Profiling and Functional Analysis of microRNA Deregulation in Cancer-Associated Fibroblasts in Oral Squamous Cell Carcinoma Depicts an Anti-Invasive Role of microRNA-204 via Regulation of Their Motility
Abstract
:1. Introduction
2. Results
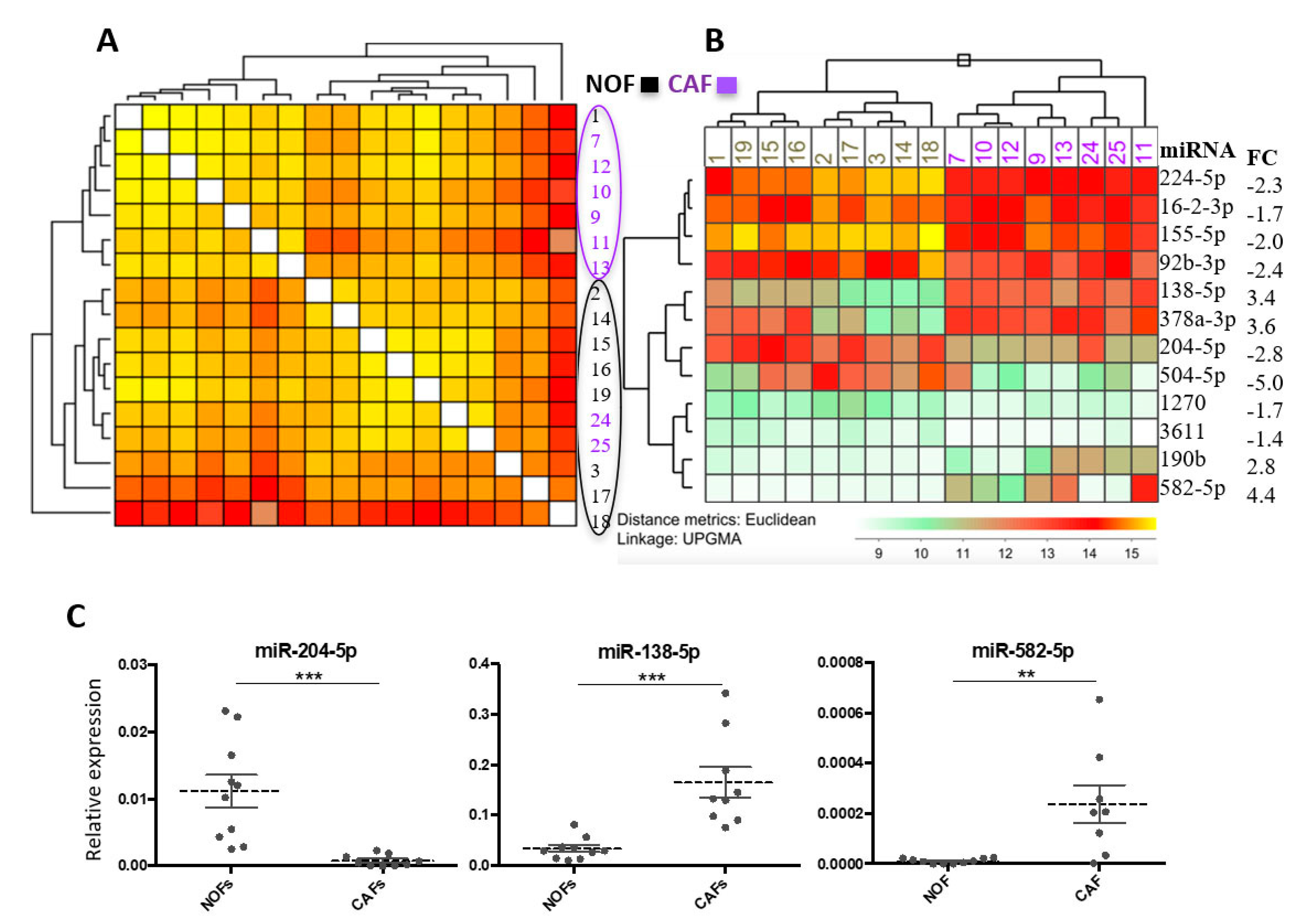
2.1. miR Array Identifies Twelve miRs with Differential Expression in CAFs versus NOFs
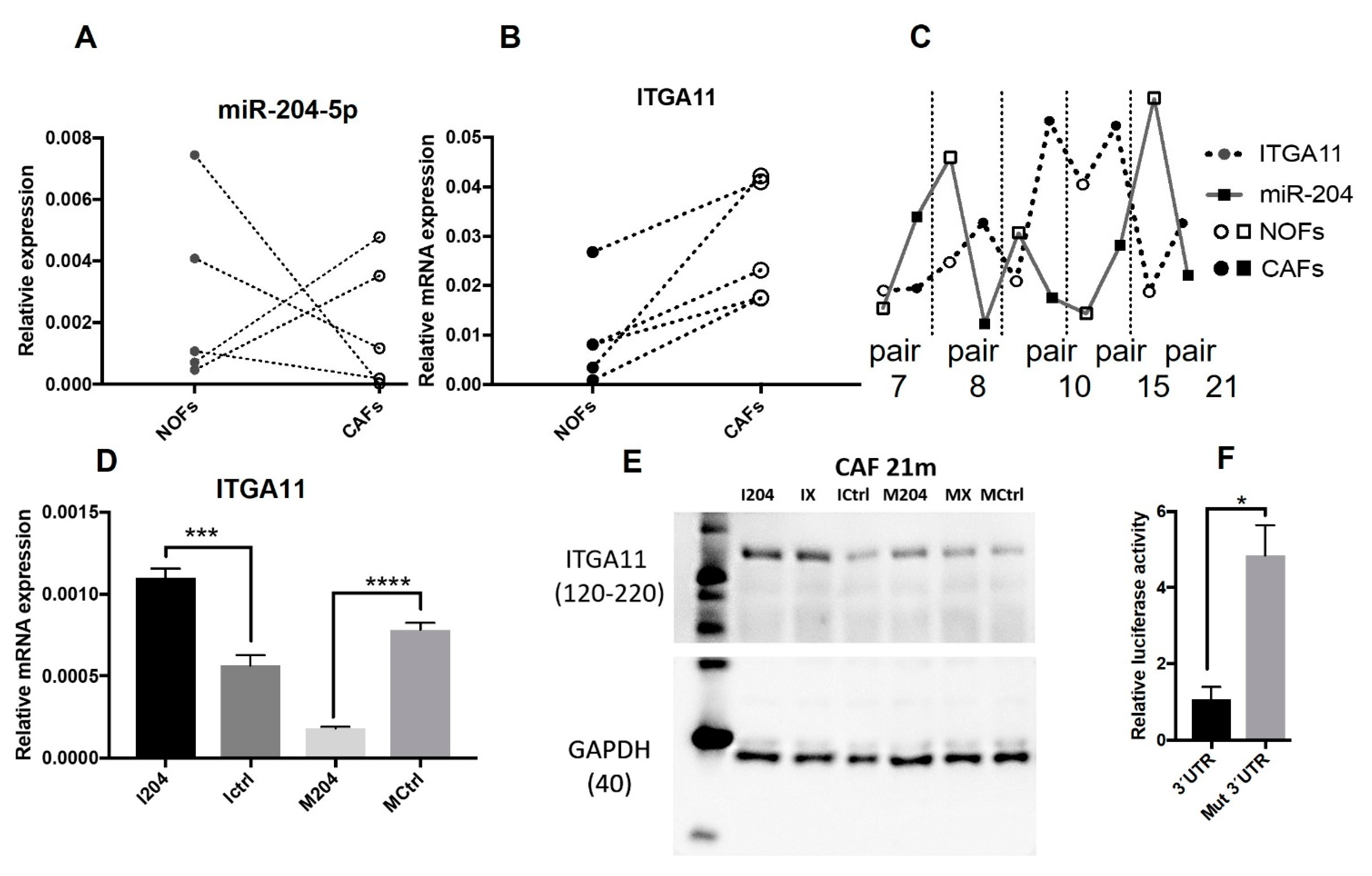
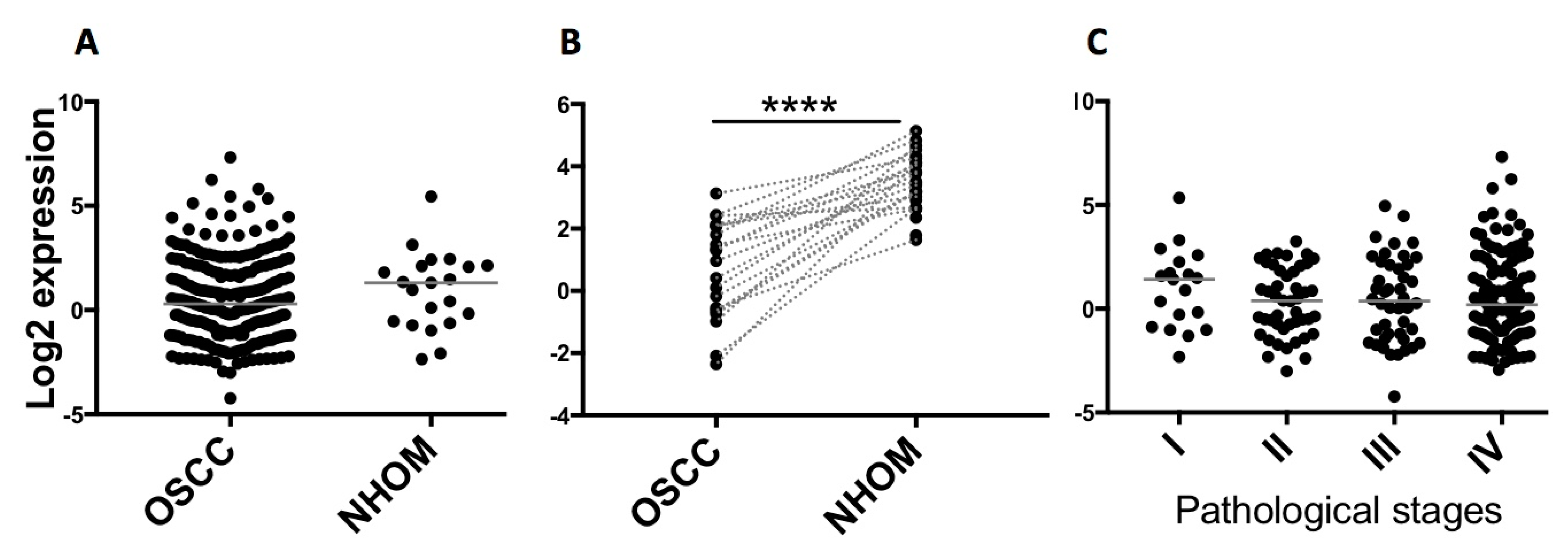
2.2. Validation of Altered Expression of miR-204 in Cultured Fibroblasts and Patient Material
2.3. ITGA11 Is a Direct Target of miR-204 in Fibroblasts
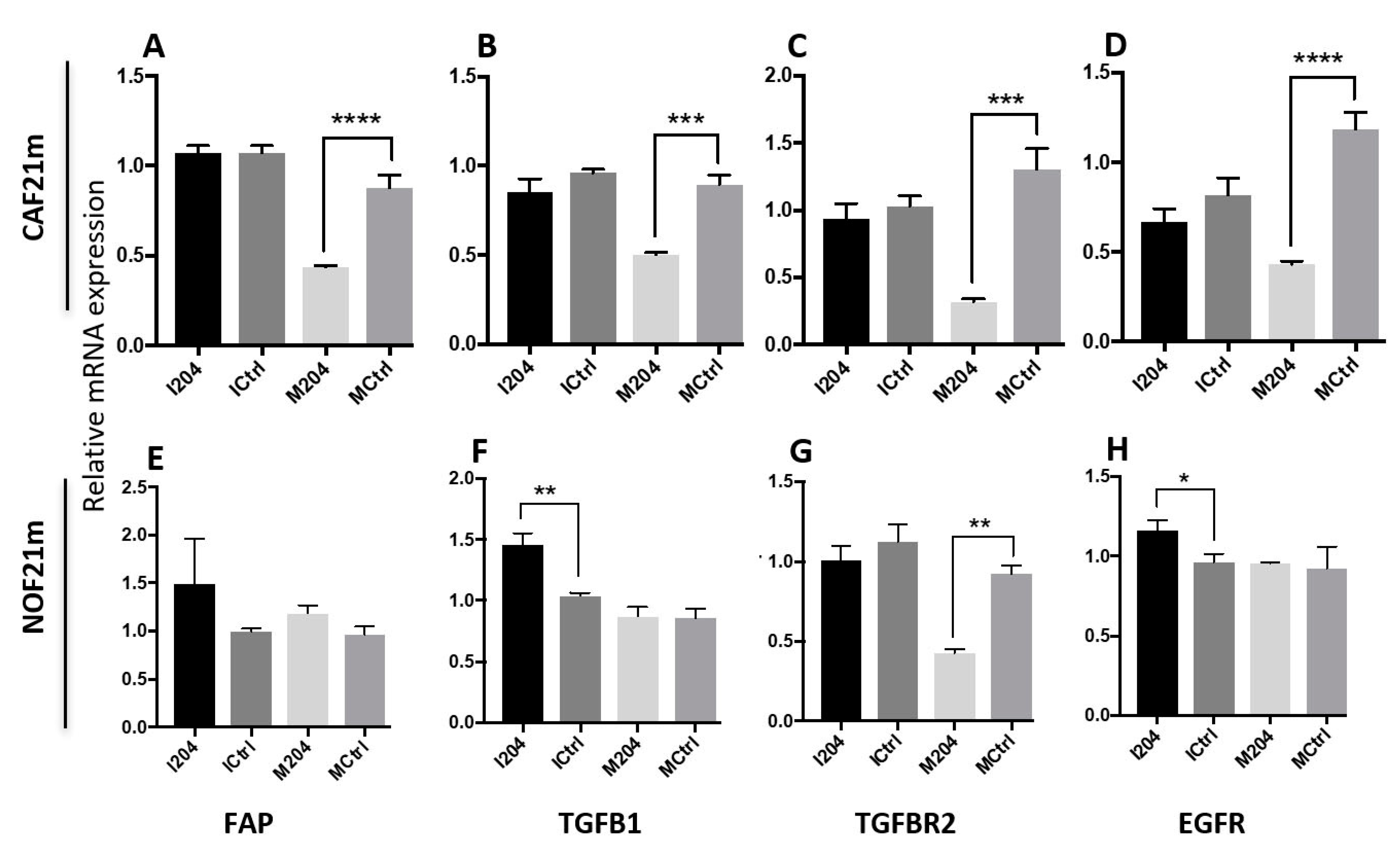
2.4. miR-204 Modulates the Expression of Several CAF-Related Molecules
2.5. miR-204 Decreases Migration and Collagen Contraction Abilities of CAFs
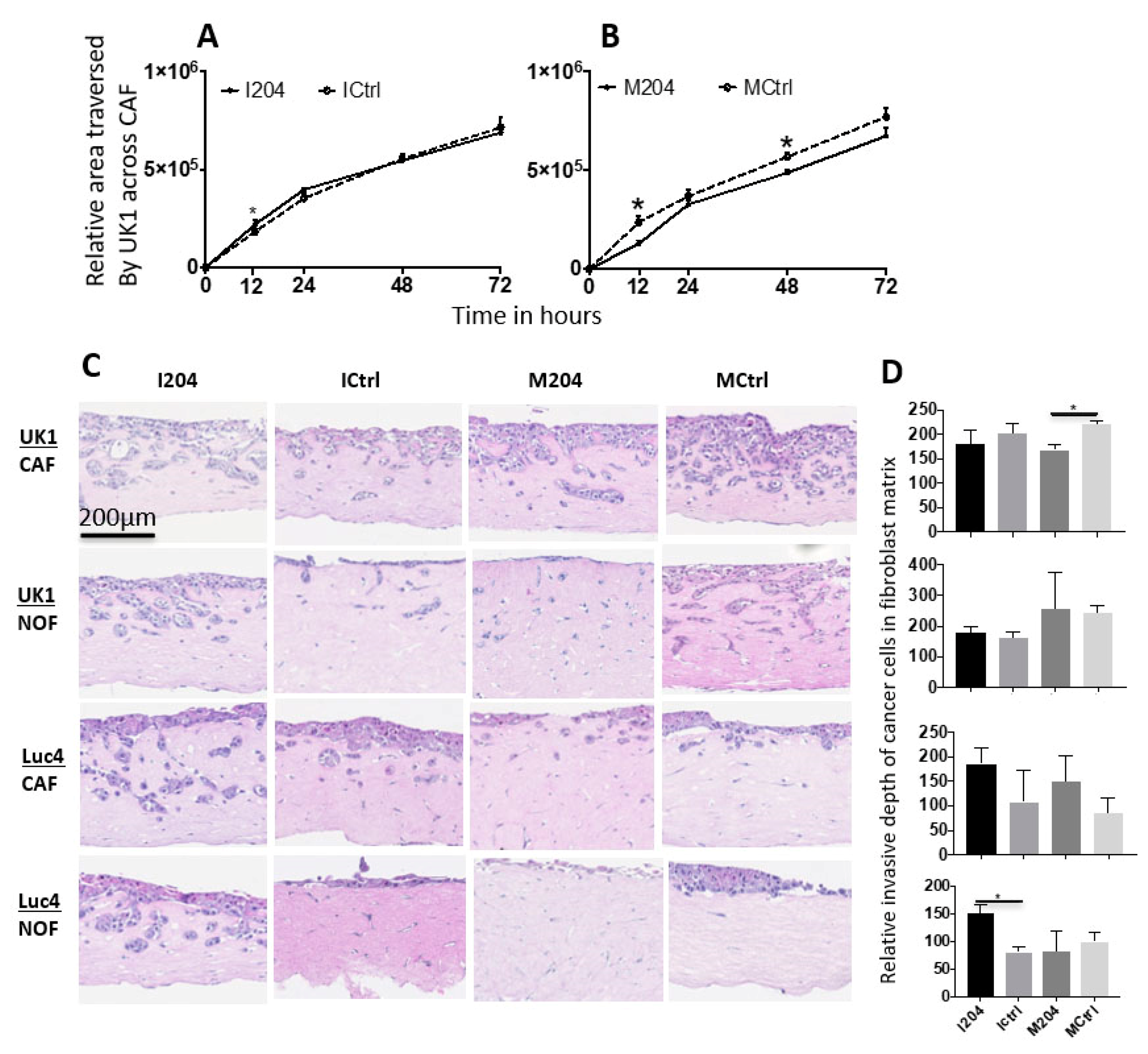
2.6. Expression of miR-204 in Fibroblasts Inhibits Invasion and Migration of Adjacent OSCC Cells
2.7. miR-204 in Fibroblasts Regulates Several Molecules Involved in Cell Migration
3. Discussion
4. Materials and Methods
4.1. Patient Material
4.2. Fibroblast Isolation and Cell Culture
4.3. Total RNA Isolation and Small RNA Enrichment for miR Microarray
4.4. miR Microarray
4.5. Reverse Transcription
4.6. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.7. TCGA Data Analysis
4.8. miR Target Identification and miR Dual Luciferase Target Reporter Assay
4.9. miR-204 Modulation in Cultured Fibroblasts
4.10. Protein Isolation and Quantification
4.11. Western Blotting
4.12. Migration Assay (2D Co-Culture)
4.13. Collagen Contraction Assay
4.14. 3D Organotypic Co-Cultures
4.15. Quantification of Invasion of OSCC Cells in 3D-Organotypic Models
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, M.; Nair, R.; Jamieson, L.; Liu, Z.; Bi, P. Incidence Trends of Lip, Oral Cavity, and Pharyngeal Cancers: Global Burden of Disease 1990–2017. J. Dent. Res. 2020, 99, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Dobrossy, L. Epidemiology of