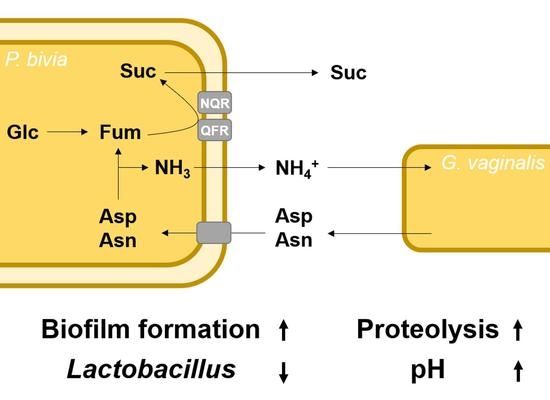
Central Carbon Metabolism, Sodium-Motive Electron Transfer, and Ammonium Formation by the Vaginal Pathogen Prevotella bivia
Abstract
:1. Introduction
2. Results
2.1. CO2-Dependent Succinate Formation by P. bivia
2.2. Ammonia Formation from L-Asparagine by P. bivia
2.3. Membrane-Bound Electron Transfer Complexes in P. bivia
2.4. NADH:Quinone and Quinol:Fumarate Oxidoreduction Activities of P. bivia Membranes
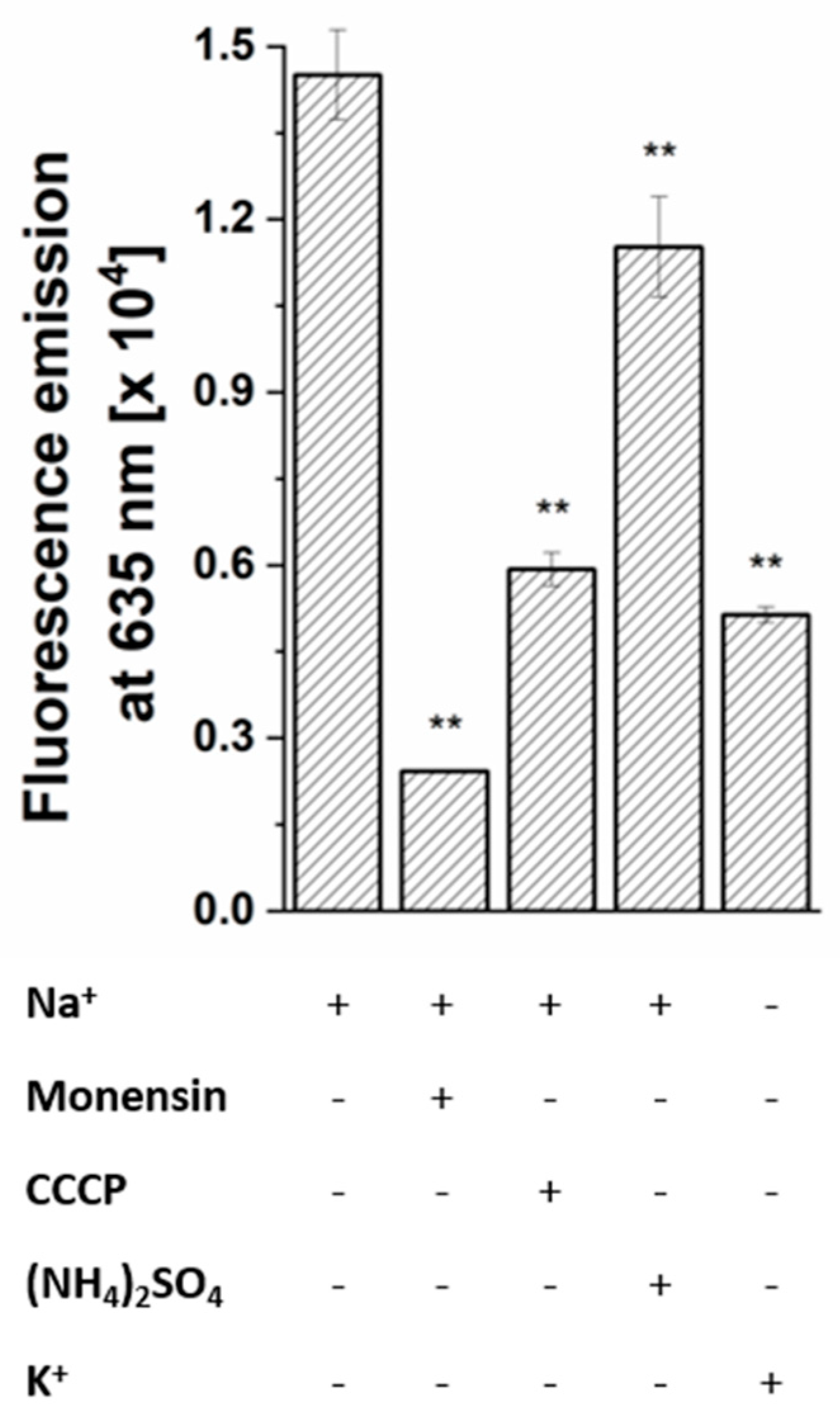
2.5. Sodium Dependent Membrane Potential in P. bivia
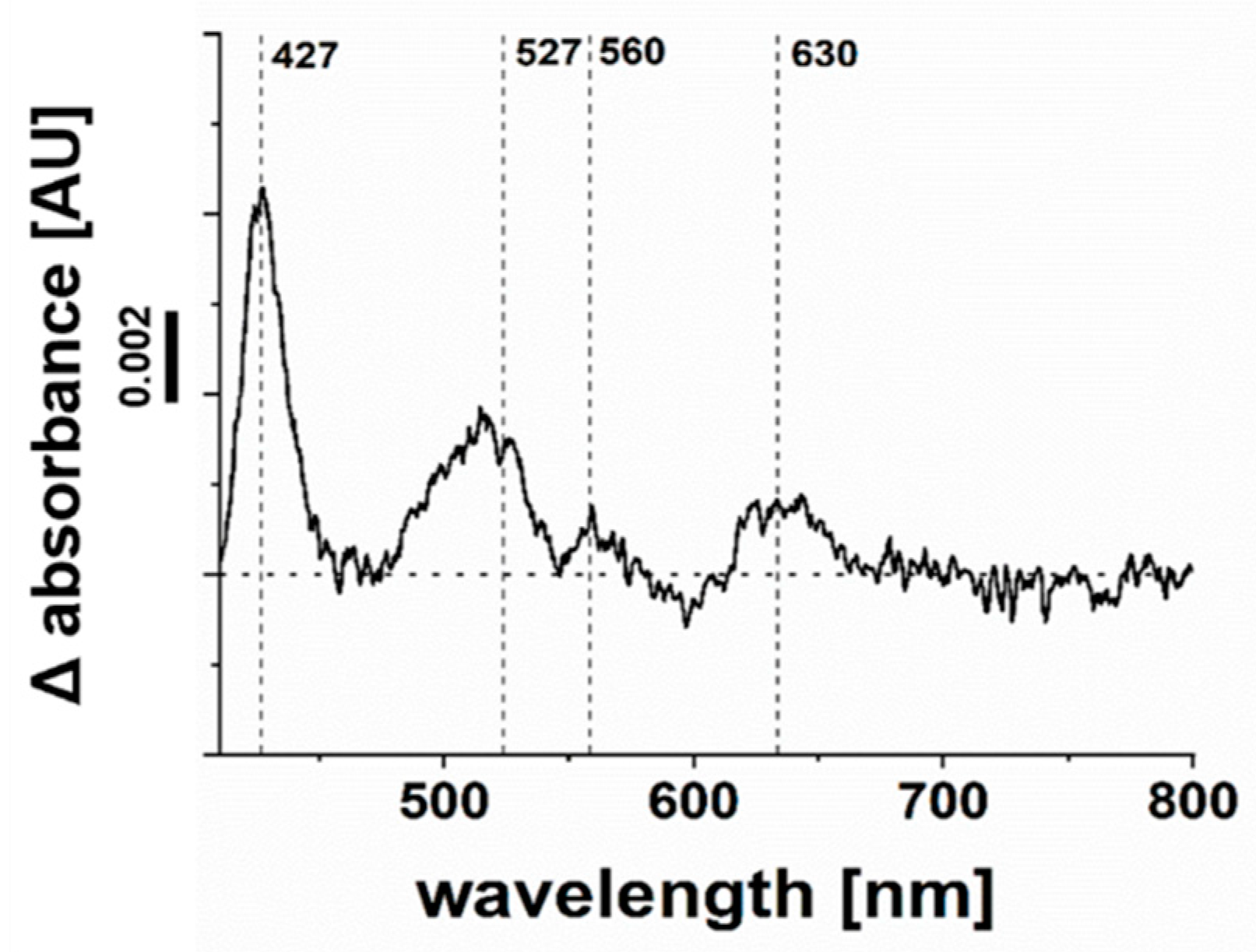
2.6. Cytochrome bd Quinol Oxidase of P. bivia
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Isolation and Solubilization of Membranes
4.3. Bacterial Growth
4.4. Analytical Methods
4.5. UV/Vis Difference Spectra of Redox Cofactors in P. bivia
4.6. Enzymatic Assays
4.7. Membrane Potential
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Onderdonk, A.B.; Delaney, M.L.; Fichorova, R.N. The human microbiome during bacterial vaginosis. Clin. Microbiol. Rev. 2016, 29, 223–238. [Google Scholar] [CrossRef] [Green Version]
- Muzny, C.A.; Taylor, C.M.; Swords, W.E.; Tamhane, A.; Chattopadhyay, D.; Cerca, N.; Schwebke, J.R. An updated conceptual model on the pathogenesis of bacterial vaginosis. J. Infect. Dis. 2019, 220, 1399–1405. [Google Scholar] [CrossRef]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.K.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4680–4687. [Google Scholar] [CrossRef] [Green Version]
- Nunn, K.L.; Clair, G.C.; Adkins, J.N.; Engbrecht, K.; Fillmore, T.; Forney, L.J. Amylases in the human vagina. mSphere 2020, 5, e00943-20. [Google Scholar] [CrossRef]
- Zozaya-Hinchliffe, M.; Lillis, R.; Martin, D.H.; Ferris, M.J. Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. J. Clin. Microbiol. 2010, 48, 1812–1819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delaney, M. Nugent score related to vaginal culture in pregnant women. Obstet. Gynecol. 2001, 98, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Aroutcheva, A.; Ling, Z.; Faro, S. Prevotella bivia as a source of lipopolysaccharide in the vagina. Anaerobe 2008, 14, 256–260. [Google Scholar] [CrossRef] [Green Version]
