Genetic Biomarkers in Chronic Myeloid Leukemia: What Have We Learned So Far?
Abstract
1. Introduction
2. Variations of the Philadelphia Chromosome
2.1. Ph-Positive Karyotype in CML
2.2. Double Ph-Positive Karyotype in CML
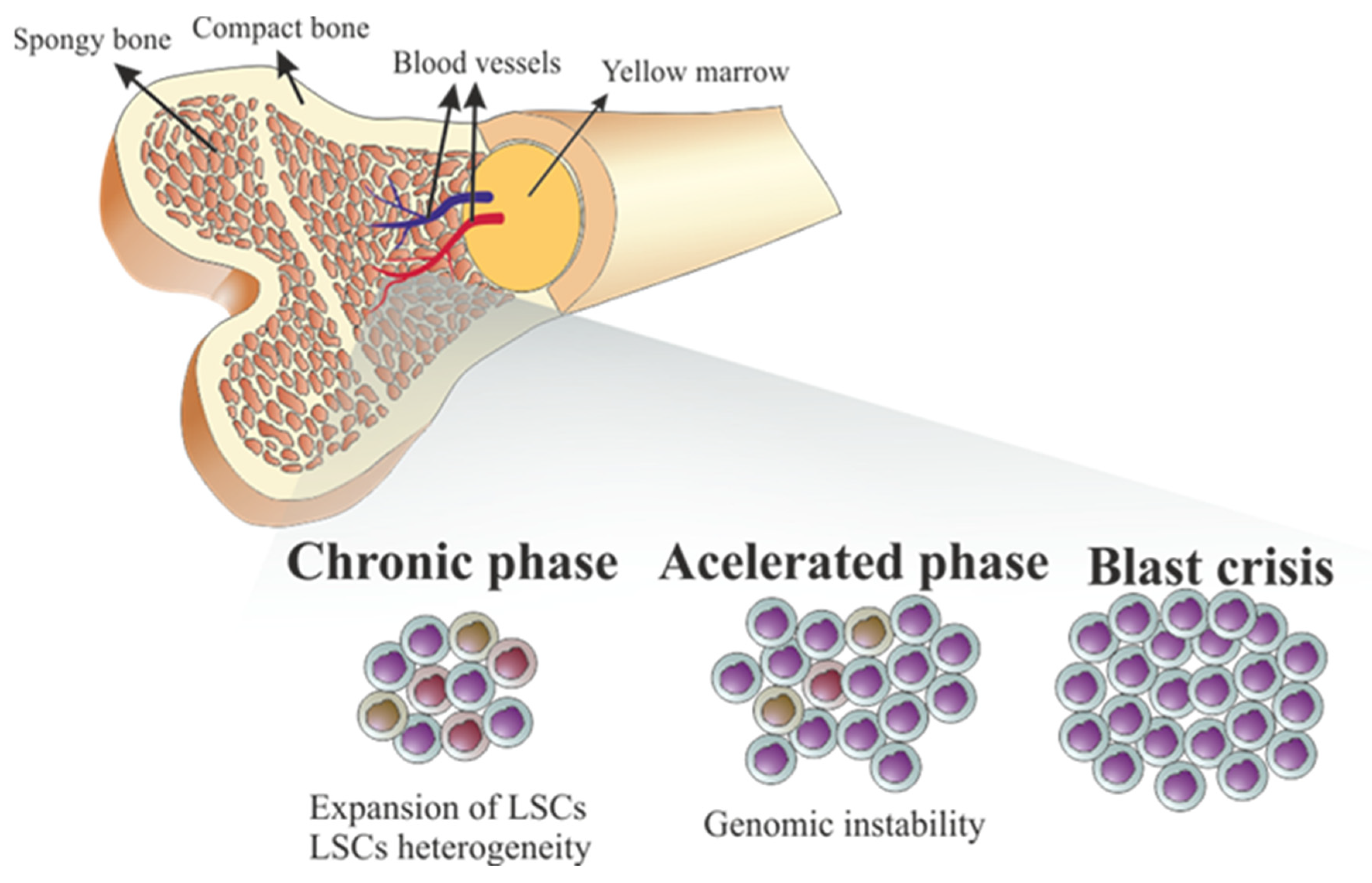
3. Genomic Instability in CML
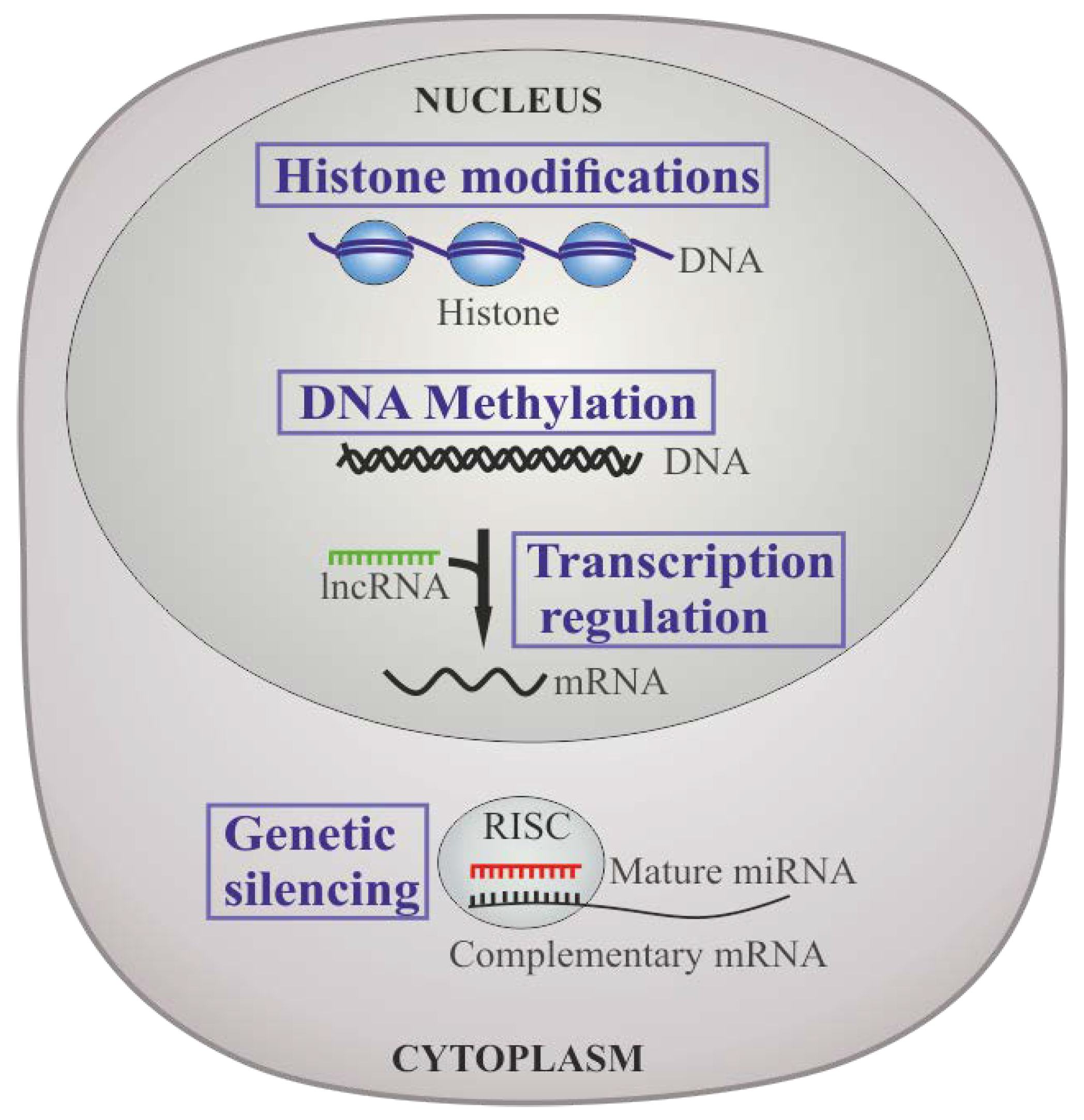
4. Epigenetic Regulation in CML
5. miRNAs Dysregulation in CML
5.1. miRNAs That Target BCR-ABL1
5.2. miRNA-21
5.3. miRNAs Role in Resistance to TKIs and Blast Crisis Progression
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2020 Update on diagnosis, therapy and monitoring. Am. J. Hematol. 2020, 95, 691–709. [Google Scholar] [CrossRef]
- Society, American Cancer. Cancer Facts & Figures 2019; American Cancer Society: Atlanta, GA, USA, 2019. [Google Scholar]
- Hochhaus, A.; Saussele, S.; Rosti, G.; Mahon, F.-X.; Janssen, J.J.W.M.; Hjorth-Hansen, H.; Richter, J.; Buske, C. Chronic myeloid leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017, 28, iv41–iv51. [Google Scholar] [CrossRef]
- Hijiya, N.; Schultz, K.R.; Metzler, M.; Millot, F.; Suttorp, M. Pediatric chronic myeloid leukemia is a unique disease that requires a different approach. Blood 2016, 127, 392–399. [Google Scholar] [CrossRef]
- Nowell, P.C.; Hungerford, D.A. A minute chromosome in human chronic granulocytic leukemia. Science 1960, 132, 1497. [Google Scholar]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2018 Update on diagnosis, therapy and monitoring. Am. J. Hematol. 2018, 93, 442–459. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.J.; Liu, Y.F.; Xu, L.Z.; Long, Z.J.; Huang, D.; Yang, Y.; Liu, B.; Feng, J.X.; Pan, Y.J.; Yan, J.S.; et al. The philadelphia chromosome in leukemogenesis. Chin. J. Cancer 2016, 35, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kaleem, B.; Shahab, S.; Ahmed, N.; Shamsi, T.S. Chronic myeloid leukemia–Prognostic value of mutations. Asian Pac. J. Cancer Prev. 2015, 16, 7415–7423. [Google Scholar] [CrossRef]
- Flis, S.; Chojnacki, T. Chronic myelogenous leukemia, a still unsolved problem: Pitfalls and new therapeutic possibilities. Drug Des. Devel. Ther. 2019, 13, 825–843. [Google Scholar] [CrossRef]
- Ayatollahi, H.; Keramati, M.R.; Shirdel, A.; Kooshyar, M.M.; Raiszadeh, M.; Shakeri, S.; Sadeghian, M.H. BCR-ABL fusion genes and laboratory findings in patients with chronic myeloid leukemia in northeast Iran. Casp. J. Intern. Med. 2018, 9, 65–70. [Google Scholar] [CrossRef]
- Avelino, K.Y.P.S.; Silva, R.R.; da Silva Junior, A.G.; Oliveira, M.D.L.; Andrade, C.A.S. Smart applications of bionanosensors for BCR/ABL fusion gene detection in leukemia. J. King Saud. Univ. Sci. 2017, 29, 413–423. [Google Scholar] [CrossRef]
- Pane, F.; Frigeri, F.; Sindona, M.; Luciano, L.; Ferrara, F.; Cimino, R.; Meloni, G.; Saglio, G.; Salvatore, F.; Rotoli, B. Neutrophilic-chronic myeloid leukemia: A distinct disease with a specific molecular marker (BCR/ABL with C3/A2 junction). Blood 1996, 88, 2410–2414. [Google Scholar] [CrossRef] [PubMed]
- Quintás-Cardama, A.; Cortes, J.E. Chronic myeloid leukemia: Diagnosis and treatment. Mayo Clin. Proc. 2006, 81, 973–988. [Google Scholar] [CrossRef]
- Marley, S.B.; Gordon, M.Y. Chronic myeloid leukaemia: Stem cell derived but progenitor cell driven. Clin. Sci. 2005, 109, 13–25. [Google Scholar] [CrossRef]
- Faderl, S.; Talpaz, M.; Estrov, Z.; O’Brien, S.; Kurzrock, R.; Kantarjian, H.M. The biology of chronic myeloid leukemia. New Engl. J. Med. 1999, 341, 164–172. [Google Scholar] [CrossRef]
- Perrotti, D.; Jamieson, C.; Goldman, J.; Skorski, T. Chronic myeloid leukemia: Mechanisms of blastic transformation. J. Clin. Investig. 2010, 120, 2254–2264. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.J.; Holyoake, T.L. Preclinical approaches in chronic myeloid leukemia: From cells to systems. Exp. Hematol. 2017, 47, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Minciacchi, V.R.; Kumar, R.; Krause, D.S. Chronic myeloid leukemia: A model disease of the past, present and future. Cells 2021, 10, 117. [Google Scholar] [CrossRef]
- Popp, H.D.; Kohl, V.; Naumann, N.; Flach, J.; Brendel, S.; Kleiner, H.; Weiss, C.; Seifarth, W.; Saussele, S.; Hofmann, W.K.; et al. DNA damage and dna damage response in chronic myeloid leukemia. Int. J. Mol. Sci. 2020, 21, 1177. [Google Scholar] [CrossRef]
- Hochhaus, A.; O’Brien, S.G.; Guilhot, F.; Druker, B.J.; Branford, S.; Foroni, L.; Goldman, J.M.; Müller, M.C.; Radich, J.P.; Rudoltz, M.; et al. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia 2009, 23, 1054–1061. [Google Scholar] [CrossRef]
- Druker, B.J.; Guilhot, F.; O’Brien, S.G.; Gathmann, I.; Kantarjian, H.; Gattermann, N.; Deininger, M.W.N.; Silver, R.T.; Goldman, J.M.; Stone, R.M.; et al. Five-Year Follow-up of Patients Receiving Imatinib for Chronic Myeloid Leukemia. N. Engl. J. Med. 2006, 355, 2408–2417. [Google Scholar] [CrossRef]
- Sasaki, K.; Strom, S.S.; O’brien, S.; Jabbour, E.; Ravandi, F.; Konopleva, M.; Borthakur, G.; Pemmaraju, N.; Daver, N.; Jain, P.; et al. Prospective Analysis: Relative Survival in Patients with Chronic Myeloid Leukemia in Chronic Phase in the Era of Tyrosine Kinase Inhibitors. Lancet Haematol. 2015, 2, e186–e193. [Google Scholar] [CrossRef]
