigs, M.; Lee, E.; Ng, M.L.; Pavy, M.; Lobigs, P. A flavivirus signal peptide balances the catalytic activity of two proteases and thereby facilitates virus morphogenesis. Virology 2010, 401, 80–89. [Google Scholar] [CrossRef] [Green Version]
- Snapp, E.L.; McCaul, N.; Quandte, M.; Cabartova, Z.; Bontjer, I.; Källgren, C.; Nilsson, I.; Land, A.; Von Heijne, G.; Sanders, R.W.; et al. Structure and topology around the cleavage site regulate post-translational cleavage of the HIV-1 gp160 signal peptide. eLife 2017, 6, e26067. [Google Scholar] [CrossRef]
- Carrère-Kremer, S.; Montpellier, C.; Lorenzo, L.; Brulin, B.; Cocquerel, L.; Belouzard, S.; Penin, F.; Dubuisson, J. Regulation of hepatitis C virus polyprotein processing by signal peptidase involves structural determinants at the p7 sequence junctions. J. Biol. Chem. 2004, 279, 41384–41392. [Google Scholar] [CrossRef] [Green Version]
- Kapp, K.; Schrempf, S.; Lemberg, M.K.; Dobberstein, B. Protein Transport into the Endoplasmic Reticulum. In Protein Transport into the Endoplasmic Reticulum; Landes Bioscience: Austin, TX, USA, 2009; Volume 1, pp. 1–16. [Google Scholar]
- McDowell, M.A.; Heimes, M.; Fiorentino, F.; Mehmood, S.; Farkas, Á.; Coy-Vergara, J.; Wu, D.; Bolla, J.R.; Schmid, V.; Heinze, R.; et al. Structural basis of tail-anchored membrane protein biogenesis by the GET insertase complex. Mol. Cell 2020, 80, 72–86. [Google Scholar] [CrossRef]
- Aviram, N.; Schuldiner, M. Embracing the void-how much do we really know about targeting and translocation to the endoplasmic reticulum? Curr. Opin. Cell Biol. 2014, 29, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Fenech, E.J.; Ben-Dor, S.; Schuldiner, M. Double the Fun, Double the Trouble: Paralogs and Homologs Functioning in the Endoplasmic Reticulum. Annu. Rev. Biochem. 2020, 89, 637–666. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Canada, C.; Kelleher, D.J.; Gilmore, R. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 2009, 136, 272–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liaci, A.M.; Förster, F. Take Me Home, Protein Roads: Structural Insights into Signal Peptide Interactions during ER Translocation. Int. J. Mol. Sci. 2021, 22, 11871. https://doi.org/10.3390/ijms222111871
Liaci AM, Förster F. Take Me Home, Protein Roads: Structural Insights into Signal Peptide Interactions during ER Translocation. International Journal of Molecular Sciences. 2021; 22(21):11871. https://doi.org/10.3390/ijms222111871
Chicago/Turabian StyleLiaci, A. Manuel, and Friedrich Förster. 2021. "Take Me Home, Protein Roads: Structural Insights into Signal Peptide Interactions during ER Translocation" International Journal of Molecular Sciences 22, no. 21: 11871. https://doi.org/10.3390/ijms222111871
APA StyleLiaci, A. M., & Förster, F. (2021). Take Me Home, Protein Roads: Structural Insights into Signal Peptide Interactions during ER Translocation. International Journal of Molecular Sciences, 22(21), 11871. https://doi.org/10.3390/ijms222111871