head and neck cancer: Magnitude of the problem. Cancer Metastasis Rev. 2005, 24, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Karnov, K.K.S.; Grønhøj, C.; Jensen, D.H.; Wessel, I.; Charabi, B.W.; Specht, L.; Kjaer, A.; von Buchwald, C. Increasing incidence and survival in oral cancer: A nationwide Danish study from 1980 to 2014. Acta Oncol. 2017, 56, 1204–1209. [Google Scholar] [CrossRef] [Green Version]
- Bennardo, L.; Bennardo, F.; Giudice, A.; Passante, M.; Dastoli, S.; Morrone, P.; Provenzano, E.; Patruno, C.; Nisticò, S. Local Chemotherapy as an Adjuvant Treatment in Unresectable Squamous Cell Carcinoma: What Do We Know So Far? Curr. Oncol. 2021, 28, 2317–2325. [Google Scholar] [CrossRef]
- Pentangelo, G.; Nisticò, S.; Provenzano, E.; Cisale, G.; Bennardo, L. Topical 5% imiquimod sequential to surgery for HPV-related squamous cell carcinoma of the lip. Medicina 2021, 57, 563. [Google Scholar] [CrossRef] [PubMed]
- Malik, U.U.; Zarina, S.; Pennington, S.R. Oral squamous cell carcinoma: Key clinical questions, biomarker discovery, and the role of proteomics. Arch. Oral Biol. 2016, 63, 53–65. [Google Scholar] [CrossRef]
- Ragin, C.C.R.; Taioli, E. Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: Review and meta-analysis. Int. J. Cancer 2007, 121, 1813–1820. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Lee, R.C.; Ambros, V. An Extensive Class of Small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.H.; Pan, X.P.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Min, A.; Zhu, C.; Peng, S.; Rajthala, S.; Costea, D.E.; Sapkota, D. MicroRNAs as Important Players and Biomarkers in Oral Carcinogenesis. Biomed. Res. Int. 2015, 2015, 186904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, N.; O’Brien, C.J.; Clark, J.; Rose, B. Potential Role of Micro-Rnas in Head and Neck Tumorigenesis. Head Neck J. Sci. Spec. Head Neck 2010, 32, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Sethi, N.; Wright, A.; Wood, H.; Rabbitts, P. MicroRNAs and head and neck cancer: Reviewing the first decade of research. Eur. J. Cancer 2014, 50, 2619–2635. [Google Scholar] [CrossRef]