- Hehlmann, R.; Lauseker, M.; Saußele, S.; Pfirrmann, M.; Krause, S.; Kolb, H.J.; Neubauer, A.; Hossfeld, D.K.; Nerl, C.; Gratwohl, A.; et al. Assessment of imatinib as first-line treatment of chronic myeloid leukemia: 10-year survival results of the randomized CML study IV and impact of non-CML determinants. Leukemia 2017, 31, 2398–2406. [Google Scholar] [CrossRef] [PubMed]
- Thielen, N.; Visser, O.; Ossenkoppele, G.; Janssen, J. Chronic myeloid leukemia in the Netherlands: A population-based study on incidence, treatment, and survival in 3585 patients from 1989 to 2012. Eur. J. Haematol. 2016, 97, 145–154. [Google Scholar] [CrossRef]
- Bower, H.; Björkholm, M.; Dickman, P.W.; Höglund, M.; Lambert, P.C.; Andersson, T.M.L. Life expectancy of patients with chronic myeloid leukemia approaches the life expectancy of the general population. J. Clin. Oncol. 2016, 34, 2851–2857. [Google Scholar] [CrossRef]
- Welch, H.G.; Kramer, B.S.; Black, W.C. Epidemiologic Signatures in Cancer. N. Engl. J. Med. 2019, 381, 1378–1386. [Google Scholar] [CrossRef] [PubMed]
- Hochhaus, A.; Baccarani, M.; Silver, R.T.; Schiffer, C.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Deininger, M.W.; Guilhot, F.; et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 2020, 34, 966–984. [Google Scholar] [CrossRef]
- Kalmanti, L.; Saussele, S.; Lauseker, M.; Müller, M.C.; Dietz, C.T.; Heinrich, L.; Hanfstein, B.; Proetel, U.; Fabarius, A.; Krause, S.W.; et al. Safety and efficacy of imatinib in CML over a period of 10 years: Data from the randomized CML-study IV. Leukemia 2015, 29, 1123–1132. [Google Scholar] [CrossRef]
- Steegmann, J.L.; Baccarani, M.; Breccia, M.; Casado, L.F.; García-Gutiérrez, V.; Hochhaus, A.; Kim, D.W.; Kim, T.D.; Khoury, H.J.; Le Coutre, P.; et al. European LeukemiaNet recommendations for the management and avoidance of adverse events of treatment in chronic myeloid leukaemia. Leukemia 2016, 30, 1648–1671. [Google Scholar] [CrossRef]
- Cross, N.C.P.; White, H.E.; Müller, M.C.; Saglio, G.; Hochhaus, A. Standardized definitions of molecular response in chronic myeloid leukemia. Leukemia 2012, 26, 2172–2175. [Google Scholar] [CrossRef]
- Hehlmann, R.; Müller, M.C.; Lauseker, M.; Hanfstein, B.; Fabarius, A.; Schreiber, A.; Proetel, U.; Pletsch, N.; Pfirrmann, M.; Haferlach, C.; et al. Deep molecular response is reached by the majority of patients treated with imatinib, predicts survival, and is achieved more quickly by optimized high-dose imatinib: Results from the randomized CML-Study IV. J. Clin. Oncol. 2014, 32, 415–423. [Google Scholar] [CrossRef]
- Mahon, F.X.; Réa, D.; Guilhot, J.; Guilhot, F.; Huguet, F.; Nicolini, F.; Legros, L.; Charbonnier, A.; Guerci, A.; Varet, B.; et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: The prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010, 11, 1029–1035. [Google Scholar] [CrossRef]
- Saussele, S.; Richter, J.; Guilhot, J.; Gruber, F.X.; Hjorth-Hansen, H.; Almeida, A.; Janssen, J.J.W.M.; Mayer, J.; Koskenvesa, P.; Panayiotidis, P.; et al. Discontinuation of tyrosine kinase inhibitor therapy in chronic myeloid leukaemia (EURO-SKI): A prespecified interim analysis of a prospective, multicentre, non-randomised, trial. Lancet Oncol. 2018, 19, 747–757. [Google Scholar] [CrossRef]
- Cortes, J.; Kantarjian, H. Chronic myeloid leukemia: Sequencing of TKI therapies. Hematology 2016, 2016, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Gorre, M.E.; Mohammed, M.; Ellwood, K.; Hsu, N.; Paquette, R.; Rao, P.N.; Sawyers, C.L. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science 2001, 293, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.P.; Rousselot, P.; Schiffer, C.; Rea, D.; Cortes, J.E.; Milone, J.; Mohamed, H.; Healey, D.; Kantarjian, H.; Hochhaus, A.; et al. Dasatinib in imatinib-resistant or -intolerant chronic-phase, chronic myeloid leukemia patients: 7-Year follow-up of study CA180-034. Am. J. Hematol. 2016, 91, 869–874. [Google Scholar] [CrossRef]
- Giles, F.J.; Le Coutre, P.D.; Pinilla-Ibarz, J.; Larson, R.A.; Gattermann, N.; Ottmann, O.G.; Hochhaus, A.; Radich, J.P.; Saglio, G.; Hughes, T.P.; et al. Nilotinib in imatinib-resistant or imatinib-intolerant patients with chronic myeloid leukemia in chronic phase: 48-month follow-up results of a phase II study. Leukemia 2013, 27, 107–112. [Google Scholar] [CrossRef]
- O’Hare, T.; Shakespeare, W.C.; Zhu, X.; Eide, C.A.; Rivera, V.M.; Wang, F.; Adrian, L.T.; Zhou, T.; Huang, W.S.; Xu, Q.; et al. AP24534, a Pan-BCR-ABL Inhibitor for Chronic Myeloid Leukemia, Potently Inhibits the T315I Mutant and Overcomes Mutation-Based Resistance. Cancer Cell 2009, 16, 401–412. [Google Scholar] [CrossRef]
- Miller, G.D.; Bruno, B.J.; Lim, C.S. Resistant mutations in CML and Ph+ALL—Role of ponatinib. Biol. Ther. 2014, 8, 243–254. [Google Scholar] [CrossRef]
- Wylie, A.A.; Schoepfer, J.; Jahnke, W.; Cowan-Jacob, S.W.; Loo, A.; Furet, P.; Marzinzik, A.L.; Pelle, X.; Donovan, J.; Zhu, W.; et al. The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature 2017, 543, 733–737. [Google Scholar] [CrossRef]
- Korski, T. BCR/ABL, DNA damage and DNA repair: Implications for new treatment concepts. Leuk. Lymphoma 2008, 49, 610–614. [Google Scholar] [CrossRef]