- Strömbeck, L.; Sandros, J.; Holst, E.; Madianos, P.; Nannmark, U.; Papapanou, P.; Mattsby-Baltzer, I. Prevotella bivia can invade human cervix epithelial (HeLa) cells. APMIS 2007, 115, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Mikamo, H.; Kawazoe, K.; Sato, Y.; Imai, A.; Tamaya, T. Preterm labor and bacterial intra-amniotic infection: Arachidonic acid liberation by phospholipase A2 of Prevotella bivia. Anaerobe 1998, 4, 209–212. [Google Scholar] [CrossRef]
- Gilbert, N.M.; Lewis, W.G.; Lewis, A.L. Clinical features of bacterial vaginosis in a murine model of vaginal infection with Gardnerella vaginalis. PLoS ONE. 2013, 8, e59539. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, N.M.; Lewis, W.G.; Li, G.; Sojka, D.K.; Lubin, J.B.; Lewis, A.L. Gardnerella vaginalis and Prevotella bivia trigger distinct and overlapping phenotypes in a mouse model of bacterial vaginosis. J. Infect. Dis. 2019, 220, 1099–1108. [Google Scholar] [CrossRef]
- Pybus, V.; Onderdonk, A.B. Evidence for a commensal, symbiotic relationship between Gardnerella vaginalis and Prevotella bivia involving ammonia: Potential significance for bacterial vaginosis. J. Infect. Dis. 1997, 175, 406–413. [Google Scholar] [CrossRef]
- Franke, T.; Deppenmeier, U. Physiology and central carbon metabolism of the gut bacterium Prevotella copri. Mol. Microbiol. 2018, 109, 528–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deusch, S.; Bok, E.; Schleicher, L.; Seifert, J.; Steuber, J. Occurrence and function of the Na+-translocating NADH:quinone oxidoreductase in Prevotella spp. Microorganisms 2019, 7, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trautmann, A.; Schleicher, L.; Deusch, S.; Gätgens, J.; Steuber, J.; Seifert, J. Short-chain fatty acids modulate metabolic pathways and membrane lipids in Prevotella bryantii B14. Proteomes 2020, 8, 28. [Google Scholar] [CrossRef]
- Pybus, V.; Onderdonk, A.B. The effect of pH on growth and succinate production by Prevotella bivia. Microb. Ecol. Health Dis. 1996, 9, 19–25. [Google Scholar] [CrossRef]
- Wolfe, A.J. The acetate switch. Microbiol. Mol. Biol. Rev. 2005, 69, 12–50. [Google Scholar] [CrossRef] [Green Version]
- Müller, T.; Walter, B.; Wirtz, A.; Burkovski, A. Ammonium toxicity in bacteria. Curr. Microbiol. 2006, 52, 400–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lancaster, C.D. Wolinella succinogenes quinol:fumarate reductase—2.2-Å resolution crystal structure and the E-pathway hypothesis of coupled transmembrane proton and electron transfer. Biochim. Biophys. Acta 2002, 1565, 215–231. [Google Scholar] [CrossRef] [Green Version]
