Insights into Intra-Tumoral Heterogeneity: Transcriptional Profiling of Chemoresistant MPM Cell Subpopulations Reveals Involvement of NFkB and DNA Repair Pathways and Contributes a Prognostic Signature
Abstract
:1. Introduction
2. Results
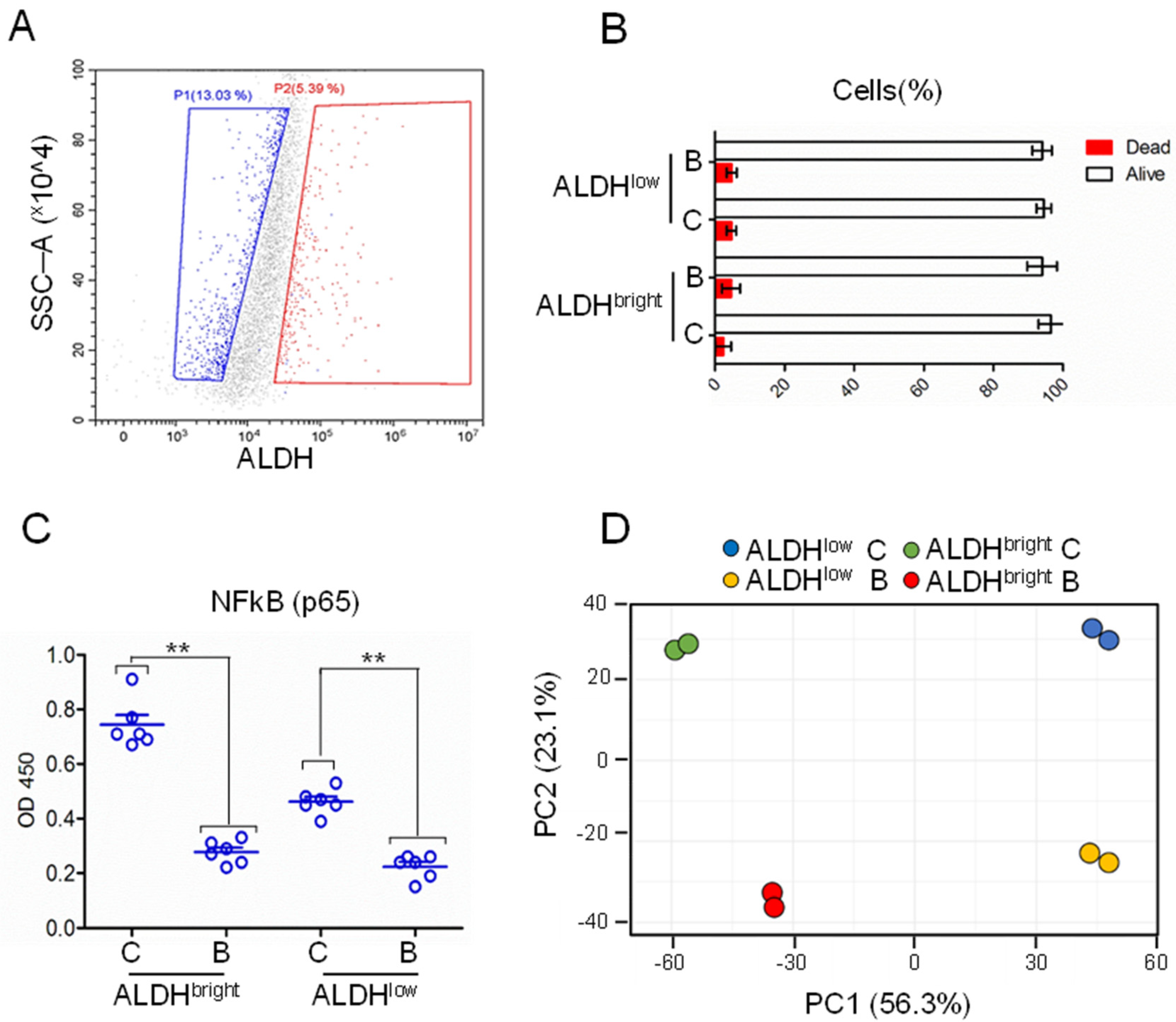
2.1. ALDH1A3 Expression May Stratify MPM Patients
2.2. Gene Expression Profile of Purified ALDHbright and ALDHlow Cells, at Steady State and after Butein Treatment
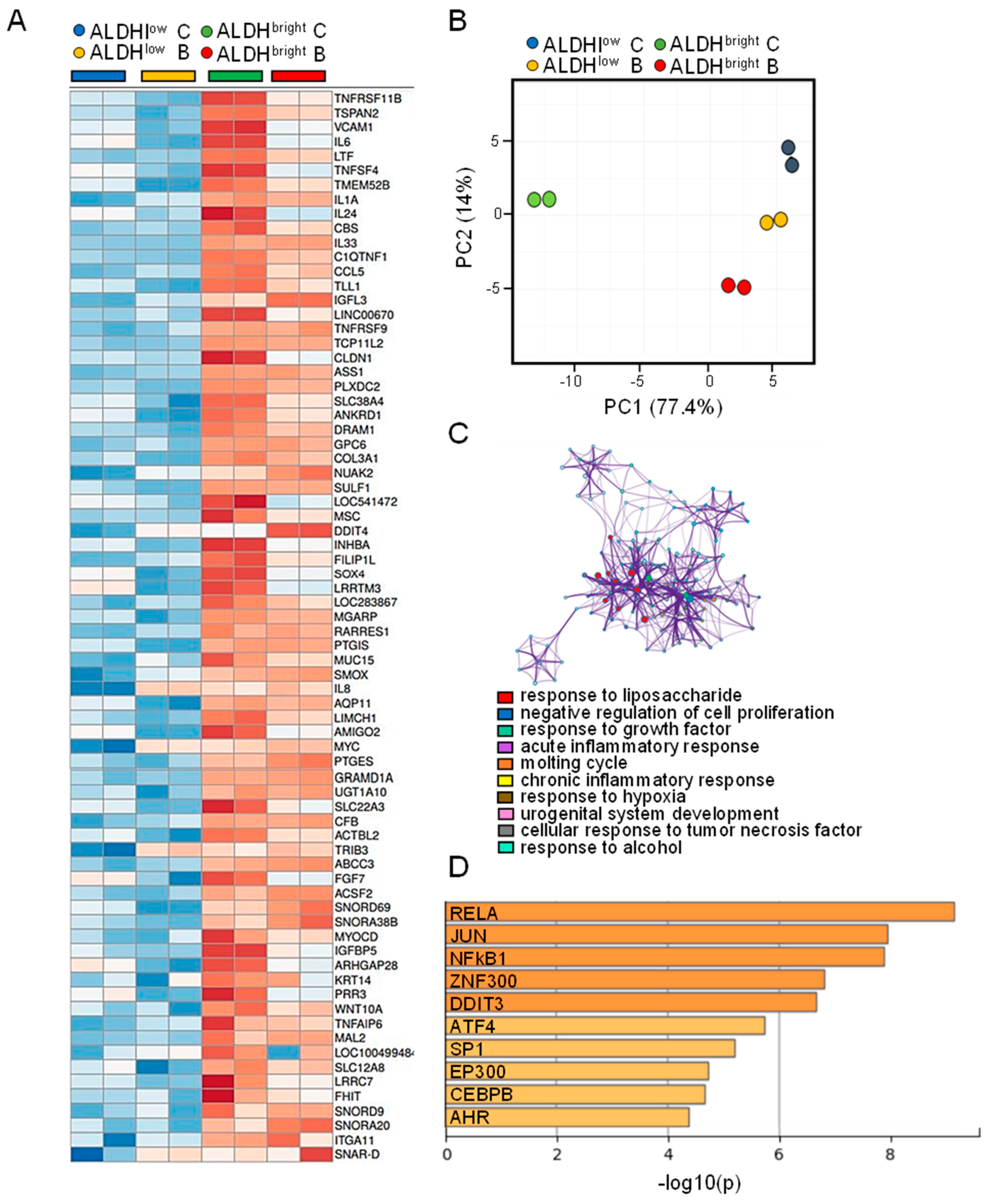
2.3. Analyzing the Gene Expression Profile of ALDHbright and ALDHlow Cell Subpopulations
2.4. The MPM ALDHbright Cells Exhibited Enrichment for NFkB-Related Pathways
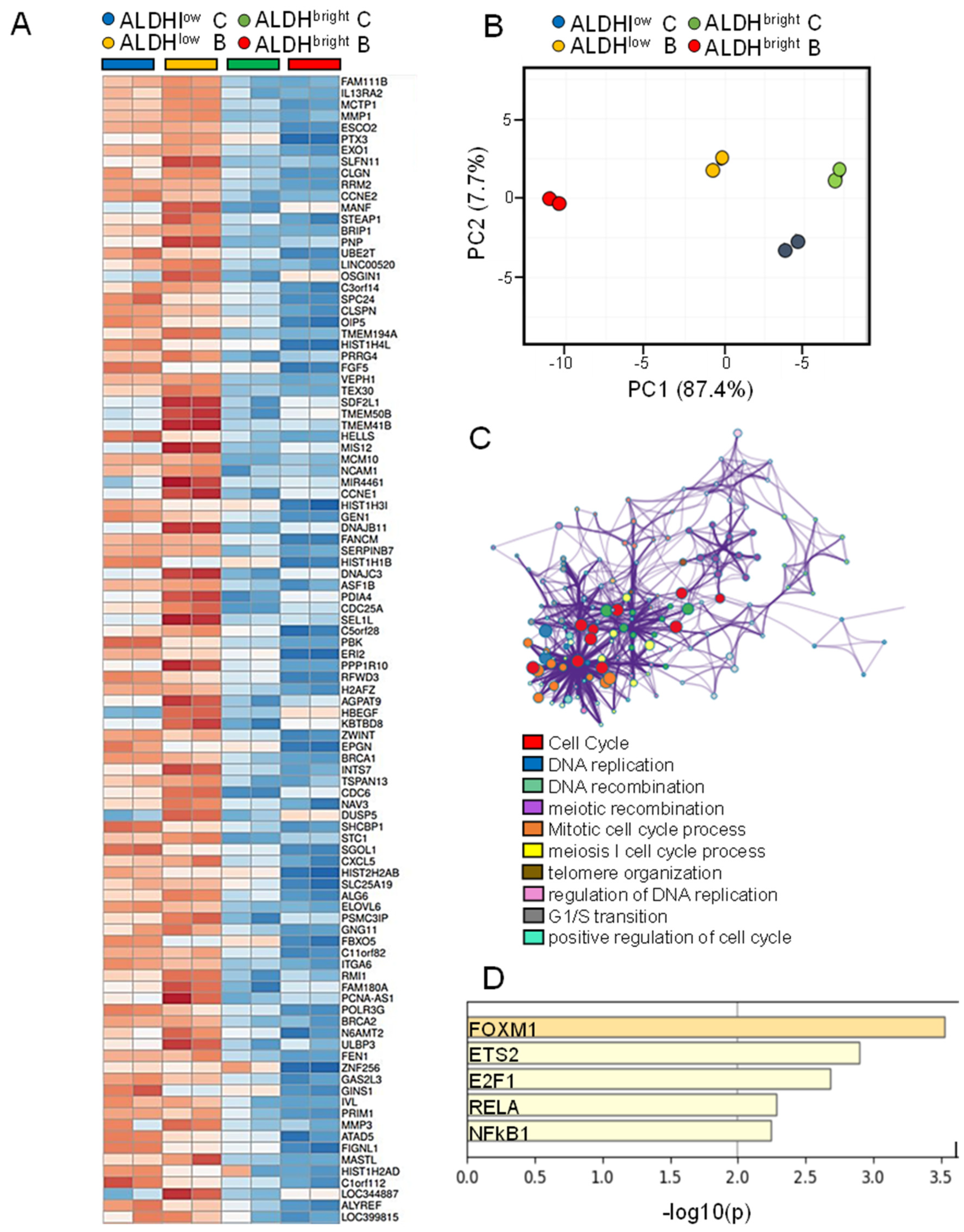
2.5. The mRNAs Downregulated More Deeply in the ALDHbright Cells by Butein Were Related to DNA Replication and Repair Functions
2.6. mRNAs Enriched in the ALDHbright Cells May Have Prognostic Potential
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Treatments
4.2. ALDH Detection
4.3. FACS-Based Purification of ALDHbright and ALDHlow Cells
4.4. RNA Extraction and Analysis
4.5. Q-RTPCR
4.6. Microarray Analysis
4.7. Differential Gene Expression Statistical Analysis
4.8. Validation of the mRNA Expression
4.9. Principal Component Analysis (PCA)
4.10. Pathway Enrichment Analysis
4.11. Identification of a Prognostic MPM Signature
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALDH | Aldehyde Dehydrogenase |
| MPM | Malignant Pleural Mesothelioma |
| ELISA | Enzyme-linked immunosorbent assay |
| NFkB | nuclear factor of kappa light polypeptide gene enhancer in B-cells |
| CSC | Cancer Stem Cell |
| EMT | Epithelial to Mesenchymal Transition |
| FACS | Fluorescence Activated Cell Sorting |
References
- Baram, T.; Rubinstein-Achiasaf, L.; Ben-Yaakov, H.; Ben-Baruch, A. Inflammation-Driven Breast Tumor Cell Plasticity: Stemness/EMT, Therapy Resistance and Dormancy. Front. Oncol. 2020, 10, 614468. [Google Scholar] [CrossRef]
- Ou, A.; Yung, W.K.A.; Majd, N. Molecular Mechanisms of Treatment Resistance in Glioblastoma. Int. J. Mol. Sci. 2020, 22, 351. [Google Scholar] [CrossRef] [PubMed]