- Radich, J.P.; Dai, H.; Mao, M.; Oehler, V.; Schelter, J.; Druker, B.; Sawyers, C.; Shah, N.; Stock, W.; Willman, C.L.; et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc. Natl. Acad. Sci. USA 2006, 103, 2794–2799. [Google Scholar] [CrossRef] [PubMed]
- Druker, B.J.; Tamura, S.; Buchdunger, E.; Ohno, S.; Segal, G.M.; Fanning, S.; Zimmermann, J.; Lydon, N.B. Effects of a selective inhibitor of the Ab1 tyrosine kinase on the growth of Bcr-Ab1 positive cells. Nat. Med. 1996, 2, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Giustacchini, A.; Thongjuea, S.; Barkas, N.; Woll, P.S.; Povinelli, B.J.; Booth, C.A.G.; Sopp, P.; Norfo, R.; Rodriguez-Meira, A.; Ashley, N.; et al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat. Med. 2017, 23, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Lakkireddy, S.; Aula, S.; Kapley, A.; Swamy, A.V.N.; Digumarti, R.R.; Kutala, V.K.; Jamil, K. Association of Vascular Endothelial Growth Factor A (VEGFA) and its Receptor (VEGFR2) Gene Polymorphisms with Risk of Chronic Myeloid Leukemia and Influence on Clinical Outcome. Mol. Diagn. Ther. 2016, 20, 33–44. [Google Scholar] [CrossRef]
- Deregowska, A.; Pepek, M.; Pruszczyk, K.; Machnicki, M.M.; Wnuk, M.; Stoklosa, T. Differential regulation of telomeric complex by bcr-abl1 kinase in human cellular models of chronic myeloid leukemia—from single cell analysis to next-generation sequencing. Genes 2020, 11, 1145. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, Y.P.; Cao, H.; Chen, Q.; Zhang, X. Integrated computational biology analysis to evaluate target genes for chronic myelogenous leukemia. Mol. Med. Rep. 2018, 18, 1766–1772. [Google Scholar] [CrossRef]
- Bergamaschi, G.; Merante, S.; Orlandi, E.; Galli, A.; Bernasconi, P.; Cazzola, M. TP53 codon 72 polymorphism in patients with chronic myeloid leukemia. Haematologica 2004, 89, 868–869. [Google Scholar]
- Camelo-Santos, J.; do Prado Barbosa, A.; de Paula Silveira-Lacerda, E.; Guillo, L.A. Arginine homozygosity in codon 72 of p53 correlates with failure to imatinib response in chronic myeloid leukemia. Biomed. Pharm. Pharm. 2013, 67, 103–107. [Google Scholar] [CrossRef]
- Weich, N.; Ferri, C.; Moiraghi, B.; Bengió, R.; Giere, I.; Pavlovsky, C.; Larripa, I.; Fundia, A. TP53 codon 72 polymorphism predicts chronic myeloid leukemia susceptibility and treatment outcome. Blood Cells Mol. Dis. 2016, 59, 129–133. [Google Scholar] [CrossRef]
- Koschmieder, S.; Vetrie, D. Epigenetic dysregulation in chronic myeloid leukaemia: A myriad of mechanisms and therapeutic options. Semin. Cancer Biol. 2018, 51, 180–197. [Google Scholar] [CrossRef]
- MacHova Polakova, K.; Koblihova, J.; Stopka, T. Role of epigenetics in chronic myeloid leukemia. Curr. Hematol. Malig. Rep. 2013, 8, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Marzocchi, G.; Castagnetti, F.; Luatti, S.; Baldazzi, C.; Stacchini, M.; Gugliotta, G.; Amabile, M.; Specchia, G.; Sessarego, M.; Giussani, U.; et al. Variant Philadelphia translocations: Molecular-cytogenetic characterization and prognostic influence on frontline imatinib therapy, a GIMEMA working party on CML analysis. Blood 2011, 117, 6793–6800. [Google Scholar] [CrossRef] [PubMed]
- Huret, J.L. Complex translocations, simple variant translocations and Ph-negative cases in chronic myelogenous leukaemia. Hum. Genet. 1990, 85, 565–568. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.M.; Strike, P.; Scott, C.; Moorman, A.V. Breakpoints of variant 9;22 translocations in chronic myeloid leukemia locate preferentially in the CG-richest regions of the genome. Genes Chromosom. Cancer 2005, 43, 383–389. [Google Scholar] [CrossRef]
- Baccarani, M.; Cortes, J.; Pane, F.; Niederwieser, D.; Saglio, G.; Apperley, J.; Cervantes, F.; Deininger, M.; Gratwohl, A.; Guilhot, F.; et al. Chronic myeloid leukemia: An update of concepts and management recommendations of European LeukemiaNet. J. Clin. Oncol. 2009, 27, 6041–6051. [Google Scholar] [CrossRef]
- Bumm, T.; Müller, C.; Al-Ali, H.K.; Krohn, K.; Shepherd, P.; Schmidt, E.; Leiblein, S.; Franke, C.; Hennig, E.; Friedrich, T.; et al. Emergence of clonal cytogenetic abnormalities in Ph− cells in some CML patients in cytogenetic remission to imatinib but restoration of polyclonal hematopoiesis in the majority. Blood 2003, 101, 1941–1949. [Google Scholar] [CrossRef]
- Jabbour, E.; Kantarjian, H.M.; Abruzzo, L.V.; O’Brien, S.; Garcia-Manero, G.; Verstovsek, S.; Shan, J.; Rios, M.B.; Cortes, J. Chromosomal abnormalities in Philadelphia chromosome-negative metaphases appearing during imatinib mesylate therapy in patients with newly diagnosed chronic myeloid leukemia in chronic phase. Blood 2007, 110, 2991–2995. [Google Scholar] [CrossRef]
- Koshiyama, D.B.; Capra, M.E.Z.; Paskulin, G.A.; Rosa, R.F.M.; Oliveira, C.A.V.; Vanelli, T.; Fogliatto, L.M.; Zen, P.R.G. Cytogenetic response to imatinib treatment in Southern Brazilian patients with chronic myelogenous leukemia and variant Philadelphia chromosome. Ann. Hematol. 2013, 92, 185–189. [Google Scholar] [CrossRef]
- El-Zimaity, M.M.T.; Kantarjian, H.; Talpaz, M.; O’Brien, S.; Giles, F.; Garcia-Manero, G.; Verstovsek, S.; Thomas, D.; Ferrajoli, A.; Hayes, K.; et al. Results of imatinib mesylate therapy in chronic myelogenous leukaemia with variant Philadelphia chromosome. Br. J. Haematol. 2004, 125, 187–195. [Google Scholar] [CrossRef]