- Borisov, V.B.; Gennis, R.B.; Hemp, J.; Verkhovsky, M.I. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta. 2011, 1807, 1398–1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schleicher, L.; Trautmann, A.; Stegmann, D.P.; Fritz, G.; Gätgens, J.; Bott, M.; Hein, S.; Simon, J.; Seifert, J.; Steuber, J. A sodium-translocating module linking succinate production to formation of a membrane potential in Prevotella bryantii. Appl. Environ. Microbiol. 2021, 87, AEM0121121. [Google Scholar] [CrossRef]
- Steuber, J.; Vohl, G.; Casutt, M.S.; Vorburger, T.; Diederichs, K.; Fritz, G. Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase. Nature 2014, 516, 62–67. [Google Scholar] [CrossRef] [Green Version]
- Steuber, J.; Vohl, G.; Muras, V.; Toulouse, C.; Claußen, B.; Vorburger, T.; Fritz, G. The structure of Na+-translocating NADH:ubiquinone oxidoreductase of Vibrio cholerae: Implications on coupling between electron transfer and Na+ transport. Biol. Chem. 2015, 396, 1015–1030. [Google Scholar] [CrossRef]
- Casutt, M.S.; Wendelspiess, S.; Steuber, J.; Fritz, G. Crystallization of the Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 1677–1679. [Google Scholar] [CrossRef]
- Nowicka, B.; Kruk, J. Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta 2010, 1797, 1587–1605. [Google Scholar] [CrossRef] [Green Version]
- Moparthi, V.K.; Hägerhäll, C. The evolution of respiratory chain complex I from a smaller last common ancestor consisting of 11 protein subunits. J. Mol. Evol. 2011, 72, 484–497. [Google Scholar] [CrossRef] [Green Version]
- Heikal, A.; Nakatani, Y.; Dunn, E.; Weimar, M.R.; Day, C.L.; Baker, E.N.; Lott, J.S.; Sazanov, L.A.; Cook, G.M. Structure of the bacterial type II NADH dehydrogenase: A monotopic membrane protein with an essential role in energy generation. Mol. Microbiol. 2014, 91, 950–964. [Google Scholar] [CrossRef]
- Steuber, J.; Krebs, W.; Dimroth, P. The Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio alginolyticus--redox states of the FAD prosthetic group and mechanism of Ag+ inhibition. Eur. J. Biochem. 1997, 249, 770–776. [Google Scholar] [CrossRef] [Green Version]
- Asano, M.; Hayashi, M.; Unemoto, T.; Tokuda, H. Ag+ -sensitive NADH dehydrogenas in the Na+ -motive respiratory chain of the marine bacterium Vibrio alginolyticus. Agric. Biol. Chem. 1985, 49, 2813–2817. [Google Scholar] [CrossRef]
- Hackmann, T.J.; Ngugi, D.K.; Firkins, J.L.; Tao, J. Genomes of rumen bacteria encode atypical pathways for fermenting hexoses to short-chain fatty acids. Environ. Microbiol. 2017, 19, 4670–4683. [Google Scholar] [CrossRef] [Green Version]
