Digging Deeper: Advancements in Visualization of Inhibitory Synapses in Neurodegenerative Disorders
Abstract
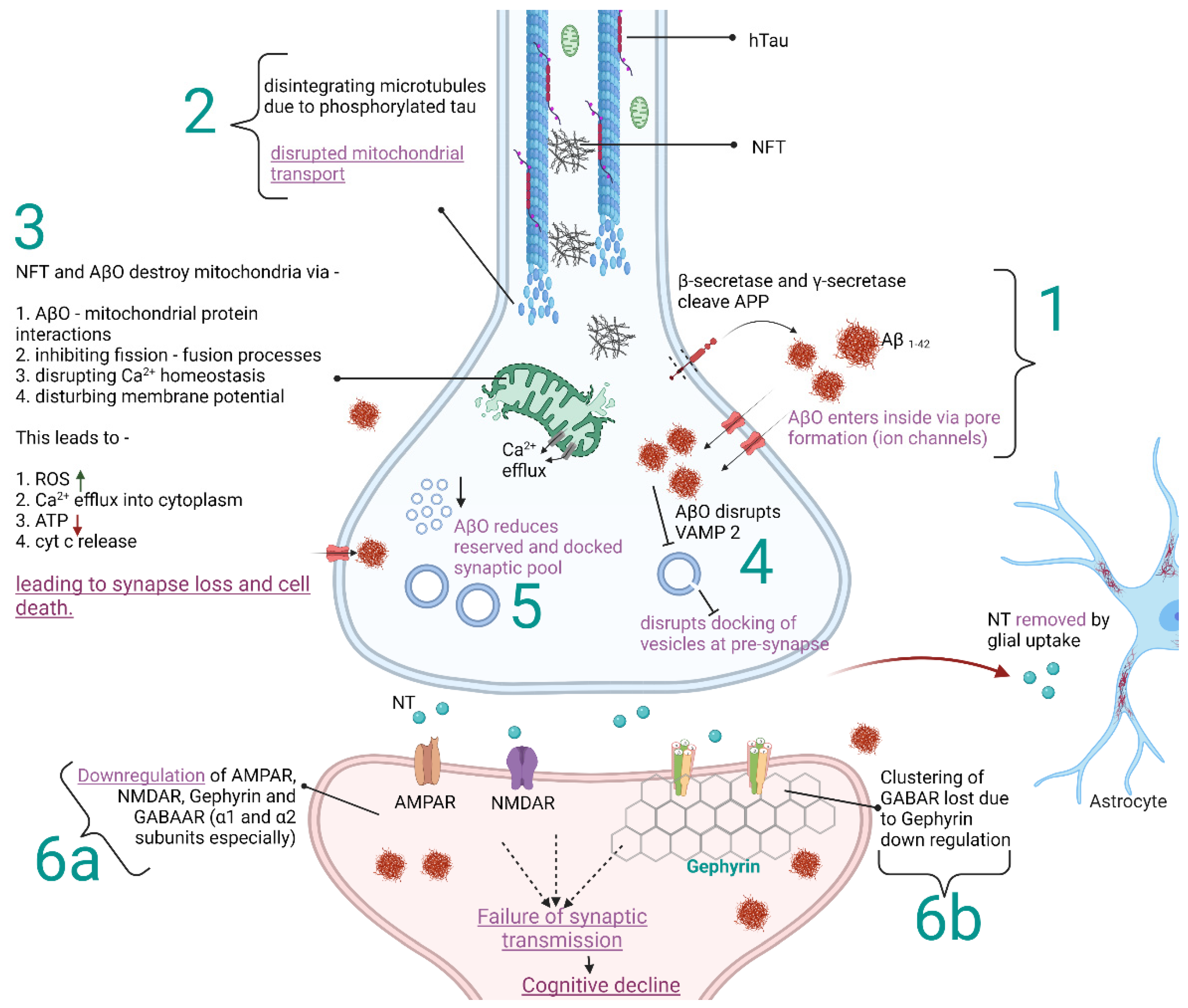
:1. Introduction
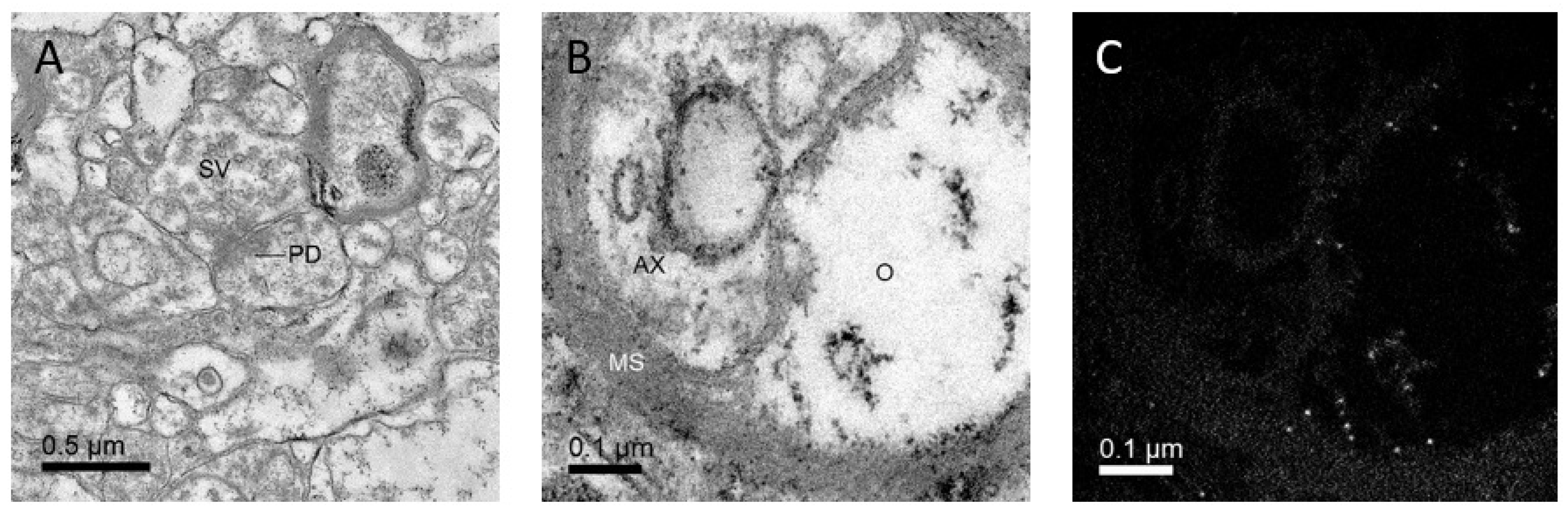
2. Discovering Details of Synapses with Electron Microscopy
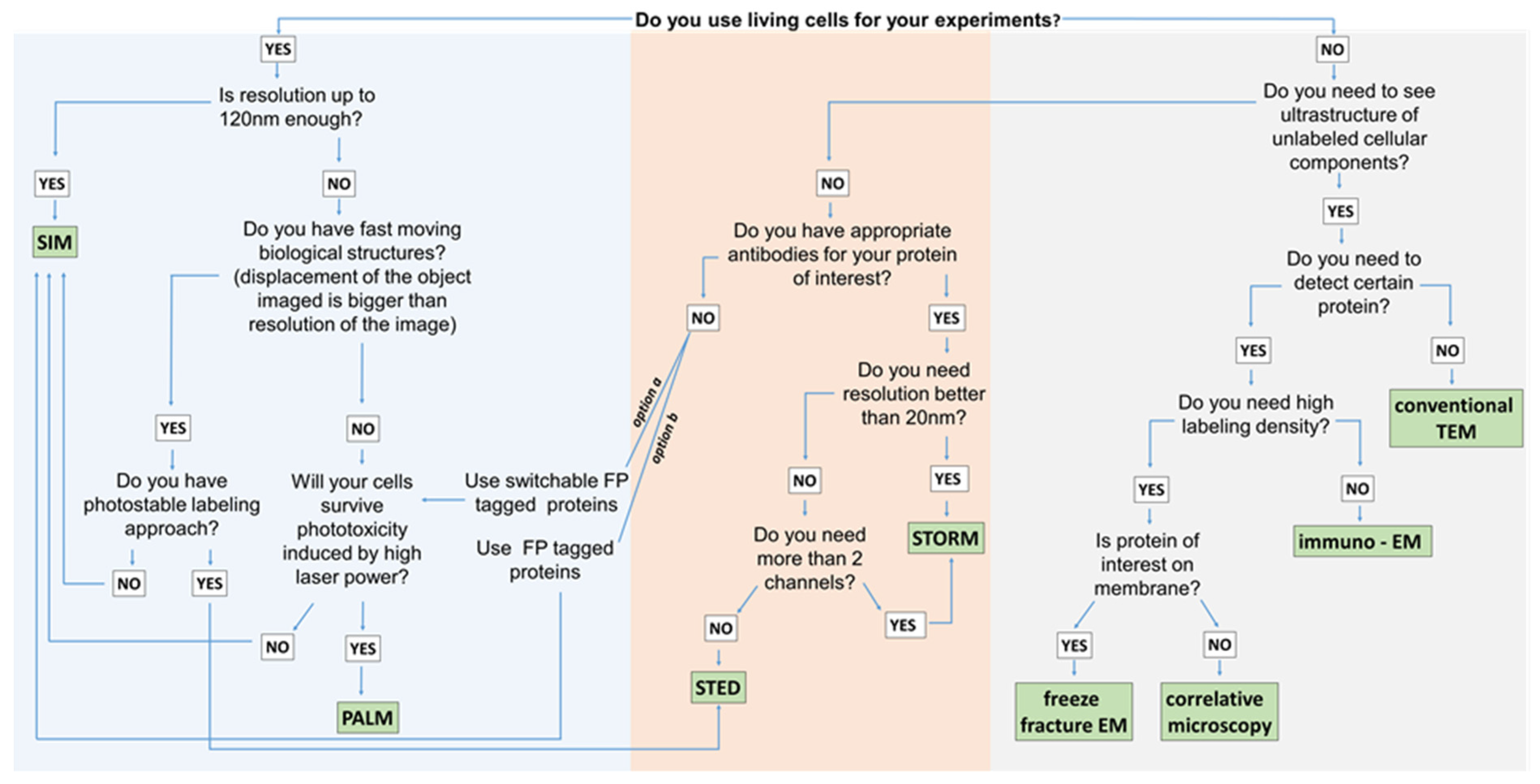
3. Super-Resolution Imaging
4. How to Visualize Iron in Neurodegenerative Disorders
5. Discussion and Conclusions
6. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Batool, S.; Raza, H.; Zaidi, J.; Riaz, S.; Hasan, S.; Syed, N.I. Synapse formation: From cellular and molecular mechanisms to neurodevelopmental and neurodegenerative disorders. J. Neurophysiol. 2019, 121, 1381–1397. [Google Scholar] [CrossRef] [PubMed]
- Terry, R.D.; Masliah, E.; Salmon, D.P.; Butters, N.; DeTeresa, R.; Hill, R.; Hansen, L.A.; Katzman, R. Physical basis of cognitive alterations in Alzheimer’s disease: Synapse loss is the major correlate of cognitive impairment. Ann. Neurol. 1991, 30, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Chakroborty, S.; Hill, E.S.; Christian, D.T.; Helfrich, R.; Riley, S.; Schneider, C.; Kapecki, N.; Mustaly-Kalimi, S.; Seiler, F.A.; Peterson, D.A.; et al. Reduced presynaptic vesicle stores mediate cellular and network plasticity defects in an early-stage mouse model of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, J.; Jambrina, E.; Li, J.; Marston, H.; Menzies, F.; Phillips, K.; Gilmour, G. Targeting the synapse in Alzheimer’s disease. Front. Neurosci. 2019, 13, 735. [Google Scholar] [CrossRef] [Green Version]
- Mele, M.; Leal, G.; Duarte, C.B. Role of GABAAR trafficking in the plasticity of inhibitory synapses. J. Neurochem. 2016, 139, 997–1018. [Google Scholar] [CrossRef]
- Jack, C.R.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA research framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Scheltens, P.; Blennow, K.; Breteler, M.M.B.; de Strooper, B.; Frisoni, G.B.; Salloway, S.; Van der Flier, W.M. Alzheimer’s disease. Lancet 2016, 388, 505–517. [Google Scholar] [CrossRef]
- Budak, M.; Zochowski, M. Synaptic failure differentially affects pattern formation in heterogenous networks. Front. Neural Circuits 2019, 13, 31. [Google Scholar] [CrossRef]
- Shimojo, M.; Takuwa, H.; Takado, Y.; Tokunaga, M.; Tsukamoto, S.; Minatohara, K.; Ono, M.; Seki, C.; Maeda, J.; Urushihata, T.; et al. Selective disruption of inhibitory synapses leading to neuronal hyperexcitability at an early stage of tau pathogenesis in a mouse model. J. Neurosci. 2020, 40, 3491–3501. [Google Scholar] [CrossRef]
- Amorim, J.A.; Canas, P.M.; Tomé, A.R.; Rolo, A.P.; Agostinho, P.; Palmeira, C.M.; Cunha, R.A. Mitochondria in excitatory and inhibitory synapses have similar susceptibility to amyloid-β peptides modeling alzheimer’s disease. J. Alzheimers Dis. 2017, 60, 525–536. [Google Scholar] [CrossRef]
- Bae, J.R.; Kim, S.H. Synapses in neurodegenerative diseases. BMB Rep. 2017, 50, 237–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakravorty, A.; Jetto, C.T.; Manjithaya, R. Dysfunctional mitochondria and mitophagy as drivers of Alzheimer’s disease pathogenesis. Front. Aging Neurosci. 2019, 11, 311. [Google Scholar] [CrossRef] [PubMed]
- Groeneweg, F.L.; Trattnig, C.; Kuhse, J.; Nawrotzki, R.A.; Kirsch, J. Gephyrin: A key regulatory protein of inhibitory synapses and beyond. Histochem. Cell Biol. 2018, 150, 489–508. [Google Scholar] [CrossRef] [PubMed]
- Kulijewicz-Nawrot, M.; Syková, E.; Chvátal, A.; Verkhratsky, A.; Rodríguez, J.J. Astrocytes and glutamate homoeostasis in Alzheimer’s disease: A decrease in glutamine synthetase, but not in glutamate transporter-1, in the prefrontal cortex. ASN Neuro 2013, 5, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, K.; Li, N.; Wang, X.; Xiao, X.; Li, L.; Li, L.; He, Y.; Zhang, J.; Wo, J.; et al. Reversible GABAergic dysfunction involved in hippocampal hyperactivity predicts early-stage Alzheimer disease in a mouse model. Alzheimers Res. Ther. 2021, 13, 114. [Google Scholar] [CrossRef]
- Limon, A.; Reyes-Ruiz, J.M.; Miledi, R. Loss of functional GABA A receptors in the Alzheimer diseased brain. Proc. Natl. Acad. Sci. USA 2012, 109, 10071–10076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, H.; Yu, E.; Pigino, G.; Hernandez, A.I.; Kim, N.; Moreira, J.E.; Sugimori, M.; Llinás, R.R. Synaptic transmission block by presynaptic injection of oligomeric amyloid beta. Proc. Natl. Acad. Sci. USA 2009, 106, 5901–5906. [Google Scholar] [CrossRef] [Green Version]