- Valencia, A.; Cervera, J.; Such, E.; Barragán, E.; Bolufer, P.; Fuster, O.; Collado, R.; Martínez, J.; Sanz, M.A. Complex variant t(9;22) chromosome translocations in five cases of chronic myeloid leukemia. Adv. Hematol. 2009, 2009, 187125. [Google Scholar] [CrossRef]
- Fabarius, A.; Leitner, A.; Hochhaus, A.; Müller, M.C.; Hanfstein, B.; Haferlach, C.; Göhring, G.; Schlegelberger, B.; Jotterand, M.; Reiter, A.; et al. Impact of additional cytogenetic aberrations at diagnosis on prognosis of CML: Long-term observation of 1151 patients from the randomized CML Study IV. Blood 2011, 118, 6760–6768. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, H.H.; Liu, Y.C.; Tsai, H.J.; Hsu, J.F.; Yang, W.C.; Chang, C.S.; Lin, S.F. Additional chromosome abnormalities in chronic myeloid leukemia. Kaohsiung J. Med. Sci. 2011, 27, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Stagno, F.; Vigneri, P.; Fabro, V. Del Stella, S.; Cupri, A.; Massimino, M.; Consoli, C.; Tambè, L.; Consoli, M.L.; Antolino, A.; et al. Influence of complex variant chromosomal translocations in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors. Acta Oncol. (Madrid) 2010, 49, 506–508. [Google Scholar] [CrossRef] [PubMed]
- Gorusu, M.; Benn, P.; Li, Z.; Fang, M. On the genesis and prognosis of variant translocations in chronic myeloid leukemia. Cancer Genet. Cytogenet. 2007, 173, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Choi, S.Y.; Bang, J.H.; Kim, S.H.; Jang, E.J.; Byeun, J.Y.; Park, J.E.; Jeon, H.R.; Oh, Y.J.; Kim, M.; et al. The long-term clinical implications of clonal chromosomal abnormalities in newly diagnosed chronic phase chronic myeloid leukemia patients treated with imatinib mesylate. Cancer Genet. 2012, 205, 563–571. [Google Scholar] [CrossRef]
- Vinhas, R.; Lourenço, A.; Santos, S.; Ribeiro, P.; Silva, M.; De Sousa, A.B.; Baptista, P.V.; Fernandes, A.R. A double Philadelphia chromosome-positive chronic myeloid leukemia patient, co-expressing P210(BCR-ABL1) and P195(BCR-ABL1) isoforms. Haematologica 2018, 103, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Onida, F.; Ball, G.; Kantarjian, H.M.; Smith, T.L.; Glassman, A.; Albitar, M.; Scappini, B.; Rios, M.B.; Keating, M.J.; Beran, M. Characteristics and outcome of patients with Philadelphia chromosome negative, bcr/abl negative chronic myelogenous leukemia. Cancer 2002, 95, 1673–1684. [Google Scholar] [CrossRef]
- Lichty, B.D.; Keating, A.; Callum, J.; Yee, K.; Croxford, R.; Corpus, G.; Nwachukwu, B.; Kim, P.; Guo, J.; Kamel-Reid, S. Expression of p210 and p190 BCR-ABL due to alternative splicing in chronic myelogenous leukaemia. Br. J. Haematol. 1998, 103, 711–715. [Google Scholar] [CrossRef]
- Arana-Trejo, R.M.; RuízSánchez, E.; Ignacio-Ibarra, G.; De La BáezFuente, E.; Garces, O.; GómezMorales, E.; Castro Granados, M.; OvillaMartínez, R.; Rubio-Borja, M.E.; SolísAnaya, L.; et al. BCR/ABL p210, p190 and p230 fusion genes in 250 Mexican patients with chronic myeloid leukaemia (CML). Clin. Lab. Haematol. 2002, 24, 145–150. [Google Scholar] [CrossRef]
- Branford, S.; Kim, D.D.H.; Apperley, J.F.; Eide, C.A.; Mustjoki, S.; Ong, S.T.; Nteliopoulos, G.; Ernst, T.; Chuah, C.; Gambacorti-Passerini, C.; et al. Laying the foundation for genomically-based risk assessment in chronic myeloid leukemia. Leukemia 2019, 33, 1835–1850. [Google Scholar] [CrossRef]
- Otero, L.; Ornellas, M.H.; Dobbin, J.; De Souza Fernandez, T. Double Philadelphia-chromosome: A resistance factor on the imatinib mesylate therapy for chronic myeloid leukemia. Int. J. Lab. Hematol. 2008, 30, 346–348. [Google Scholar] [CrossRef]
- Langabeer, S.E.; Crampe, M.; Kelly, J.; Fadalla, K.; Connaghan, G.; Conneally, E. Nilotinib and allogeneic stem cell transplantation in a chronic myeloid leukemia patient with e6a2 and e1a2 BCR-ABL transcripts. Leuk. Res. 2010, 34, 204–205. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, C.H.M.; Weissman, I.L.; Passegué, E. Chronic versus acute myelogenous leukemia: A question of self-renewal. Cancer Cell 2004, 6, 531–533. [Google Scholar] [CrossRef]
- Ohyashiki, K.; Iwama, H.; Tauchi, T.; Shimamoto, T.; Hayashi, S.; Ando, K.; Kawakubo, K.; Ohyashiki, J.H. Telomere dynamics and genetic instability in disease progression of chronic myeloid leukemia. Leuk. Lymphoma 2000, 40, 49–56. [Google Scholar] [CrossRef]
- Loscocco, F.; Visani, G.; Galimberti, S.; Curti, A.; Isidori, A. BCR-ABL independent mechanisms of resistance in chronic myeloid leukemia. Front. Oncol. 2019, 9, 939. [Google Scholar] [CrossRef] [PubMed]
- Saikia, T. The Cure of Chronic Myeloid Leukemia: Are We There Yet? Curr. Oncol. Rep. 2018, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Skorski, T. Chronic myeloid leukemia cells refractory/resistant to tyrosine kinase inhibitors are genetically unstable and may cause relapse and malignant progression to the terminal diesease state. Leuk. Lymphoma 2011, 52 (Suppl. 1), 23–29. [Google Scholar] [CrossRef]
- Nieborowska-Skorska, M.; Kopinski, P.K.; Ray, R.; Hoser, G.; Ngaba, D.; Flis, S.; Cramer, K.; Reddy, M.M.; Koptyra, M.; Penserga, T.; et al. Rac2-MRC-cIII-generated ROS cause genomic instability in chronic myeloid leukemia stem cells and primitive progenitors. Blood 2012, 119, 4253–4263. [Google Scholar] [CrossRef]