- Macklaim, J.M.; Fernandes, A.D.; Di Bella, J.M.; Hammond, J.; Reid, G.; Gloor, G.B. Comparative meta-RNA-seq of the vaginal microbiota and differential expression by Lactobacillus iners in health and dysbiosis. Microbiome 2013, 1, 12. [Google Scholar] [CrossRef] [Green Version]
- Dibrov, P.; Dibrov, E.; Maddaford, T.G.; Kenneth, M.; Nelson, J.; Resch, C.; Pierce, G.N. Development of a novel rationally designed antibiotic to inhibit a nontraditional bacterial target. Can. J. Physiol. Pharmacol. 2017, 95, 595–603. [Google Scholar] [CrossRef] [Green Version]
- Meier, T.; Polzer, P.; Diederichs, K.; Welte, W.; Dimroth, P. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science 2005, 308, 659–662. [Google Scholar] [CrossRef] [Green Version]
- Amabebe, E.; Anumba, D.O.C. The vaginal microenvironment: The physiologic role of Lactobacilli. Front. Med. 2018, 5, 181. [Google Scholar] [CrossRef] [Green Version]
- Schleicher, L.; Fritz, G.; Seifert, J.; Steuber, J. Anoxic cell rupture of Prevotella bryantii by high-pressure homogenization protects the Na+-translocating NADH:quinone oxidoreductase from oxidative damage. Arch. Microbiol. 2020, 202, 1263–1266. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.; Ehlers, M.M.; Lombaard, H.; Redelinghuys, M.J.; Kock, M.M. Etiology of bacterial vaginosis and polymicrobial biofilm formation. Crit. Rev. Microbiol. 2017, 43, 651–667. [Google Scholar] [CrossRef] [Green Version]
- Schwebke, J.R.; Muzny, C.A.; Josey, W.E. Role of Gardnerella vaginalis in the pathogenesis of bacterial vaginosis: A conceptual model. J. Infect. Dis. 2014, 210, 338–343. [Google Scholar] [CrossRef]
- Harwich, M.D.; Alves, J.M.; Buck, G.A.; Strauss, J.F.; Patterson, J.L.; Oki, A.T.; Girerd, P.H.; Jefferson, K.K. Drawing the line between commensal and pathogenic Gardnerella vaginalis through genome analysis and virulence studies. BMC Genom. 2010, 11, 375. [Google Scholar] [CrossRef] [Green Version]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Ito, M.; Cooperberg, B.; Krulwich, T.A. Diverse genes of alkaliphilic Bacillus firmus OF4 that complement K+-uptake-deficient Escherichia coli include an ftsH homologue. Extremophiles 1997, 1, 22–28. [Google Scholar] [CrossRef]