- Pickett, E.K.; Koffie, R.M.; Wegmann, S.; Henstridge, C.M.; Herrmann, A.G.; Colom-Cadena, M.; Lleo, A.; Kay, K.R.; Vaught, M.; Soberman, R.; et al. Non-fibrillar oligomeric amyloid-β within synapses. J. Alzheimers Dis. 2016, 53, 787–800. [Google Scholar] [CrossRef] [Green Version]
- Tönnies, E.; Trushina, E. Oxidative stress, synaptic dysfunction, and Alzheimer’s disease. J. Alzheimers Dis. 2017, 57, 1105–1121. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhao, M.; Han, Y.; Zhang, H. GABAergic inhibitory interneuron deficits in Alzheimer’s disease: Implications for treatment. Front. Neurosci. 2020, 14, 660. [Google Scholar] [CrossRef]
- Zhang, W.; Xiong, B.R.; Zhang, L.Q.; Huang, X.; Yuan, X.; Tian, Y.K.; Tian, X.B. The role of the GABAergic system in diseases of the central nervous system. Neuroscience 2021, 470, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Kurucu, H.; Colom-Cadena, M.; Davies, C.; Wilkins, L.; King, D.; Rose, J.; Tzioras, M.; Tulloch, J.H.; Smith, C.; Spires-Jones, T.L. Inhibitory synapse loss and accumulation of amyloid beta in inhibitory presynaptic terminals in Alzheimer’s disease. Eur. J. Neurol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Pickett, E.K.; Herrmann, A.G.; McQueen, J.; Abt, K.; Dando, O.; Tulloch, J.; Jain, P.; Dunnett, S.; Sohrabi, S.; Fjeldstad, M.P.; et al. Amyloid beta and tau cooperate to cause reversible behavioral and transcriptional deficits in a model of Alzheimer’s disease. Cell Rep. 2019, 29, 3592–3604.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willén, K.; Sroka, A.; Takahashi, R.H.; Gouras, G.K. Heterogeneous association of Alzheimer’s disease-linked amyloid-β and amyloid-β protein precursor with synapses. J. Alzheimers Dis. 2017, 60, 511–524. [Google Scholar] [CrossRef] [Green Version]
- Folch, J.; Busquets, O.; Ettcheto, M.; Sánchez-López, E.; Castro-Torres, R.D.; Verdaguer, E.; Garcia, M.L.; Olloquequi, J.; Casadesús, G.; Beas-Zarate, C.; et al. Memantine for the treatment of dementia: A review on its current and future applications. J. Alzheimers Dis. 2018, 62, 1223–1240. [Google Scholar] [CrossRef] [Green Version]
- Gong, N.J.; Dibb, R.; Bulk, M.; van der Weerd, L.; Liu, C. Imaging beta amyloid aggregation and iron accumulation in Alzheimer’s disease using quantitative susceptibility mapping MRI. NeuroImage 2019, 191, 176–185. [Google Scholar] [CrossRef]
- Zoupi, L.; Booker, S.A.; Eigel, D.; Werner, C.; Kind, P.C.; Spires-Jones, T.L.; Newland, B.; Williams, A.C. Selective vulnerability of inhibitory networks in multiple sclerosis. Acta Neuropathol. 2021, 141, 415–429. [Google Scholar] [CrossRef]
- Bos, M.A.J.V.d.; Higashihara, M.; Geevasinga, N.; Menon, P.; Kiernan, M.C.; Vucic, S. Imbalance of cortical facilitatory and inhibitory circuits underlies hyperexcitability in ALS. Neurology 2018, 91, e1669–e1676. [Google Scholar] [CrossRef]
- Diana, A.; Pillai, R.; Bongioanni, P.; O’Keeffe, A.G.; Miller, R.G.; Moore, D.H. Gamma aminobutyric acid (GABA) modulators for amyotrophic lateral sclerosis/motor neuron disease. Cochrane Database Syst. Rev. 2017, 1, CD006049. [Google Scholar] [CrossRef] [Green Version]
- Van Es, M.A.; Hardiman, O.; Chio, A.; Al-Chalabi, A.; Pasterkamp, R.J.; Veldink, J.H.; van den Berg, L.H. Amyotrophic lateral sclerosis. Lancet 2017, 390, 2084–2098. [Google Scholar] [CrossRef]
- Fu, Z.; Yang, H.; Xiao, Y.; Zhao, G.; Huang, H. The gamma-aminobutyric acid type B (GABAB) receptor agonist baclofen inhibits morphine sensitization by decreasing the dopamine level in rat nucleus accumbens. Behav. Brain Funct. 2012, 8, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardiman, O.; Al-Chalabi, A.; Chio, A.; Corr, E.M.; Logroscino, G.; Robberecht, W.; Shaw, P.J.; Simmons, Z.; van den Berg, L.H. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers 2017, 3, 17071. [Google Scholar] [CrossRef] [PubMed]
- Benedict, R.H.B.; Amato, M.P.; DeLuca, J.; Geurts, J.J.G. Cognitive impairment in multiple sclerosis: Clinical management, MRI, and therapeutic avenues. Lancet Neurol. 2020, 19, 860–871. [Google Scholar] [CrossRef]
- Filippini, G.; Giovane, C.D.; Vacchi, L.; D’Amico, R.; Pietrantonj, C.D.; Beecher, D.; Salanti, G. Immunomodulators and immunosuppressants for multiple sclerosis: A network meta-analysis. Cochrane Database Syst. Rev. 2011, 6, CD008933. [Google Scholar] [CrossRef] [Green Version]