- Swayden, M.; Chhouri, H.; Anouar, Y.; Grumolato, L. Tolerant/Persister Cancer Cells and the Path to Resistance to Targeted Therapy. Cells 2020, 9, 2601. [Google Scholar] [CrossRef]
- Zhu, X.; Li, S.; Xu, B.; Luo, H. Cancer evolution: A means by which tumors evade treatment. Biomed. Pharm. 2021, 133, 111016. [Google Scholar] [CrossRef] [PubMed]
- Moris, N.; Pina, C.; Arias, A.M. Transition states and cell fate decisions in epigenetic landscapes. Nat. Rev. Genet. 2016, 17, 693–703. [Google Scholar] [CrossRef]
- Quagliano, A.; Gopalakrishnapillai, A.; Barwe, S.P. Understanding the Mechanisms by Which Epigenetic Modifiers Avert Therapy Resistance in Cancer. Front. Oncol. 2020, 10, 992. [Google Scholar] [CrossRef] [PubMed]
- Saini, A.; Gallo, J.M. Epigenetic instability may alter cell state transitions and anticancer drug resistance. PLoS Comput. Biol. 2021, 17, e1009307. [Google Scholar] [CrossRef]
- Ramon, Y.C.S.; Sese, M.; Capdevila, C.; Aasen, T.; De Mattos-Arruda, L.; Diaz-Cano, S.J.; Hernandez-Losa, J.; Castellvi, J. Clinical implications of intratumor heterogeneity: Challenges and opportunities. J. Mol. Med. (Berl.) 2020, 98, 161–177. [Google Scholar] [CrossRef] [Green Version]
- Vessoni, A.T.; Filippi-Chiela, E.C.; Lenz, G.; Batista, L.F.Z. Tumor propagating cells: Drivers of tumor plasticity, heterogeneity, and recurrence. Oncogene 2020, 39, 2055–2068. [Google Scholar] [CrossRef]
- Heppner, G.H. Tumor heterogeneity. Cancer Res. 1984, 44, 2259–2265. [Google Scholar]
- Brims, F. Epidemiology and Clinical Aspects of Malignant Pleural Mesothelioma. Cancers 2021, 13, 4194. [Google Scholar] [CrossRef]
- Disselhorst, M.J.; Quispel-Janssen, J.; Lalezari, F.; Monkhorst, K.; de Vries, J.F.; van der Noort, V.; Harms, E.; Burgers, S.; Baas, P. Ipilimumab and nivolumab in the treatment of recurrent malignant pleural mesothelioma (INITIATE): Results of a prospective, single-arm, phase 2 trial. Lancet Respir. Med. 2019, 7, 260–270. [Google Scholar] [CrossRef]