- Fidler, I.J. Timeline-The pathogenesis of cancer metastasis: The ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer 2003, 3, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costea, D.E.; Hills, A.; Osman, A.H.; Thurlow, J.; Kalna, G.; Huang, X.; Pena Murillo, C.; Parajuli, H.; Suliman, S.; Kulasekara, K.K.; et al. Identification of two distinct carcinoma-associated fibroblast subtypes with differential tumor-promoting abilities in oral squamous cell carcinoma. Cancer Res. 2013, 73, 3888–3901. [Google Scholar] [CrossRef] [Green Version]
- Gaggioli, C.; Hooper, S.; Hidalgo-Carcedo, C.; Grosse, R.; Marshall, J.F.; Harrington, K.; Sahai, E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007, 9, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Daly, A.J.; McIlreavey, L.; Irwin, C.R. Regulation of HGF and SDF-1 expression by oral fibroblasts-Implications for invasion of oral cancer. Oral Oncol. 2008, 44, 646–651. [Google Scholar] [CrossRef]
- Kawashiri, S.; Tanaka, A.; Noguchi, N.; Hase, T.; Nakaya, H.; Ohara, T.; Kato, K.; Yamamoto, E. Significance of stromal desmoplasia and myofibroblast appearance at the invasive front in squamous cell carcinoma of the oral cavity. Head Neck J. Sci. Spec. Head Neck 2009, 31, 1346–1353. [Google Scholar] [CrossRef] [PubMed]
- Kellermann, M.G.; Sobral, L.M.; da Silva, S.D.; Zecchin, K.G.; Graner, E.; Lopes, M.A.; Nishimoto, I.; Kowalski, L.P.; Coletta, R.D. Myofibroblasts in the stroma of oral squamous cell carcinoma are associated with poor prognosis. Histopathology 2007, 51, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, H.; Teh, M.T.; Abrahamsen, S.; Christoffersen, I.; Neppelberg, E.; Lybak, S.; Osman, T.; Johannessen, A.C.; Gullberg, D.; Skarstein, K.; et al. Integrin alpha11 is overexpressed by tumour stroma of head and neck squamous cell carcinoma