- Pawlowska, E.; Blasiak, J. DNA repair—A double-edged sword in the genomic stability of cancer cells—The case of chronic myeloid leukemia. Int. J. Mol. Sci. 2015, 16, 27535–27549. [Google Scholar] [CrossRef]
- Nowicki, M.O.; Falinski, R.; Koptyra, M.; Slupianek, A.; Stoklosa, T.; Gloc, E.; Nieborowska-Skorska, M.; Blasiak, J.; Skorski, T. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood 2004, 104, 3746–3753. [Google Scholar] [CrossRef]
- Koptyra, M.; Falinski, R.; Nowicki, M.O.; Stoklosa, T.; Majsterek, I.; Nieborowska-Skorska, M.; Blasiak, J.; Skorski, T. BCR/ABL kinase induces self-mutagenesis via reactive oxygen species to encode imatinib resistance. Blood 2006, 108, 319–327. [Google Scholar] [CrossRef]
- Raynaud, C.M.; Sabatier, L.; Philipot, O.; Olaussen, K.A.; Soria, J.C. Telomere length, telomeric proteins and genomic instability during the multistep carcinogenic process. Crit. Rev. Oncol. Hematol. 2008, 66, 99–117. [Google Scholar] [CrossRef]
- Samassekou, O.; Hebert, J.; Mai, S.; Yan, J. Nuclear remodeling of telomeres in chronic myeloid leukemia. Genes Chromosomes Cancer 2013, 52, 495–502. [Google Scholar] [CrossRef]
- Bennour, A.; Saad, A.; Sennana, H. Chronic myeloid leukemia: Relevance of cytogenetic and molecular assays. Crit. Rev. Oncol. Hematol. 2016, 97, 263–274. [Google Scholar] [CrossRef]
- Cannella, L.; Loglisci, G.; Nanni, M.; De Cuia, M.R.; Colafigli, G.; Salaroli, A.; Serrao, A.; Alimena, G.; Breccia, M. Trisomy 8 in Philadelphia chromosome negative cell preceding the evolution of a Philadelphia chromosome positive clone with the same additional change during imatinib treatment: Revisiting the role of genetic instability in chronic myeloid leukemia. Leuk. Lymphoma 2012, 53, 497–498. [Google Scholar] [CrossRef] [PubMed]
- Dkhissi, F.; Aggoune, D.; Pontis, J.; Sorel, N.; Piccirilli, N.; LeCorf, A.; Guilhot, F.; Chomel, J.C.; Ait-Si-Ali, S.; Turhan, A.G. The downregulation of BAP1 expression by BCR-ABL reduces the stability of BRCA1 in chronic myeloid leukemia. Exp. Hematol. 2015, 43, 775–780. [Google Scholar] [CrossRef]
- Villuendas, R.; Steegmann, J.L.; Pollán, M.; Tracey, L.; Granda, A.; Fernández-Ruiz, E.; Casado, L.F.; Martínez, J.; Martínez, P.; Lombardía, L.; et al. Identification of genes involved in imatinib resistance in CML: A gene-expression profiling approach. Leukemia 2006, 20, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Sankar, M.; Tanaka, K.; Kumaravel, T.S.; Arif, M.; Shintani, T.; Yagi, S.; Kyo, T.; Dohy, H.; Kamada, N. Identification of a commonly deleted region at 17p13.3 in leukemia and lymphoma associated with 17p abnormality. Leukemia 1998, 12, 510–516. [Google Scholar] [CrossRef]
- Peller, S.; Yona, R.; Kopilova, Y.; Prokocimer, M.; Goldfinger, N.; Uysal, A.; Karabulut, H.G.; Tukun, A.; Bokesoy, I.; Tuncman, G.; et al. Molecular alterations in the TP53 gene of peripheral blood cells of patients with chronic myeloid leukemia. Genes Chromosom. Cancer 1998, 21, 2–7. [Google Scholar] [CrossRef]
- Elkhouly, E.A.; Khalifa, K.A.; Radwan, W.M.; Younis, Y.A. 17PTumor suppressor P53 gene codon 72 polymorphism and imatinib cytogenetic response in chronic myeloid leukemia. Ann. Oncol. 2018, 29, vi6. [Google Scholar] [CrossRef]
- Kokate, P.; Dalvi, R.; Mandava, S. A complex three-way translocation with deletion of the TP53 gene in a blast crisis chronic myeloid leukemia patient. J. Cancer Res. Ther. 2015, 11, 1239–1241. [Google Scholar] [CrossRef]
- Sill, H.; Goldman, J.M.; Cross, N.C.P. Homozygous deletions of the p16 tumor-suppressor gene are associated with lymphoid transformation of chronic myeloid leukemia. Blood 1995, 85, 2013–2016. [Google Scholar] [CrossRef] [PubMed]
- Dierov, J.; Dierova, R.; Carroll, M. BCR/ABL translocates to the nucleus and disrupts an ATR-dependent intra-S phase checkpoint. Cancer Cell 2004, 5, 275–285. [Google Scholar] [CrossRef]
- Rumpold, H.; Webersinke, G. Molecular Pathogenesis of Philadelphia-Positive Chronic Myeloid Leukemia–is it all BCR-ABL? Curr. Cancer Drug Targets 2011, 11, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Mead, A.J. Single cell genomics in chronic myeloid leukemia. HemaSphere 2018, 2, 54–55. [Google Scholar] [CrossRef]
- Fu, S.; Hu, Y.; Fu, Y.; Chen, F.; Liu, X.; Zhang, M.; Wang, X.; Tu, S.; Zhang, J. Novel BCR-ABL1 fusion and leukemic mutations of SETBP1, PAX5, and TP53 detected by next generation sequencing in chronic myeloid leukemia. Cancer Biol. Ther. 2016, 17, 1003–1009. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zilfou, J.T.; Lowe, S.W. Tumor suppressive functions of p53. Cold Spring Harb. Perspect. Biol. 2009, 1, a001883. [Google Scholar] [CrossRef]
- Aubrey, B.J.; Strasser, A.; Kelly, G.L. Tumor-Suppressor Functions of the TP53 Pathway. Cold Spring Harb. Perspect. Med. 2016, 6, a026062. [Google Scholar] [CrossRef]
- Leroy, B.; Anderson, M.; Soussi, T. TP53 mutations in human cancer: Database reassessment and prospects for the next decade. Hum. Mutat. 2014, 35, 672–688. [Google Scholar] [CrossRef]
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019, 26, 199–212. [Google Scholar] [CrossRef]