- Mohamed, S.A.; Elshal, M.F.; Kumosani, T.A.; Aldahlawi, A.M. Purification and characterization of asparaginase from Phaseolus vulgaris seeds. Evid.-Based Complement. Alternat. Med. 2015, 2015, 309214. [Google Scholar] [CrossRef] [Green Version]
- Leutbecher, H.; Greiner, G.; Amann, R.; Stolz, A.; Beifuss, U.; Conrad, J. Laccase-catalyzed phenol oxidation. Rapid assignment of ring-proton deficient polycyclic benzofuran regioisomers by experimental 1H-13C long-range coupling constants and DFT-predicted product formation. Org. Biomol. Chem. 2011, 9, 2667–2673. [Google Scholar] [CrossRef] [PubMed]
- Schägger, H.; von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- Vohl, G.; Nedielkov, R.; Claussen, B.; Casutt, M.S.; Vorburger, T.; Diederichs, K.; Möller, H.M.; Steuber, J.; Fritz, G. Crystallization and preliminary analysis of the NqrA and NqrC subunits of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio cholerae. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2014, 70, 987–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juárez, O.; Athearn, K.; Gillespie, P.; Barquera, B. Acid residues in the transmembrane helices of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae involved in sodium translocation. Biochemistry 2009, 48, 9516–9524. [Google Scholar] [CrossRef] [Green Version]
- Kröger, A.; Innerhofer, A. The function of menaquinone, covalently bound FAD and iron-sulfur protein in the electron transport from formate to fumarate of Vibrio succinogenes. Eur. J. Biochem. 1976, 69, 487–495. [Google Scholar] [CrossRef]
- Al-Attar, S.; Yu, Y.; Pinkse, M.; Hoeser, J.; Friedrich, T.; Bald, D.; de Vries, S. Cytochrome bd displays significant quinol peroxidase activity. Sci. Rep. 2016, 6, 27631. [Google Scholar] [CrossRef] [Green Version]
- Trumpower, B.L.; Edwards, C.A. Purification of a reconstitutively active iron-sulfur protein (oxidation factor) from succinate cytochrome c reductase complex of bovine heart mitochondria. J. Biol. Chem. 1979, 254, 8697–8706. [Google Scholar] [CrossRef]
- Geisler, V.; Ullmann, R.; Kröger, A. The direction of the proton exchange associated with the redox reactions of menaquinone during electron transport in Wolinella succinogenes. Biochim. Biophys. Acta 1994, 1184, 219–226. [Google Scholar] [CrossRef]