and correlates positively with alpha smooth muscle actin expression. J. Oral Pathol. Med. 2017, 46, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Fujii, N.; Shomori, K.; Shiomi, T.; Nakabayashi, M.; Takeda, C.; Ryoke, K.; Ito, H. Cancer-associated fibroblasts and CD163-positive macrophages in oral squamous cell carcinoma: Their clinicopathological and prognostic significance. J. Oral Pathol. Med. 2012, 41, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Marsh, D.; Suchak, K.; Moutasim, K.A.; Vallath, S.; Hopper, C.; Jerjes, W.; Upile, T.; Kalavrezos, N.; Violette, S.M.; Weinreb, P.H.; et al. Stromal features are predictive of disease mortality in oral cancer patients. J. Pathol. 2011, 223, 470–481. [Google Scholar] [CrossRef]
- Xia, Y.; Zhu, Y.; Ma, T.; Pan, C.F.; Wang, J.; He, Z.C.; Li, Z.; Qi, X.T.; Chen, Y.J. miR-204 functions as a tumor suppressor by regulating SIX1 in NSCLC. Febs Lett. 2014, 588, 3703–3712. [Google Scholar] [CrossRef] [PubMed]
- Sacconi, A.; Biagioni, F.; Canu, V.; Mori, F.; Di Benedetto, A.; Lorenzon, L.; Ercolani, C.; Di Agostino, S.; Cambria, A.M.; Germoni, S.; et al. miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis. 2012, 3, e423. [Google Scholar] [CrossRef]
- Hong, B.S.; Ryu, H.S.; Kim, N.; Kim, J.; Lee, E.; Moon, H.; Kim, K.H.; Jin, M.S.; Kwon, N.H.; Kim, S.; et al. Tumor suppressor miRNA-204-5p regulates growth, metastasis, and immune microenvironment remodeling in breast cancer. Cancer Res. 2019, 79, 1520–1534. [Google Scholar]
- Bao, W.; Wang, H.H.; Tian, F.J.; He, X.Y.; Qiu, M.T.; Wang, J.Y.; Zhang, H.J.; Wang, L.H.; Wan, X.P. A TrkB-STAT3-miR-204-5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol. Cancer 2013, 12, 155. [Google Scholar] [CrossRef] [Green Version]
- Imam, J.S.; Plyler, J.R.; Bansal, H.; Prajapati, S.; Bansal, S.; Rebeles, J.; Chen, H.I.H.; Chang, Y.F.; Panneerdoss, S.; Zoghi, B.; et al. Genomic loss of tumor suppressor miRNA-204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PLoS ONE 2012, 7, e52397. [Google Scholar]