- Jasek, Ł.; Śmigielski, J.; Siger, M. Late onset multiple sclerosis—Multiparametric MRI characteristics. Neurol. Neurochir. Pol. 2020, 54, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Kaskow, B.J.; Baecher-Allan, C. Effector T cells in multiple sclerosis. Cold Spring Harb. Perspect. Med. 2018, 8, a029025. [Google Scholar] [CrossRef]
- Ziemssen, T.; Akgün, K.; Brück, W. Molecular biomarkers in multiple sclerosis. J. Neuroinflamm. 2019, 16, 272. [Google Scholar] [CrossRef] [Green Version]
- Miller, R.G.; Mitchell, J.D.; Moore, D.H. Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND). Cochrane Database Syst Rev. 2012, 3, CD001447. [Google Scholar] [CrossRef]
- Sugiyama, A.; Saitoh, A.; Yamada, M.; Oka, J.I.; Yamada, M. Administration of riluzole into the basolateral amygdala has an anxiolytic-like effect and enhances recognition memory in the rat. Behav. Brain Res. 2017, 327, 98–102. [Google Scholar] [CrossRef]
- Zucchi, E.; Bonetto, V.; Sorarù, G.; Martinelli, I.; Parchi, P.; Liguori, R.; Mandrioli, J. Neurofilaments in motor neuron disorders: Towards promising diagnostic and prognostic biomarkers. Mol. Neurodegener. 2020, 15, 58. [Google Scholar] [CrossRef]
- Garret, M.; Du, Z.; Chazalon, M.; Cho, Y.H.; Baufreton, J. Alteration of GABAergic neurotransmission in Huntington’s disease. CNS Neurosci. Ther. 2018, 24, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-T.; Chang, Y.-G.; Chern, Y. Insights into GABAAergic system alteration in Huntington’s disease. Open Biol. 2018, 8, 180165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McColgan, P.; Tabrizi, S.J. Huntington’s disease: A clinical review. Eur. J. Neurol. 2018, 25, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Arellano, A.; Tejeda-Guzmán, C.; Lorca-Ponce, E.; Palma-Tirado, L.; Mantellero, C.A.; Rojas, P.; Missirlis, F.; Castro, M.A. Huntington’s disease leads to decrease of GABA-A tonic subunits in the D2 neostriatal pathway and their relocalization into the synaptic cleft. Neurobiol. Dis. 2018, 110, 142–153. [Google Scholar] [CrossRef]
- Wyant, K.J.; Ridder, A.J.; Dayalu, P. Huntington’s disease—Update on treatments. Curr. Neurol. Neurosci. Rep. 2017, 17, 33. [Google Scholar] [CrossRef]
- Abbe, E. Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung. Arch. Für Mikrosk. Anat. 1873, 9, 413–468. [Google Scholar] [CrossRef]
- Klemann, C.J.; Roubos, E.W. The gray area between synapse structure and function-Gray’s synapse types I and II revisited. Synapse 2011, 65, 1222–1230. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, Y.; Peng, Y.; Zhao, Z.; Zhou, Y.; Wang, X.; Peng, Y. Protective effect of potassium 2-(l-hydroxypentyl)-benzoate on hippocampal neurons, synapses and dystrophic axons in APP/PS1 mice. Psychopharmacology 2019, 236, 2761–2771. [Google Scholar] [CrossRef]
- Sterio, D.C. The unbiased estimation of number and sizes of arbitrary particles using the disector. J. Microsc. 1984, 134, 127–136. [Google Scholar] [CrossRef]
- Napper, R.M.A. Total number is important: Using the disector method in design-based stereology to understand the structure of the rodent brain. Front. Neuroanat. 2018, 12, 16. [Google Scholar] [CrossRef] [Green Version]
- Omelchenko, N.; Roy, P.; Balcita-Pedicino, J.J.; Poloyac, S.; Sesack, S.R. Impact of prenatal nicotine on the structure of midbrain dopamine regions in the rat. Brain Struct. Funct. 2016, 221, 1939–1953. [Google Scholar] [CrossRef]
- Reichmann, F.; Painsipp, E.; Holzer, P.; Kummer, D.; Bock, E.; Leitinger, G. A novel unbiased counting method for the quantification of synapses in the mouse brain. J. Neurosci. Methods 2015, 240, 13–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reichmann, F.; Wegerer, V.; Jain, P.; Mayerhofer, R.; Hassan, A.M.; Frohlich, E.E.; Bock, E.; Pritz, E.; Herzog, H.; Holzer, P.; et al. Environmental enrichment induces behavioural disturbances in neuropeptide Y knockout mice. Sci. Rep. 2016, 6, 28182. [Google Scholar] [CrossRef] [PubMed]
- Ardalan, M.; Wegener, G.; Polsinelli, B.; Madsen, T.M.; Nyengaard, J.R. Neurovascular plasticity of the hippocampus one week after a single dose of ketamine in genetic rat model of depression. Hippocampus 2016, 26, 1414–1423. [Google Scholar] [CrossRef] [PubMed]
- Ardalan, M.; Wegener, G.; Rafati, A.H.; Nyengaard, J.R. S-ketamine rapidly reverses synaptic and vascular deficits of hippocampus in genetic animal model of depression. Int. J. Neuropsychopharmacol. 2017, 20, 247–256. [Google Scholar] [CrossRef]