- Scherpereel, A.; Mazieres, J.; Greillier, L.; Lantuejoul, S.; Do, P.; Bylicki, O.; Monnet, I.; Corre, R.; Audigier-Valette, C.; Locatelli-Sanchez, M.; et al. Nivolumab or nivolumab plus ipilimumab in patients with relapsed malignant pleural mesothelioma (IFCT-1501 MAPS2): A multicentre, open-label, randomised, non-comparative, phase 2 trial. Lancet Oncol. 2019, 20, 239–253. [Google Scholar] [CrossRef]
- Vogelzang, N.J.; Rusthoven, J.J.; Symanowski, J.; Denham, C.; Kaukel, E.; Ruffie, P.; Gatzemeier, U.; Boyer, M.; Emri, S.; Manegold, C.; et al. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J. Clin. Oncol. 2003, 21, 2636–2644. [Google Scholar] [CrossRef]
- Nicolini, F.; Bocchini, M.; Bronte, G.; Delmonte, A.; Guidoboni, M.; Crino, L.; Mazza, M. Malignant Pleural Mesothelioma: State-of-the-Art on Current Therapies and Promises for the Future. Front. Oncol. 2019, 9, 1519. [Google Scholar] [CrossRef] [PubMed]
- Mujoomdar, A.A.; Tilleman, T.R.; Richards, W.G.; Bueno, R.; Sugarbaker, D.J. Prevalence of in vitro chemotherapeutic drug resistance in primary malignant pleural mesothelioma: Result in a cohort of 203 resection specimens. J. Thorac. Cardiovasc. Surg. 2010, 140, 352–355. [Google Scholar] [CrossRef] [Green Version]
- Clark, D.W.; Palle, K. Aldehyde dehydrogenases in cancer stem cells: Potential as therapeutic targets. Ann. Transl. Med. 2016, 4, 518. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, T.; Zhu, J.; Zhang, H.; Jiang, H.; Sun, H. High ALDH activity defines ovarian cancer stem-like cells with enhanced invasiveness and EMT progress which are responsible for tumor invasion. Biochem. Biophys. Res. Commun. 2018, 495, 1081–1088. [Google Scholar] [CrossRef]
- Voronkova, M.A.; Rojanasakul, L.W.; Kiratipaiboon, C.; Rojanasakul, Y. The SOX9-Aldehyde Dehydrogenase Axis Determines Resistance to Chemotherapy in Non-Small-Cell Lung Cancer. Mol. Cell Biol. 2020, 40, e00307-19. [Google Scholar] [CrossRef]
- Brocker, C.; Vasiliou, M.; Carpenter, S.; Carpenter, C.; Zhang, Y.; Wang, X.; Kotchoni, S.O.; Wood, A.J.; Kirch, H.H.; Kopecny, D.; et al. Aldehyde dehydrogenase (ALDH) superfamily in plants: Gene nomenclature and comparative genomics. Planta 2013, 237, 189–210. [Google Scholar] [CrossRef] [Green Version]