- Schneider, A.; Victoria, B.; Lopez, Y.N.; Suchorska, W.; Barczak, W.; Sobecka, A.; Golusinski, W.; Masternak, M.M.; Golusinski, P. Tissue and serum microRNA profile of oral squamous cell carcinoma patients. Sci. Rep. 2018, 8, 675. [Google Scholar] [CrossRef] [Green Version]
- Tsai, S.C.; Huang, S.F.; Chiang, J.H.; Chen, Y.F.; Huang, C.C.; Tsai, M.H.; Tsai, F.J.; Kao, M.C.; Yang, J.S. The differential regulation of microRNAs is associated with oral cancer. Oncol. Rep. 2017, 38, 1613–1620. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, E.; Singh, R.; Ray, A.; Roy, R.; De Sarkar, N.; Paul, R.R.; Pal, M.; Aich, R.; Roy, B. Expression deregulation of mir31 and CXCL12 in two types of oral precancers and cancer: Importance in progression of precancer and cancer. Sci. Rep. 2016, 6, 32735. [Google Scholar] [CrossRef]
- Yu, C.C.; Chen, P.N.; Peng, C.Y.; Yu, C.H.; Chou, M.Y. Suppression of miR-204 enables oral squamous cell carcinomas to promote cancer stemness, EMT traits, and lymph node metastasis. Oncotarget 2016, 7, 20180–20192. [Google Scholar] [CrossRef] [Green Version]
- Rajthala, S.; Dongre, H.; Parajuli, H.; Min, A.; Nginamau, E.S.; Kvalheim, A.; Lybak, S.; Sapkota, D.; Johannessen, A.C.; Costea, D.E. Combined In situ hybridization and immunohistochemistry on archival tissues reveals stromal microRNA-204 as prognostic biomarker for oral squamous cell carcinoma. Cancers 2021, 13, 1307. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabor, M.P.; Brakenhoff, R.H.; van Houten, V.M.M.; Kummer, J.A.; Snel, M.H.J.; Snijders, P.J.F.; Snow, G.B.; Leemans, C.R.; Braakhuis, B.J.M. Persistence of genetically altered fields in head and neck cancer patients Biological and clinical implications. Clin. Cancer Res. 2001, 7, 1523–1532. [Google Scholar]