- Blasco, L.; Kahala, M.; Tupasela, T.; Joutsjoki, V. Determination of aspartase activity in dairy Propionibacterium strains. FEMS Microbiol. Lett. 2011, 321, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Bzura, J.; Koncki, R. A mechanized urease activity assay. Enzyme Microb. Technol. 2019, 123, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vorburger, T.; Nedielkov, R.; Brosig, A.; Bok, E.; Schunke, E.; Steffen, W.; Mayer, S.; Götz, F.; Möller, H.M.; Steuber, J. Role of the Na+-translocating NADH:quinone oxidoreductase in voltage generation and Na+ extrusion in Vibrio cholerae. Biochim. Biophys. Acta 2016, 1857, 473–482. [Google Scholar] [CrossRef] [PubMed]









| Growth Condition | OD600 (t = 7 d) | Doubling Time (h) | NH4+ (mM; t = 0) | NH4+ (mM; t = 7) | Net (NH3 + NH4+) Formed (mM) | Rate of Net (NH3 + NH4+) Formation (nmol min−1 mg−1) |
|---|---|---|---|---|---|---|
| pH 5 + Asn | 0.5 ± 0.1 | 91 | 14.7 ± 0.2 | 62.9 ± 0.8 | 48.2 ± 4.4 | 265.7 ± 24.1 |
| pH 5 − Asn | 0.5 ± 0.1 | 101 | 14.8 ± 0.1 | 27.9 ± 0.2 | 13.1 ± 0.1 | 59.7 ± 0.4 |
| pH 6 + Asn | 0.4 ± 0.1 | 17 | 16.9 ± 2.1 | 92.4 ± 1.1 | 75.5 ± 5.6 | 416.4 ± 31.4 |
| pH 6 − Asn | 1.1 ± 0.4 | 13 | 12.1 ± 0.5 | 25.5 ± 0.2 | 13.1 ± 0.6 | 21.7 ± 2.1 |
| pH 7 + Asn | 1.6 ± 0.4 | 10 | 18.1 ± 0.8 | 101.5 ± 0.5 | 83.4 ± 4.1 | 119.1 ± 2.5 |
| pH 7 − Asn | 1.8 ± 0.2 | 8 | 16.5 ± 0.7 | 37.6 ± 0.03 | 21.1 ± 0.25 | 25.2 ± 0.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schleicher, L.; Herdan, S.; Fritz, G.; Trautmann, A.; Seifert, J.; Steuber, J. Central Carbon Metabolism, Sodium-Motive Electron Transfer, and Ammonium Formation by the Vaginal Pathogen Prevotella bivia. Int. J. Mol. Sci. 2021, 22, 11925. https://doi.org/10.3390/ijms222111925
Schleicher L, Herdan S, Fritz G, Trautmann A, Seifert J, Steuber J. Central Carbon Metabolism, Sodium-Motive Electron Transfer, and Ammonium Formation by the Vaginal Pathogen Prevotella bivia. International Journal of Molecular Sciences. 2021; 22(21):11925. https://doi.org/10.3390/ijms222111925
Chicago/Turabian StyleSchleicher, Lena, Sebastian Herdan, Günter Fritz, Andrej Trautmann, Jana Seifert, and Julia Steuber. 2021. "Central Carbon Metabolism, Sodium-Motive Electron Transfer, and Ammonium Formation by the Vaginal Pathogen Prevotella bivia" International Journal of Molecular Sciences 22, no. 21: 11925. https://doi.org/10.3390/ijms222111925
APA StyleSchleicher, L., Herdan, S., Fritz, G., Trautmann, A., Seifert, J., & Steuber, J. (2021). Central Carbon Metabolism, Sodium-Motive Electron Transfer, and Ammonium Formation by the Vaginal Pathogen Prevotella bivia. International Journal of Molecular Sciences, 22(21), 11925. https://doi.org/10.3390/ijms222111925