- Sailaja, K.; Rao, V.R.; Yadav, S.; Rajasekhar Reddy, R.; Surekha, D.; Nageswara Rao, D.; Raghunadharao, D.; Vishnupriya, S. Intronic SNPs of TP53 gene in chronic myeloid leukemia: Impact on drug response. J. Nat. Sci. Biol. Med. 2012, 3, 182–185. [Google Scholar] [CrossRef]
- Abraham, S.A.; Hopcroft, L.E.M.; Carrick, E.; Drotar, M.E.; Dunn, K.; Williamson, A.J.K.; Korfi, K.; Baquero, P.; Park, L.E.; Scott, M.T.; et al. Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature 2016, 534, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Neviani, P.; Harb, J.G.; Oaks, J.J.; Santhanam, R.; Walker, C.J.; Ellis, J.J.; Ferenchak, G.; Dorrance, A.M.; Paisie, C.A.; Eiring, A.M.; et al. Neviani P (Jour ClincInv, 2013)–PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. J. Clin. Investig. 2013, 123, 4144–4157. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.X.; Riggs, A.D. DNA methylation and demethylation in mammal. J. Biol. Chem. 2011, 286, 18347–18353. [Google Scholar] [CrossRef]
- Bonifer, C.; Cockerill, P.N. Chromatin mechanisms regulating gene expression in health and disease. Adv. Exp. Med. Biol. 2011, 711, 12–25. [Google Scholar] [CrossRef]
- Schemionek, M.; Herrmann, O.; Reher, M.M.; Chatain, N.; Schubert, C.; Costa, I.G.; Hnzelmann, S.; Gusmao, E.G.; Kintsler, S.; Braunschweig, T.; et al. Mtss1 is a critical epigenetically regulated tumor suppressor in CML. Leukemia 2016, 30, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.C.; Wang, D.H.; RayWhay, C.Y.; Luo, J.; Gu, W.; Baylin, S.B. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 2005, 123, 437–448. [Google Scholar] [CrossRef]
- Issa, J.P.J.; Zehnbauer, B.A.; Kaufmann, S.H.; Biel, M.A.; Baylin, S.B. HIC1 hypermethylation is a late event in hematopoietic neoplasms. Cancer Res. 1997, 57, 1678–1681. [Google Scholar]
- Riether, C.; Schürch, C.M.; Flury, C.; Hinterbrandner, M.; Drück, L.; Huguenin, A.L.; Baerlocher, G.M.; Radpour, R.; Ochsenbein, A.F. Tyrosine kinase inhibitor-induced CD70 expression mediates drug resistance in leukemia stem cells by activating Wnt signaling. Sci. Transl. Med. 2015, 7, 298ra119. [Google Scholar] [CrossRef] [PubMed]
- Felsenfeld, G.; Groudine, M. Controlling the double helix. Nature 2003, 421, 448–453. [Google Scholar] [CrossRef]
- Shahbazian, M.D.; Grunstein, M. Functions of Site-Specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100. [Google Scholar] [CrossRef]
- Zhang, L.Q.; Fan, G.L.; Liu, J.J.; Liu, L.; Li, Q.Z.; Lin, H. Identification of Key Histone Modifications and Their Regulatory Regions on Gene Expression Level Changes in Chronic Myelogenous Leukemia. Front. Cell Dev. Biol. 2021, 8, 621578. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Q.; Li, Q.Z. Estimating the effects of transcription factors binding and histone modifications on gene expression levels in human cells. Oncotarget 2017, 8, 40090–40103. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, L.; Liu, X.; Nie, Z.; Wang, X.; Pan, Y.; Luo, J. Research on the epigenetic regulation mechanism of the PTPN6 gene in advanced chronic myeloid leukaemia. Br. J. Haematol. 2017, 178, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Q.; Li, Q.Z.; Jin, W.; Zuo, Y.; Guo, S.C. Genome-wide analysis of H3K36me3 and its regulations to cancer-related genes expression in human cell lines. BioSystems 2018, 171, 59–65. [Google Scholar] [CrossRef]
- Ma, J.; Wang, J.D.; Zhang, W.J.; Zou, B.; Chen, W.J.; Lam, C.S.C.; Chen, M.-H.; Pang, R.; Tan, V.P.Y.; Hung, I.F.; et al. Promoter hypermethylation and histone hypoacetylation contribute to pancreatic-duodenal homeobox 1 silencing in gastric cancer. Carcinogenesis 2010, 31, 1552–1560. [Google Scholar] [CrossRef] [PubMed]
- Nimmanapalli, R.; Fuino, L.; Stobaugh, C.; Richon, V.; Bhalla, K. Cotreatment with the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) enhances imatinib-induced apoptosis of Bcr-Abl-positive human acute leukemia cells. Blood 2003, 101, 3236–3239. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Lafontaine, D.L.J. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 2015, 22, 11–19. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigó, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T.; Thurman, R.E.; et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- McManus, M.T. MicroRNAs and cancer. Semin. Cancer Biol. 2003, 13, 253–258. [Google Scholar] [CrossRef]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.R.B.; Moraes, L.N. de Cury, S.S.; Dadalto, J.; Capannacci, J.; Carvalho, R.F.; Nogueira, C.R.; Hokama, N.K.; Hokama, P.; de Oliveira Montandon Hokama, P. Comparison of microRNA Expression Profile in Chronic Myeloid Leukemia Patients Newly Diagnosed and Treated by Allogeneic Hematopoietic Stem Cell Transplantation. Front. Oncol. 2020, 10, 1544. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.H.; Moles, R.; Nicot, C. Clinical significance of microRNAs in chronic and acute human leukemia. Mol. Cancer 2016, 15, 37. [Google Scholar] [CrossRef]
- Di Stefano, C.; Mirone, G.; Perna, S.; Marfe, G. The roles of microRNAs in the pathogenesis and drug resistance of chronic myelogenous leukemia (Review). Oncol. Rep. 2016, 35, 614–624. [Google Scholar] [CrossRef]