- Dinavahi, S.S.; Bazewicz, C.G.; Gowda, R.; Robertson, G.P. Aldehyde Dehydrogenase Inhibitors for Cancer Therapeutics. Trends Pharm. Sci. 2019, 40, 774–789. [Google Scholar] [CrossRef]
- Durinikova, E.; Kozovska, Z.; Poturnajova, M.; Plava, J.; Cierna, Z.; Babelova, A.; Bohovic, R.; Schmidtova, S.; Tomas, M.; Kucerova, L.; et al. ALDH1A3 upregulation and spontaneous metastasis formation is associated with acquired chemoresistance in colorectal cancer cells. BMC Cancer 2018, 18, 848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcato, P.; Dean, C.A.; Giacomantonio, C.A.; Lee, P.W. Aldehyde dehydrogenase: Its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle 2011, 10, 1378–1384. [Google Scholar] [CrossRef] [PubMed]
- Canino, C.; Luo, Y.; Marcato, P.; Blandino, G.; Pass, H.I.; Cioce, M. A STAT3-NFkB/DDIT3/CEBPbeta axis modulates ALDH1A3 expression in chemoresistant cell subpopulations. Oncotarget 2015, 6, 12637–12653. [Google Scholar] [CrossRef] [Green Version]
- Vassalli, G. Aldehyde Dehydrogenases: Not Just Markers, but Functional Regulators of Stem Cells. Stem Cells Int. 2019, 2019, 3904645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cioce, M.; Canino, C.; Pulito, C.; Muti, P.; Strano, S.; Blandino, G. Butein impairs the protumorigenic activity of malignant pleural mesothelioma cells. Cell Cycle 2012, 11, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.X.; Siu, M.K.; Wang, J.J.; Mo, X.T.; Leung, T.H.; Chan, D.W.; Cheung, A.N.; Ngan, H.Y.; Chan, K.K. Ascites-derived ALDH+CD44+ tumour cell subsets endow stemness, metastasis and metabolic switch via PDK4-mediated STAT3/AKT/NF-kappaB/IL-8 signalling in ovarian cancer. Br. J. Cancer 2020, 123, 275–287. [Google Scholar] [CrossRef]
- Arnold, K.M.; Opdenaker, L.M.; Flynn, N.J.; Appeah, D.K.; Sims-Mourtada, J. Radiation induces an inflammatory response that results in STAT3-dependent changes in cellular plasticity and radioresistance of breast cancer stem-like cells. Int. J. Radiat. Biol. 2020, 96, 434–447. [Google Scholar] [CrossRef]