- Ganci, F.; Sacconi, A.; Manciocco, V.; Covello, R.; Benevolo, M.; Rollo, F.; Strano, S.; Valsoni, S.; Bicciato, S.; Spriano, G.; et al. Altered peritumoral microRNA expression predicts head and neck cancer patients with a high risk of recurrence. Mod. Pathol. 2017, 30, 1387–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cukierman, E.; Pankov, R.; Stevens, D.R.; Yamada, K.M. Taking cell-matrix adhesions to the third dimension. Science 2001, 294, 1708–1712. [Google Scholar] [CrossRef] [PubMed]
- Doldi, V.; Callari, M.; Giannoni, E.; D’Aiuto, F.; Maffezzini, M.; Valdagni, R.; Chiarugi, P.; Gandellini, P.; Zaffaroni, N. Integrated gene and miRNA expression analysis of prostate cancer associated fibroblasts supports a prominent role for interleukin-6 in fibroblast activation. Oncotarget 2015, 6, 31441–31460. [Google Scholar] [CrossRef] [Green Version]
- Negrete-Garcia, M.C.; Ramirez-Rodriguez, S.L.; Rangel-Escareno, C.; Munoz-Montero, S.; Kelly-Garcia, J.; Vazquez-Manriquez, M.E.; Santillan, P.; Ramirez, M.M.; Ramirez-Martinez, G.; Ramirez-Venegas, A.; et al. Deregulated MicroRNAs in Cancer-Associated Fibroblasts from Front Tumor Tissues of Lung Adenocarcinoma as Potential Predictors of Tumor Promotion. Tohoku J. Exp. Med. 2018, 246, 107–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Z.; Qin, X.; Yan, M.; Li, R.; Chen, G.; Zhang, J.; Chen, W. Cancer-associated fibroblasts promote cancer cell growth through a miR-7-RASSF2-PAR-4 axis in the tumor microenvironment. Oncotarget 2017, 8, 1290–1303. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.S.; Yang, X.H.; Chen, X.; Wang, X.D.; Hua, J.; Zhou, D.L.; Zhou, B.; Song, Z.S. MicroRNA-106b in cancer-associated fibroblasts from gastric cancer promotes cell migration and invasion by targeting PTEN. FEBS Lett. 2014, 588, 2162–2169. [Google Scholar] [CrossRef] [Green Version]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015, 43, D153–D159. [Google Scholar] [CrossRef] [PubMed]