- Mollaei, H.; Safaralizadeh, R.; Rostami, Z. MicroRNA replacement therapy in cancer. J. Cell. Physiol. 2019, 234, 12369–12384. [Google Scholar] [CrossRef]
- Takahashi, R.U.; Prieto-Vila, M.; Kohama, I.; Ochiya, T. Development of miRNA-based therapeutic approaches for cancer patients. Cancer Sci. 2019, 110, 1140–1147. [Google Scholar] [CrossRef] [PubMed]
- Buhagiar, A.; Borg, J.; Ayers, D. Overview of current microRNA biomarker signatures as potential diagnostic tools for leukaemic conditions. Non-Coding RNA Res. 2020, 5, 22–26. [Google Scholar] [CrossRef]
- Wang, F.Y.; Gu, Z.Y.; Gao, C.J. Emerging role of long non-coding RNAs in normal and malignant hematopoiesis. Chin. Med. J. 2020, 133, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, K.; Guo, G.; Chen, J.L. Noncoding RNAs and their functional involvement in regulation of chronic myeloid leukemia. Brief. Funct. Genom. 2016, 15, 239–248. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J. Long non-coding RNAs Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef]
- Bonasio, R.; Shiekhattar, R. Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 2014, 48, 433–455. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, Y.; Wu, G.; Chen, X.; Liu, Y.; Wang, X.; Yu, J.; Li, C.; Chen, X.; Jose, P.A.; et al. Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin. Sci. 2015, 129, 675–685. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Martens-Uzunova, E.S.; Böttcher, R.; Croce, C.M.; Jenster, G.; Visakorpi, T.; Calin, G.A. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 2014, 65, 1140–1151. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yan, I.; Haga, H.; Patel, T. Long noncoding RNA in liver diseases. Hepatology 2014, 60, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Spizzo, R.; Almeida, M.I.; Colombatti, A.; Calin, G.A. Long non-coding RNAs and cancer: A new frontier of translational research. Oncogene 2012, 31, 4577–4587. [Google Scholar] [CrossRef] [PubMed]
- Martelli, A.M.; Evangelisti, C.; Chappell, W.; Abrams, S.L.; Bäsecke, J.; Stivala, F.; Donia, M.; Fagone, P.; Nicoletti, F.; Libra, M.; et al. Targeting the translational apparatus to improve leukemia therapy: Roles of the PI3K/PTEN/Akt/mTOR pathway. Leukemia 2011, 25, 1064–1079. [Google Scholar] [CrossRef]
- Guo, G.; Kang, Q.; Zhu, X.; Chen, Q.; Wang, X.; Chen, Y.; Ouyang, J.; Zhang, L.; Tan, H.; Chen, R.; et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene 2014, 34, 1768–1779. [Google Scholar] [CrossRef]
- Peláez, N.; Carthew, R.W. Biological robustness and the role of microRNAs: A network perspective. Curr. Top. Dev. Biol. 2012, 99, 237–255. [Google Scholar] [CrossRef]
- Nam, J.; Rissland, O.S.; Koppstein, D.; Abreu-goodger, C.; Jan, C.H.; Agarwal, V.; Yildirim, M.A.; Rodriguez, A.; Bartel, D.P. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell 2014, 53, 1031–1043. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Alves, R.; Gonçalves, A.C.; Jorge, J.; Marques, G.; Luís, D.; Ribeiro, A.B.; Freitas-Tavares, P.; Oliveiros, B.; Almeida, A.M.; Sarmento-Ribeiro, A.B. MicroRNAsignature refine response prediction in CML. Sci. Rep. 2019, 9, 9666. [Google Scholar] [CrossRef]
- Feng, Y.H.; Tsao, C.J. Emerging role of microRNA-21 in cancer (Review). Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.A.B.; Guru, S.A.; Abdullah, S.M.; Rizvi, A.; Saxena, A. microRNA-21 expression as prognostic and therapeutic response marker in chronic myeloid leukaemia patients. Asian Pac. J. Cancer Prev. 2019, 20, 2379–2383. [Google Scholar] [CrossRef] [PubMed]
- Seca, H.; Lima, R.; Lopes-Rodrigues, V.; Guimaraes, J.; Gabriela, G.; Vasconcelos, M. Targeting miR-21 Induces Autophagy and Chemosensitivity of Leukemia Cells. Curr. Drug Targets 2013, 14, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, H.; Tao, K.; Xiao, Q.; Huang, Z.; Zhong, L.; Cao, W.; Wen, J.; Feng, W. MiR-29b suppresses CML cell proliferation and induces apoptosis via regulation of BCR/ABL1 protein. Exp. Cell Res. 2013, 319, 1094–1101. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, L.; Zhao, M.; Zhu, S.; Kang, R.; Vernon, P.; Tang, D.; Cao, L. Targeting microRNA-30a-mediated autophagy enhances imatinib activity against human chronic myeloid leukemia cells. Leukemia 2012, 26, 1752–1760. [Google Scholar] [CrossRef] [PubMed]
- Khalil, N.A.; Desouky, M.N.; Diab, I.H.; Hamed, N.A.M.; Mannaa, H.F. MicroRNA 30a Mediated Autophagy and Imatinib Response in Egyptian Chronic Myeloid Leukemia Patients. Indian J. Hematol. Blood Transfus. 2020, 36, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Fu, H.; Gao, L.; Wang, L.; Wang, W.; Li, J.; Li, Y.; Dou, L.; Gao, X.; Luo, X.; et al. BCR-ABL/GATA1/miR-138 mini circuitry contributes to the leukemogenesis of chronic myeloid leukemia. Oncogene 2014, 33, 44–54. [Google Scholar] [CrossRef]
- Bueno, M.J.; Pérez de Castro, I.; Gómez de Cedrón, M.; Santos, J.; Calin, G.A.; Cigudosa, J.C.C.; Croce, C.M.; Fernández-Piqueras, J.; Malumbres, M. Genetic and Epigenetic Silencing of MicroRNA-203 Enhances ABL1 and BCR-ABL1 Oncogene Expression. Cancer Cell 2008, 13, 496–506. [Google Scholar] [CrossRef]