- Shiraiwa, K.; Matsuse, M.; Nakazawa, Y.; Ogi, T.; Suzuki, K.; Saenko, V.; Xu, S.; Umezawa, K.; Yamashita, S.; Tsukamoto, K.; et al. JAK/STAT3 and NF-kappaB Signaling Pathways Regulate Cancer Stem-Cell Properties in Anaplastic Thyroid Cancer Cells. Thyroid 2019, 29, 674–682. [Google Scholar] [CrossRef]
- Moreira, M.P.; da Conceicao Braga, L.; Cassali, G.D.; Silva, L.M. STAT3 as a promising chemoresistance biomarker associated with the CD44(+/high)/CD24(-/low)/ALDH(+) BCSCs-like subset of the triple-negative breast cancer (TNBC) cell line. Exp. Cell Res. 2018, 363, 283–290. [Google Scholar] [CrossRef]
- Lin, L.; Jou, D.; Wang, Y.; Ma, H.; Liu, T.; Fuchs, J.; Li, P.K.; Lu, J.; Li, C.; Lin, J. STAT3 as a potential therapeutic target in ALDH+ and CD44+/CD24+ stem cell-like pancreatic cancer cells. Int. J. Oncol. 2016, 49, 2265–2274. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Fuchs, J.; Li, C.; Olson, V.; Bekaii-Saab, T.; Lin, J. STAT3 signaling pathway is necessary for cell survival and tumorsphere forming capacity in ALDH(+)/CD133(+) stem cell-like human colon cancer cells. Biochem. Biophys. Res. Commun. 2011, 416, 246–251. [Google Scholar] [CrossRef]
- Canino, C.; Mori, F.; Cambria, A.; Diamantini, A.; Germoni, S.; Alessandrini, G.; Borsellino, G.; Galati, R.; Battistini, L.; Blandino, R.; et al. SASP mediates chemoresistance and tumor-initiating-activity of mesothelioma cells. Oncogene 2012, 31, 3148–3163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, E.; Webster, J.; Do, T.; Kline, R.; Snider, L.; Hauser, Q.; Higginbottom, G.; Campbell, A.; Ma, L.; Paula, S. Hydroxylated chalcones with dual properties: Xanthine oxidase inhibitors and radical scavengers. Bioorg. Med. Chem. 2016, 24, 578–587. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.X.; Sun, S.W.; Liu, Y.; Li, Y.; Liu, X.H.; Wang, W.; Zhang, S.; Wang, W. An Improved Method for the Synthesis of Butein Using SOCl2/EtOH as Catalyst and Deciphering Its Inhibition Mechanism on Xanthine Oxidase. Molecules 2019, 24, 1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zona, S.; Bella, L.; Burton, M.J.; de Moraes, G.N.; Lam, E.W. FOXM1: An emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim. Biophys. Acta 2014, 1839, 1316–1322. [Google Scholar] [CrossRef] [Green Version]