- Zeltz, C.; Alam, J.; Liu, H.; Erusappan, P.M.; Hoschuetzky, H.; Molven, A.; Parajuli, H.; Cukierman, E.; Costea, D.E.; Lu, N.; et al. alpha11beta1 Integrin is Induced in a Subset of Cancer-Associated Fibroblasts in Desmoplastic Tumor Stroma and Mediates In Vitro Cell Migration. Cancers 2019, 11, 765. [Google Scholar] [CrossRef] [Green Version]
- Velling, T.; Kusche-Gullberg, M.; Sejersen, T.; Gullberg, D. cDNA cloning and chromosomal localization of human alpha(11) integrin: A collagen-binding, I domain-containing, beta(1)-associated integrin alpha-chain present in muscle tissues. J. Biol. Chem. 1999, 274, 25735–25742. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K. FAK-Dependent Cell Motility and Cell Elongation. Cells 2020, 9, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dourado, M.R.; de Oliveira, C.E.; Sawazaki-Calone, I.; Sundquist, E.; Coletta, R.D.; Salo, T. Clinicopathologic significance of ROCK2 expression in oral squamous cell carcinomas. J. Oral Pathol. Med. 2018, 47, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Ying, Z.; Li, Y.; Wu, J.; Zhu, X.; Yang, Y.; Tian, H.; Li, W.; Hu, B.; Cheng, S.Y.; Li, M. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 2013, 73, 990–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locke, M.; Heywood, M.; Fawell, S.; Mackenzie, I.C. Retention of intrinsic stem cell hierarchies in carcinoma-derived cell lines. Cancer Res. 2005, 65, 8944–8950. [Google Scholar] [CrossRef] [Green Version]
- Biddle, A.; Liang, X.; Gammon, L.; Fazil, B.; Harper, L.J.; Emich, H.; Costea, D.E.; Mackenzie, I.C. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res. 2011, 71, 5317–5326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stavrum, A.K.; Petersen, K.; Jonassen, I.; Dysvik, B. Analysis of gene-expression data using J-Express. Curr. Protoc. Bioinform. 2008, 21, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Vejnar, C.E.; Zdobnov, E.M. MiRmap: Comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012, 40, 11673–11683. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [Green Version]