- Lin, J.Y.; He, Y.N.; Zhu, N.; Peng, B. Metformin attenuates increase of synaptic number in the rat spinal dorsal horn with painful diabetic neuropathy induced by type 2 diabetes: A stereological study. Neurochem. Res. 2018, 43, 2232–2239. [Google Scholar] [CrossRef]
- Kim, H.W.; Oh, S.H.; Lee, S.J.; Na, J.E.; Rhyu, I.J. Differential synapse density between Purkinje cell dendritic spine and parallel fiber varicosity in the rat cerebellum among the phylogenic lobules. Appl. Microsc. 2020, 50, 6. [Google Scholar] [CrossRef] [Green Version]
- Ohgomori, T.; Iinuma, K.; Yamada, J.; Jinno, S. A unique subtype of ramified microglia associated with synapses in the rat hippocampus. Eur. J. Neurosci. 2021, 54, 4740–4754. [Google Scholar] [CrossRef]
- Gray, E.G. Axo-somatic and axo-dendritic synapses of the cerebral cortex: An electron microscope study. J. Anat. 1959, 93, 420–433. [Google Scholar]
- Tao, C.L.; Liu, Y.T.; Sun, R.; Zhang, B.; Qi, L.; Shivakoti, S.; Tian, C.L.; Zhang, P.; Lau, P.M.; Zhou, Z.H.; et al. Differentiation and characterization of excitatory and inhibitory synapses by cryo-electron tomography and correlative microscopy. J. Neurosci. 2018, 38, 1493–1510. [Google Scholar] [CrossRef] [Green Version]
- Dubochet, J. High-pressure freezing for cryoelectron microscopy. Trends Cell Biol. 1995, 5, 366–368. [Google Scholar] [CrossRef] [Green Version]
- Sele, M.; Wernitznig, S.; Lipovsek, S.; Radulovic, S.; Haybaeck, J.; Birkl-Toeglhofer, A.M.; Wodlej, C.; Kleinegger, F.; Sygulla, S.; Leoni, M.; et al. Optimization of ultrastructural preservation of human brain for transmission electron microscopy after long post-mortem intervals. Acta Neuropathol. Commun. 2019, 7, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, S.; Liu, Q.; Davis, M.W.; Hollopeter, G.; Thomas, N.; Jorgensen, N.B.; Jorgensen, E.M. Ultrafast endocytosis at Caenorhabditis elegans neuromuscular junctions. eLife 2013, 2, e00723. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Rost, B.R.; Camacho-Perez, M.; Davis, M.W.; Sohl-Kielczynski, B.; Rosenmund, C.; Jorgensen, E.M. Ultrafast endocytosis at mouse hippocampal synapses. Nature 2013, 504, 242–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges-Merjane, C.; Kim, O.; Jonas, P. Functional electron microscopy, “flash and freeze”, of identified cortical synapses in acute brain slices. Neuron 2020, 108, 1011. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Trimbuch, T.; Rosenmund, C. Synaptotagmin-1 drives synchronous Ca2+-triggered fusion by C2B-domain-mediated synaptic-vesicle-membrane attachment. Nat. Neurosci. 2018, 21, 33–40. [Google Scholar] [CrossRef]
- Imig, C.; Cooper, B.H. 3D analysis of synaptic ultrastructure in arganotypic hippocampal slice culture by high-pressure freezing and electron tomography. Methods Mol. Biol. 2017, 1538, 215–231. [Google Scholar] [CrossRef] [PubMed]
- Imig, C.; Lopez-Murcia, F.J.; Maus, L.; Garcia-Plaza, I.H.; Mortensen, L.S.; Schwark, M.; Schwarze, V.; Angibaud, J.; Nagerl, U.V.; Taschenberger, H.; et al. Ultrastructural imaging of activity-dependent synaptic membrane-trafficking events in cultured brain slices. Neuron 2020, 108, 843–860.e8. [Google Scholar] [CrossRef]
- Maus, L.; Lee, C.; Altas, B.; Sertel, S.M.; Weyand, K.; Rizzoli, S.O.; Rhee, J.; Brose, N.; Imig, C.; Cooper, B.H. Ultrastructural correlates of presynaptic functional heterogeneity in hippocampal synapses. Cell Rep. 2020, 30, 3632–3643.e8. [Google Scholar] [CrossRef]
- Fujimoto, K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J. Cell Sci. 1995, 108, 3443–3449. [Google Scholar] [CrossRef]
- Masugi-Tokita, M.; Shigemoto, R. High-resolution quantitative visualization of glutamate and GABA receptors at central synapses. Curr. Opin. Neurobiol. 2007, 17, 387–393. [Google Scholar] [CrossRef]
- Mobius, W.; Cooper, B.; Kaufmann, W.A.; Imig, C.; Ruhwedel, T.; Snaidero, N.; Saab, A.S.; Varoqueaux, F. Electron microscopy of the mouse central nervous system. Methods Cell Biol. 2010, 96, 475–512. [Google Scholar] [CrossRef]
- Martin-Belmonte, A.; Aguado, C.; Alfaro-Ruiz, R.; Itakura, M.; Moreno-Martinez, A.E.; de la Ossa, L.; Molnar, E.; Fukazawa, Y.; Lujan, R. Age-dependent shift of AMPA receptors from synapses to intracellular compartments in Alzheimer’s disease: Immunocytochemical analysis of the CA1 hippocampal region in APP/PS1 transgenic mouse model. Front. Aging Neurosci. 2020, 12, 577996. [Google Scholar] [CrossRef] [PubMed]