- Choi, E.H.; Kim, K.P. E2F1 facilitates DNA break repair by localizing to break sites and enhancing the expression of homologous recombination factors. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pandey, M.K.; Sandur, S.K.; Sung, B.; Sethi, G.; Kunnumakkara, A.B.; Aggarwal, B.B. Butein, a tetrahydroxychalcone, inhibits nuclear factor (NF)-kappaB and NF-kappaB-regulated gene expression through direct inhibition of IkappaBalpha kinase beta on cysteine 179 residue. J. Biol. Chem. 2007, 282, 17340–17350. [Google Scholar] [CrossRef] [Green Version]
- Moon, D.O.; Choi, Y.H.; Moon, S.K.; Kim, W.J.; Kim, G.Y. Butein suppresses the expression of nuclear factor-kappa B-mediated matrix metalloproteinase-9 and vascular endothelial growth factor in prostate cancer cells. Toxicol. Vitr. 2010, 24, 1927–1934. [Google Scholar] [CrossRef]
- Cortes-Dericks, L.; Froment, L.; Boesch, R.; Schmid, R.A.; Karoubi, G. Cisplatin-resistant cells in malignant pleural mesothelioma cell lines show ALDH(high)CD44(+) phenotype and sphere-forming capacity. BMC Cancer 2014, 14, 304. [Google Scholar] [CrossRef] [Green Version]
- Kamble, D.; Mahajan, M.; Dhat, R.; Sitasawad, S. Keap1-Nrf2 Pathway Regulates ALDH and Contributes to Radioresistance in Breast Cancer Stem Cells. Cells 2021, 10, 83. [Google Scholar] [CrossRef]
- Liu, P.; Brown, S.; Goktug, T.; Channathodiyil, P.; Kannappan, V.; Hugnot, J.P.; Guichet, P.O.; Bian, X.; Armesilla, A.L.; Darling, J.L.; et al. Cytotoxic effect of disulfiram/copper on human glioblastoma cell lines and ALDH-positive cancer-stem-like cells. Br. J. Cancer 2012, 107, 1488–1497. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, Y.; Li, J.; Chen, X.; Fu, X.; Sun, S.; Wu, Q. Chalcone Derivatives: Role in Anticancer Therapy. Biomolecules 2021, 11, 894. [Google Scholar] [CrossRef]
- Jayasooriya, R.; Molagoda, I.M.N.; Park, C.; Jeong, J.W.; Choi, Y.H.; Moon, D.O.; Kim, M.O.; Kim, G.Y. Molecular chemotherapeutic potential of butein: A concise review. Food Chem. Toxicol. 2018, 112, 1–10. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, W.; Li, X. A novel anticancer effect of butein: Inhibition of invasion through the ERK1/2 and NF-kappa B signaling pathways in bladder cancer cells. FEBS Lett. 2008, 582, 1821–1828. [Google Scholar] [CrossRef] [Green Version]