- Dysvik, B.; Jonassen, I. J-Express: Exploring gene expression data using Java. Bioinformatics 2001, 17, 369–370. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajthala, S.; Min, A.; Parajuli, H.; Debnath, K.C.; Ljøkjel, B.; Hoven, K.M.; Kvalheim, A.; Lybak, S.; Neppelberg, E.; Vintermyr, O.K.; et al. Profiling and Functional Analysis of microRNA Deregulation in Cancer-Associated Fibroblasts in Oral Squamous Cell Carcinoma Depicts an Anti-Invasive Role of microRNA-204 via Regulation of Their Motility. Int. J. Mol. Sci. 2021, 22, 11960. https://doi.org/10.3390/ijms222111960
Rajthala S, Min A, Parajuli H, Debnath KC, Ljøkjel B, Hoven KM, Kvalheim A, Lybak S, Neppelberg E, Vintermyr OK, et al. Profiling and Functional Analysis of microRNA Deregulation in Cancer-Associated Fibroblasts in Oral Squamous Cell Carcinoma Depicts an Anti-Invasive Role of microRNA-204 via Regulation of Their Motility. International Journal of Molecular Sciences. 2021; 22(21):11960. https://doi.org/10.3390/ijms222111960
Chicago/Turabian StyleRajthala, Saroj, Anjie Min, Himalaya Parajuli, Kala Chand Debnath, Borghild Ljøkjel, Kristin Marie Hoven, Arild Kvalheim, Stein Lybak, Evelyn Neppelberg, Olav Karsten Vintermyr, and et al. 2021. "Profiling and Functional Analysis of microRNA Deregulation in Cancer-Associated Fibroblasts in Oral Squamous Cell Carcinoma Depicts an Anti-Invasive Role of microRNA-204 via Regulation of Their Motility" International Journal of Molecular Sciences 22, no. 21: 11960. https://doi.org/10.3390/ijms222111960
APA StyleRajthala, S., Min, A., Parajuli, H., Debnath, K. C., Ljøkjel, B., Hoven, K. M., Kvalheim, A., Lybak, S., Neppelberg, E., Vintermyr, O. K., Johannessen, A. C., Sapkota, D., & Costea, D. E. (2021). Profiling and Functional Analysis of microRNA Deregulation in Cancer-Associated Fibroblasts in Oral Squamous Cell Carcinoma Depicts an Anti-Invasive Role of microRNA-204 via Regulation of Their Motility. International Journal of Molecular Sciences, 22(21), 11960. https://doi.org/10.3390/ijms222111960