- Martin-Belmonte, A.; Aguado, C.; Alfaro-Ruiz, R.; Moreno-Martinez, A.E.; de la Ossa, L.; Martinez-Hernandez, J.; Buisson, A.; Fruh, S.; Bettler, B.; Shigemoto, R.; et al. Reduction in the neuronal surface of post and presynaptic GABAB receptors in the hippocampus in a mouse model of Alzheimer.s disease. Brain Pathol. 2020, 30, 554–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin-Belmonte, A.; Aguado, C.; Alfaro-Ruiz, R.; Moreno-Martinez, A.E.; de la Ossa, L.; Martinez-Hernandez, J.; Buisson, A.; Shigemoto, R.; Fukazawa, Y.; Lujan, R. Density of GABAB receptors is reduced in granule cells of the hippocampus in a mouse model of Alzheimer’s disease. Int. J. Mol. Sci. 2020, 21, 2459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dittmayer, C.; Volcker, E.; Wacker, I.; Schroder, R.R.; Bachmann, S. Modern field emission scanning electron microscopy provides new perspectives for imaging kidney ultrastructure. Kidney Int. 2018, 94, 625–631. [Google Scholar] [CrossRef]
- Titze, B.; Genoud, C. Volume scanning electron microscopy for imaging biological ultrastructure. Biol. Cell 2016, 108, 307–323. [Google Scholar] [CrossRef] [PubMed]
- Hylton, R.K.; Swulius, M.T. Challenges and triumphs in cryo-electron tomography. iScience 2021, 24, 102959. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Busnadiego, R. Cryo-Electron Tomography of the Mammalian Synapse. Methods Mol. Biol. 2018, 1847, 217–224. [Google Scholar] [CrossRef] [Green Version]
- Gipson, P.; Fukuda, Y.; Danev, R.; Lai, Y.; Chen, D.H.; Baumeister, W.; Brunger, A.T. Morphologies of synaptic protein membrane fusion interfaces. Proc. Natl. Acad. Sci. USA 2017, 114, 9110–9115. [Google Scholar] [CrossRef] [Green Version]
- Lucic, V.; Fernandez-Busnadiego, R.; Laugks, U.; Baumeister, W. Hierarchical detection and analysis of macromolecular complexes in cryo-electron tomograms using Pyto software. J. Struct. Biol. 2016, 196, 503–514. [Google Scholar] [CrossRef]
- Schrod, N.; Vanhecke, D.; Laugks, U.; Stein, V.; Fukuda, Y.; Schaffer, M.; Baumeister, W.; Lucic, V. Pleomorphic linkers as ubiquitous structural organizers of vesicles in axons. PLoS ONE 2018, 13, e0197886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.T.; Tao, C.L.; Lau, P.M.; Zhou, Z.H.; Bi, G.Q. Postsynaptic protein organization revealed by electron microscopy. Curr. Opin. Struct. Biol. 2019, 54, 152–160. [Google Scholar] [CrossRef]
- Zuber, B.; Lucic, V. Molecular architecture of the presynaptic terminal. Curr. Opin. Struct. Biol. 2019, 54, 129–138. [Google Scholar] [CrossRef]
- Orlando, M.; Ravasenga, T.; Petrini, E.M.; Falqui, A.; Marotta, R.; Barberis, A. Correlating Fluorescence and High-Resolution Scanning Electron Microscopy (HRSEM) for the study of GABAA receptor clustering induced by inhibitory synaptic plasticity. Sci. Rep. 2017, 7, 13768. [Google Scholar] [CrossRef] [PubMed]
- Yip, K.M.; Fischer, N.; Paknia, E.; Chari, A.; Stark, H. Atomic-resolution protein structure determination by cryo-EM. Nature 2020, 587, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.S. The 2017 Nobel Prize in Chemistry: Cryo-EM comes of age. Anal. Bioanal. Chem. 2018, 410, 2053–2057. [Google Scholar] [CrossRef]
- Liu, S.; Xu, L.; Guan, F.; Liu, Y.T.; Cui, Y.; Zhang, Q.; Zheng, X.; Bi, G.Q.; Zhou, Z.H.; Zhang, X.; et al. Cryo-EM structure of the human alpha5beta3 GABAA receptor. Cell Res. 2018, 28, 958–961. [Google Scholar] [CrossRef] [Green Version]
- Phulera, S.; Zhu, H.; Yu, J.; Claxton, D.P.; Yoder, N.; Yoshioka, C.; Gouaux, E. Cryo-EM structure of the benzodiazepine-sensitive alpha1beta1gamma2S tri-heteromeric GABAA receptor in complex with GABA. eLife 2018, 7, e39383. [Google Scholar] [CrossRef]
- Zhu, S.; Noviello, C.M.; Teng, J.; Walsh, R.M., Jr.; Kim, J.J.; Hibbs, R.E. Structure of a human synaptic GABAA receptor. Nature 2018, 559, 67–72. [Google Scholar] [CrossRef]