- Moon, D.O.; Kim, M.O.; Lee, J.D.; Choi, Y.H.; Kim, G.Y. Butein suppresses c-Myc-dependent transcription and Akt-dependent phosphorylation of hTERT in human leukemia cells. Cancer Lett. 2009, 286, 172–179. [Google Scholar] [CrossRef]
- Brcic, L.; Kern, I. Clinical significance of histologic subtyping of malignant pleural mesothelioma. Transl. Lung Cancer Res. 2020, 9, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Pass, H.I.; Stevens, E.J.; Oie, H.; Tsokos, M.G.; Abati, A.D.; Fetsch, P.A.; Mew, D.J.; Pogrebniak, H.W.; Matthews, W.J. Characteristics of nine newly derived mesothelioma cell lines. Ann. Thorac. Surg. 1995, 59, 835–844. [Google Scholar] [CrossRef]
- Colacino, J.A.; Azizi, E.; Brooks, M.D.; Harouaka, R.; Fouladdel, S.; McDermott, S.P.; Lee, M.; Hill, D.; Madden, J.; Boerner, J.; et al. Heterogeneity of Human Breast Stem and Progenitor Cells as Revealed by Transcriptional Profiling. Stem. Cell Rep. 2018, 10, 1596–1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, Y.; Meiller, C.; Quetel, L.; Elarouci, N.; Ayadi, M.; Tashtanbaeva, D.; Armenoult, L.; Montagne, F.; Tranchant, R.; Renier, A.; et al. Dissecting heterogeneity in malignant pleural mesothelioma through histo-molecular gradients for clinical applications. Nat. Commun. 2019, 10, 1333. [Google Scholar] [CrossRef] [PubMed]
- Tsochantaridis, I.; Roupas, A.; Voulgaridou, G.P.; Giatromanolaki, A.; Koukourakis, M.I.; Panayiotidis, M.I.; Pappa, A. Aldehyde Dehydrogenase 1B1 Is Associated with Altered Cell Morphology, Proliferation, Migration and Chemosensitivity in Human Colorectal Adenocarcinoma Cells. Biomedicines 2021, 9, 44. [Google Scholar] [CrossRef]
- Zhang, M.; Luo, J.L.; Sun, Q.; Harber, J.; Dawson, A.G.; Nakas, A.; Busacca, S.; Sharkey, A.J.; Waller, D.; Sheaff, M.T.; et al. Clonal architecture in mesothelioma is prognostic and shapes the tumour microenvironment. Nat. Commun. 2021, 12, 1751. [Google Scholar] [CrossRef] [PubMed]