- Kim, J.J.; Gharpure, A.; Teng, J.; Zhuang, Y.; Howard, R.J.; Zhu, S.; Noviello, C.M.; Walsh, R.M., Jr.; Lindahl, E.; Hibbs, R.E. Shared structural mechanisms of general anaesthetics and benzodiazepines. Nature 2020, 585, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Hibbs, R.E. Direct structural insights into GABAA receptor pharmacology. Trends Biochem. Sci. 2021, 46, 502–517. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Shen, C.; Li, C.; Shen, D.D.; Xu, C.; Zhang, S.; Zhou, R.; Shen, Q.; Chen, L.N.; Jiang, Z.; et al. Cryo-EM structures of inactive and active GABAB receptor. Cell Res. 2020, 30, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Fu, Z.; Frangaj, A.; Liu, J.; Mosyak, L.; Shen, T.; Slavkovich, V.N.; Ray, K.M.; Taura, J.; Cao, B.; et al. Structure of human GABAB receptor in an inactive state. Nature 2020, 584, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Shaye, H.; Ishchenko, A.; Lam, J.H.; Han, G.W.; Xue, L.; Rondard, P.; Pin, J.P.; Katritch, V.; Gati, C.; Cherezov, V. Structural basis of the activation of a metabotropic GABA receptor. Nature 2020, 584, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Papasergi-Scott, M.M.; Robertson, M.J.; Seven, A.B.; Panova, O.; Mathiesen, J.M.; Skiniotis, G. Structures of metabotropic GABAB receptor. Nature 2020, 584, 310–314. [Google Scholar] [CrossRef]
- Evenseth, L.S.M.; Gabrielsen, M.; Sylte, I. The GABAB receptor-structure, ligand binding and drug development. Molecules 2020, 25, 3093. [Google Scholar] [CrossRef] [PubMed]
- Biermann, B.; Ivankova-Susankova, K.; Bradaia, A.; Abdel Aziz, S.; Besseyrias, V.; Kapfhammer, J.P.; Missler, M.; Gassmann, M.; Bettler, B. The Sushi domains of GABAB receptors function as axonal targeting signals. J. Neurosci. 2010, 30, 1385–1394. [Google Scholar] [CrossRef] [Green Version]
- Rice, H.C.; de Malmazet, D.; Schreurs, A.; Frere, S.; Van Molle, I.; Volkov, A.N.; Creemers, E.; Vertkin, I.; Nys, J.; Ranaivoson, F.M.; et al. Secreted amyloid-beta precursor protein functions as a GABABR1a ligand to modulate synaptic transmission. Science 2019, 363, 6423. [Google Scholar] [CrossRef]
- Igarashi, M.; Nozumi, M.; Wu, L.G.; Zanacchi, F.C.; Katona, I.; Barna, L.; Xu, P.; Zhang, M.; Xue, F.; Boyden, E. New observations in neuroscience using superresolution microscopy. J. Neurosci. 2018, 38, 9459–9467. [Google Scholar] [CrossRef]
- Gustafsson, M.G. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 2000, 198, 82–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demmerle, J.; Innocent, C.; North, A.J.; Ball, G.; Muller, M.; Miron, E.; Matsuda, A.; Dobbie, I.M.; Markaki, Y.; Schermelleh, L. Strategic and practical guidelines for successful structured illumination microscopy. Nat. Protoc. 2017, 12, 988–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schurmann, B.; Bermingham, D.P.; Kopeikina, K.J.; Myczek, K.; Yoon, S.; Horan, K.E.; Kelly, C.J.; Martin-de-Saavedra, M.D.; Forrest, M.P.; Fawcett-Patel, J.M.; et al. A novel role for the late-onset Alzheimer’s disease (LOAD)-associated protein Bin1 in regulating postsynaptic trafficking and glutamatergic signaling. Mol. Psychiatry 2020, 25, 2000–2016. [Google Scholar] [CrossRef] [PubMed]
- Crosby, K.C.; Gookin, S.E.; Garcia, J.D.; Hahm, K.M.; Dell’Acqua, M.L.; Smith, K.R. Nanoscale subsynaptic domains underlie the organization of the inhibitory synapse. Cell Rep. 2019, 26, 3284–3297.e3283. [Google Scholar] [CrossRef] [Green Version]
- Hell, S.W.; Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 1994, 19, 780–782. [Google Scholar] [CrossRef]
- Hell, S.W. Far-field optical nanoscopy. Science 2007, 316, 1153–1158. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Gao, Y.; Winblad, B.; Tjernberg, L.O.; Schedin-Weiss, S. A super-resolved view of the Alzheimer’s disease-related amyloidogenic pathway in hippocampal neurons. J. Alzheimers Dis. 2021, 83, 833–852. [Google Scholar] [CrossRef]
- De Rossi, P.; Nomura, T.; Andrew, R.J.; Masse, N.Y.; Sampathkumar, V.; Musial, T.F.; Sudwarts, A.; Recupero, A.J.; Le Metayer, T.; Hansen, M.T.; et al. Neuronal BIN1 Regulates Presynaptic Neurotransmitter Release and Memory Consolidation. Cell Rep. 2020, 30, 3520–3535.e7. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Jans, D.C.; Winblad, B.; Tjernberg, L.O.; Schedin-Weiss, S. Neuronal Aβ42 is enriched in small vesicles at the presynaptic side of synapses. Life Sci. Alliance 2018, 1, e201800028. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Sengupta, P.; Lippincott-Schwartz, J.; Verkhusha, V.V. Photocontrollable fluorescent proteins for superresolution imaging. Annu. Rev. Biophys. 2014, 43, 303–329. [Google Scholar] [CrossRef] [Green Version]
- Schedin-Weiss, S.; Caesar, I.; Winblad, B.; Blom, H.; Tjernberg, L.O. Super-resolution microscopy reveals gamma-secretase at both sides of the neuronal synapse. Acta Neuropathol. Commun. 2016, 4, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanguneri, S.; Pramod, R.T.; Efimova, N.; Das, D.; Jose, M.; Svitkina, T.; Nair, D. Characterization of nanoscale arganization of F-actin in morphologically distinct dendritic spines in vitro using supervised learning. eNeuro 2019, 6. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Le Corronc, H.; Legendre, P.; Triller, A.; Specht, C.G. Differential regulation of glycinergic and GABAergic nanocolumns at mixed inhibitory synapses. EMBO Rep. 2021, 22, e52154. [Google Scholar] [CrossRef]
- Grabner, C.P.; Jansen, I.; Neef, J.; Weiss, T.; Schmidt, R.; Riedel, D.; Wurm, C.A.; Moser, T. Resolving the molecular architecture of the photoreceptor active zone by MINFLUX nanoscopy. bioRxiv 2021. [Google Scholar] [CrossRef]
- Paasila, P.J.; Fok, S.Y.Y.; Flores-Rodriguez, N.; Sajjan, S.; Svahn, A.J.; Dennis, C.V.; Holsinger, R.M.D.; Kril, J.J.; Becker, T.S.; Banati, R.B.; et al. Ground state depletion microscopy as a tool for studying microglia-synapse interactions. J. Neurosci. Res. 2021, 99, 1515–1532. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Weihs, T.; Wurm, C.A.; Jansen, I.; Rehman, J.; Sahl, S.J.; Hell, S.W. MINFLUX nanometer-scale 3D imaging and microsecond-range tracking on a common fluorescence microscope. Nat. Commun. 2021, 12, 1478. [Google Scholar] [CrossRef]
- Manley, S.; Gillette, J.M.; Lippincott-Schwartz, J. Single-particle tracking photoactivated localization microscopy for mapping single-molecule dynamics. Methods Enzym. 2010, 475, 109–120. [Google Scholar] [CrossRef]
- Sauerbeck, A.D.; Gangolli, M.; Reitz, S.J.; Salyards, M.H.; Kim, S.H.; Hemingway, C.; Gratuze, M.; Makkapati, T.; Kerschensteiner, M.; Holtzman, D.M.; et al. SEQUIN multiscale imaging of mammalian central synapses reveals loss of synaptic connectivity resulting from diffuse traumatic brain injury. Neuron 2020, 107, 257–273.e5. [Google Scholar] [CrossRef]
- Dev, S.; Babitt, J.L. Overview of iron metabolism in health and disease. Hemodial. Int. 2017, 21, S6–S20. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J.R.; Ayton, S.; Bush, A.I. Iron and Alzheimer’s disease: An update on emerging mechanisms. J. Alzheimers Dis. 2018, 64, S379–S395. [Google Scholar] [CrossRef]
- Ndayisaba, A.; Kaindlstorfer, C.; Wenning, G.K. Iron in neurodegeneration—Cause or consequence? Front. Neurosci. 2019, 13, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulk, M.; Abdelmoula, W.M.; Nabuurs, R.J.A.; van der Graaf, L.M.; Mulders, C.W.H.; Mulder, A.A.; Jost, C.R.; Koster, A.J.; van Buchem, M.A.; Natte, R.; et al. Postmortem MRI and histology demonstrate differential iron accumulation and cortical myelin organization in early- and late-onset Alzheimer’s disease. Neurobiol. Aging 2018, 62, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Bulk, M.; Van Der Weerd, L.; Breimer, W.; Lebedev, N.; Webb, A.; Goeman, J.J.; Ward, R.J.; Huber, M.; Oosterkamp, T.H.; Bossoni, L. Quantitative comparison of different iron forms in the temporal cortex of Alzheimer patients and control subjects. Sci. Rep. 2018, 8, 6898. [Google Scholar] [CrossRef] [PubMed]
- Yumoto, S.; Nagai, H.; Matsuzaki, H.; Matsumura, H.; Tada, W.; Nagatsuma, E.; Kobayashi, K. Aluminium incorporation into the brain of rat fetuses and sucklings. Brain Res. Bull. 2001, 55, 229–234. [Google Scholar] [CrossRef]
- Madsen, S.J.; DiGiacomo, P.S.; Zeng, Y.; Goubran, M.; Chen, Y.; Rutt, B.K.; Born, D.; Vogel, H.; Sinclair, R.; Zeineh, M.M. Correlative microscopy to localize and characterize iron deposition in Alzheimer’s disease. J. Alzheimers Dis. Rep. 2020, 4, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.T.; Randall, J.D.; Cahill, C.M.; Eder, P.S.; Huang, X.; Gunshin, H.; Leiter, L.; McPhee, J.; Sarang, S.S.; Utsuki, T.; et al. An iron-responsive element type II in the 5′-untranslated region of the Alzheimer’s amyloid precursor protein transcript. J. Biol. Chem. 2002, 277, 45518–45528. [Google Scholar] [CrossRef] [Green Version]
- Boopathi, S.; Kolandaivel, P. Fe2+ binding on amyloid beta-peptide promotes aggregation. Proteins 2016, 84, 1257–1274. [Google Scholar] [CrossRef]
- Duce, J.A.; Tsatsanis, A.; Cater, M.A.; James, S.A.; Robb, E.; Wikhe, K.; Leong, S.L.; Perez, K.; Johanssen, T.; Greenough, M.A.; et al. Iron-export ferroxidase activity of β-amyloid precursor protein is inhibited by zinc in Alzheimer’s disease. Cell 2010, 142, 857–867. [Google Scholar] [CrossRef] [Green Version]