- Comertpay, S.; Pastorino, S.; Tanji, M.; Mezzapelle, R.; Strianese, O.; Napolitano, A.; Baumann, F.; Weigel, T.; Friedberg, J.; Sugarbaker, P.; et al. Evaluation of clonal origin of malignant mesothelioma. J. Transl. Med. 2014, 12, 301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oey, H.; Daniels, M.; Relan, V.; Chee, T.M.; Davidson, M.R.; Yang, I.A.; Ellis, J.J.; Fong, K.M.; Krause, L.; Bowman, R.V. Whole-genome sequencing of human malignant mesothelioma tumours and cell lines. Carcinogenesis 2019, 40, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, 566–570. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Han, H.; Shim, H.; Shin, D.; Shim, J.E.; Ko, Y.; Shin, J.; Kim, H.; Cho, A.; Kim, E.; Lee, T.; et al. TRRUST: A reference database of human transcriptional regulatory interactions. Sci. Rep. 2015, 5, 11432. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cioce, M.; Sacconi, A.; Pass, H.I.; Canino, C.; Strano, S.; Blandino, G.; Fazio, V.M. Insights into Intra-Tumoral Heterogeneity: Transcriptional Profiling of Chemoresistant MPM Cell Subpopulations Reveals Involvement of NFkB and DNA Repair Pathways and Contributes a Prognostic Signature. Int. J. Mol. Sci. 2021, 22, 12071. https://doi.org/10.3390/ijms222112071
Cioce M, Sacconi A, Pass HI, Canino C, Strano S, Blandino G, Fazio VM. Insights into Intra-Tumoral Heterogeneity: Transcriptional Profiling of Chemoresistant MPM Cell Subpopulations Reveals Involvement of NFkB and DNA Repair Pathways and Contributes a Prognostic Signature. International Journal of Molecular Sciences. 2021; 22(21):12071. https://doi.org/10.3390/ijms222112071
Chicago/Turabian StyleCioce, Mario, Andrea Sacconi, Harvey I. Pass, Claudia Canino, Sabrina Strano, Giovanni Blandino, and Vito Michele Fazio. 2021. "Insights into Intra-Tumoral Heterogeneity: Transcriptional Profiling of Chemoresistant MPM Cell Subpopulations Reveals Involvement of NFkB and DNA Repair Pathways and Contributes a Prognostic Signature" International Journal of Molecular Sciences 22, no. 21: 12071. https://doi.org/10.3390/ijms222112071