- Everett, J.; Brooks, J.; Lermyte, F.; O’Connor, P.B.; Sadler, P.J.; Dobson, J.; Collingwood, J.F.; Telling, N.D. Iron stored in ferritin is chemically reduced in the presence of aggregating Abeta(1-42). Sci Rep. 2020, 10, 10332. [Google Scholar] [CrossRef]
- Belaidi, A.A.; Bush, A.I. Iron neurochemistry in Alzheimer’s disease and Parkinson’s disease: Targets for therapeutics. J. Neurochem. 2016, 139 (Suppl. S1), 179–197. [Google Scholar] [CrossRef] [Green Version]
- Masaldan, S.; Bush, A.I.; Devos, D.; Rolland, A.S.; Moreau, C. Striking while the iron is hot: Iron metabolism and ferroptosis in neurodegeneration. Free Radic. Biol Med. 2019, 133, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Langkammer, C.; Ropele, S.; Pirpamer, L.; Fazekas, F.; Schmidt, R. MRI for iron mapping in Alzheimer’s disease. Neurodegener. Dis. 2014, 13, 189–191. [Google Scholar] [CrossRef] [PubMed]
- Ropele, S.; Langkammer, C. Iron quantification with susceptibility. NMR Biomed. 2017, 30, e3534. [Google Scholar] [CrossRef]
- Yumoto, S.; Kakimi, S.; Ishikawa, A. Colocalization of aluminum and iron in nuclei of nerve cells in brains of patients with Alzheimer’s disease. J. Alzheimers Dis. 2018, 65, 1267–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lal, A. Iron in health and disease: An update. Indian J. Pediatr. 2020, 87, 58–65. [Google Scholar] [CrossRef]
| Type | Profile | Major Symptoms | Impact on IS | Treatment | Drug Target Site | Reference |
|---|---|---|---|---|---|---|
| Alzheimer’s disease | MRI AβO hTau NFT | Dementia cognitive impairment | Loss of GABAergic neurons | Memantine | NMDAR antagonist—postsynapse of EN | [6,7,20,22,25,26] |
| Parkinson’s disease | MRI α—Synuclein Lewy neurites Lewy bodies | Dementia Bradykinesia Rigidity Rest tremors | Loss of dopaminergic neurons | Levodopa combined with dopamine agonists | Presynaptic nerve terminals | [27,28,29,30,31,32] |
| Multiple Sclerosis | MRI scarring of tissue demyelination oligoclonal bands Neurofilaments | cognitive impairment defects in vision muscle spasms fatigue | loss of motor neurons loss of selective inhibitory neurons | Immunosuppressants Cytokines | Myelin sheath Axon fibers | [27,33,34,35,36,37] |
| Amyotrophic Lateral Sclerosis | Neurofilaments | cognitive impairment frontotemporal dementia muscle spasms and atrophy | loss of inhibitory cortical interneurons | Riluzole Baclofen | blocks NMDAR–postsynapse inhibits glutamate release—pre-synapse GABABR agonist–postsynapse | [28,29,30,31,32,38,39,40] |
| Huntington’s disease | MRI mHTT protein Neurofilament light protein | cognitive impairment dementia chorea | loss of GABAAR | Tetrabenazine Antipsychotics | inhibits VMAT-2—presynapse | [41,42,43,44,45] |
| SIM | STED | PALM/STORM | |
|---|---|---|---|
| Resolution | x/y = 60–140 nm z = 120–250 nm | x/y = 2–40 nm z > 4300 nm MINFLUX has 2 nm resolution; others have resolution of 20–80 nm; very much dependent on the device | x/y = 1–40 nm z = 20–50 nm strongly dependent on chemical method of on/off switching |
| Live imaging | 240 fr/s Lattice SIM, Zeiss | 10–20 fr/s depending on area | 0.2 fr/s |
| Laser power | 1–10 W/cm2 | 100 MW/cm2 | 1–25 kW/cm2 |
| Colours | 4 | Max 2 | 2–4 |
| Dyes | typical fluorescent dyes | Atto647N, Chromeo 494 | AF647, mEos2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radulović, S.; Sunkara, S.; Maurer, C.; Leitinger, G. Digging Deeper: Advancements in Visualization of Inhibitory Synapses in Neurodegenerative Disorders. Int. J. Mol. Sci. 2021, 22, 12470. https://doi.org/10.3390/ijms222212470
Radulović S, Sunkara S, Maurer C, Leitinger G. Digging Deeper: Advancements in Visualization of Inhibitory Synapses in Neurodegenerative Disorders. International Journal of Molecular Sciences. 2021; 22(22):12470. https://doi.org/10.3390/ijms222212470
Chicago/Turabian StyleRadulović, Snježana, Sowmya Sunkara, Christa Maurer, and Gerd Leitinger. 2021. "Digging Deeper: Advancements in Visualization of Inhibitory Synapses in Neurodegenerative Disorders" International Journal of Molecular Sciences 22, no. 22: 12470. https://doi.org/10.3390/ijms222212470
APA StyleRadulović, S., Sunkara, S., Maurer, C., & Leitinger, G. (2021). Digging Deeper: Advancements in Visualization of Inhibitory Synapses in Neurodegenerative Disorders. International Journal of Molecular Sciences, 22(22), 12470. https://doi.org/10.3390/ijms222212470