- Shibuta, T.; Honda, E.; Shiotsu, H.; Tanaka, Y.; Vellasamy, S.; Shiratsuchi, M.; Umemura, T. Imatinib induces demethylation of miR-203 gene: An epigenetic mechanism of anti-tumor effect of imatinib. Leuk. Res. 2013, 37, 1278–1286. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, Y.; Han, X.; Roy, M.; Liu, W.; Zhao, X.; Liu, J. Differential expression profiles and functional analysis of plasma miRNAs associated with chronic myeloid leukemia phases. Future Oncol. 2019, 15, 763–776. [Google Scholar] [CrossRef]
- Lopotová, T.; Žáčková, M.; Klamová, H.; Moravcová, J. MicroRNA-451 in chronic myeloid leukemia: MiR-451-BCR-ABL regulatory loop? Leuk. Res. 2011, 35, 974–977. [Google Scholar] [CrossRef]
- Khalifa, K.; Abbas, O.; Abdlfattah, R.; El-Hassanein, S.; Shehata, A.F.; MandourEsawy, S. Decreased expression of microRNA-451 is associated with imatinib mesylate resistance in patients with chronic myeloid leukemia. Menoufia Med. J. 2019, 32, 1004. [Google Scholar] [CrossRef]
- Scholl, V.; Hassan, R.; Zalcberg, I.R. MiRNA-451: A putative predictor marker of Imatinib therapy response in chronic myeloid leukemia. Leuk. Res. 2012, 36, 119–121. [Google Scholar] [CrossRef]
- Liu, L.; Wang, S.; Chen, R.; Wu, Y.; Zhang, B.; Huang, S.; Zhang, J.; Xiao, F.; Wang, M.; Liang, Y. Myc induced miR-144/451 contributes to the acquired imatinib resistance in chronic myelogenous leukemia cell K562. Biochem. Biophys. Res. Commun. 2012, 425, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Ninawe, A.; Guru, S.A.; Yadav, P.; Masroor, M.; Samadhiya, A.; Bhutani, N.; Gupta, N.; Gupta, R.; Saxena, A. MiR-486-5p: A Prognostic Biomarker for Chronic Myeloid Leukemia. ACS Omega 2021, 6, 7711–7718. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Wu, S. Mir-451: A novel biomarker and potential therapeutic target for cancer. Onco. Targets Ther. 2019, 12, 11069–11082. [Google Scholar] [CrossRef]
- Pan, X.; Wang, R.; Wang, Z.X. The potential role of miR-451 in cancer diagnosis, prognosis, and therapy. Mol. Cancer Ther. 2013, 12, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Bandres, E.; Bitarte, N.; Arias, F.; Agorreta, J.; Fortes, P.; Agirre, X.; Zarate, R.; Diaz-Gonzalez, J.A.; Ramirez, N.; Sola, J.J.; et al. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin. Cancer Res. 2009, 15, 2281–2290. [Google Scholar] [CrossRef] [PubMed]
- MachováPolaková, K.; Lopotová, T.; Klamová, H.; Burda, P.; Trněný, M.; Stopka, T.; Moravcová, J. Expression patterns of microRNAs associated with CML phases and their disease related targets. Mol. Cancer 2011, 10, 41. [Google Scholar] [CrossRef]
- Li, Y.; Yuan, Y.; Tao, K.; Wang, X.; Xiao, Q.; Huang, Z.; Zhong, L.; Cao, W.; Wen, J.; Feng, W. Inhibition of BCR/ABL Protein Expression by miR-203 Sensitizes for Imatinib Mesylate. PLoS ONE 2013, 8, e61858. [Google Scholar] [CrossRef]
- Chan, J.A.; Krichevsky, A.M.; Kosik, K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005, 65, 6029–6033. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, X.; Gu, J.; Dong, D.; Yao, J.; Lin, C.; Huang, K.; Fei, J. Anti-miR-21 oligonucleotide sensitizes leukemic K562 cells to arsenic trioxide by inducing apoptosis. Cancer Sci. 2010, 101, 948–954. [Google Scholar] [CrossRef]
- Bai, H.; Xu, R.; Cao, Z.; Wei, D.; Wang, C. Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Lett. 2011, 585, 402–408. [Google Scholar] [CrossRef]
- Wang, W.; Pu, Q.; Lin, X.; Liu, M.; Wu, L.; Wu, Q.; Chen, Y.; Liao, F.; Zhu, J.; Jin, X. Silencing of Mir-21 Sensitizes CML CD34+ Stem/Progenitor Cells to Imatinib-Induced Apoptosis by Blocking PI3K/AKT Pathway. Leuk. Res. 2015, 39, 1117–1124. [Google Scholar] [CrossRef]
- Soverini, S.; De Santis, S.; Monaldi, C.; Bruno, S.; Mancini, M. Targeting Leukemic Stem Cells in Chronic Myeloid Leukemia: Is It Worth the Effort? Int. J. Mol. Sci. 2021, 22, 7093. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Nguyen, L.; Li, L.; Zhao, D.; Kumar, B.; Wu, H.; Lin, A.; Pellicano, F.; Hopcroft, L.; Su, Y.; et al. Bone Marrow Niche Trafficking of Mir-126 Controls the Self-Renewal of Leukemia Stem Cells in Chronic Myelogenous Leukemia. Nat. Med. 2018, 24, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Eiring, A.; Harb, J.; Neviani, P.; Garton, C.; Oaks, J.; Spizzo, R.; Liu, S.; Schwind, S.; Santhanam, R.; Hickey, C.; et al. Mir-328 Functions as an RNA Decoy to Modulate hnRNP E2 Regulation of mRNA Translation in Leukemic Blasts. Cell 2010, 140, 652–665. [Google Scholar] [CrossRef] [PubMed]
- Lovat, F.; Gasparini, P.; Nigita, G.; Larkin, K.; Byrd, J.; Minden, M.; Andreeff, M.; Carter, B.; Croce, C. Loss of Expression of Both Mir-15/16 Loci in CML Transition to Blast Crisis. Proc. Natl. Acad. Sci. USA 2021, 118, e2101566118. [Google Scholar] [CrossRef]


| Gene (Protein) | Description | NCBI Gene ID | Protein Function | CML Analysis | References |
|---|---|---|---|---|---|
| ABL1 (ABL1) | ABL proto-oncogene 1, non-receptor tyrosine | 25 | Cell division, adhesion, differentiation, and response to stress | Chronic phase Blast crisis | [47] |
| AKT1 (AKT1) | AKT serine/threonine kinase 1 | 207 | Regulation of cell proliferation, survival, metabolism, and angiogenesis | Chronic phase Blast crisis | [47] |
| ATM (ATM) | Ataxia-telangiectasia mutated serine/threonine kinase | 472 | Cell cycle checkpoint | Blast crisis | [44] |
| BAP1 | BRCA1 associated protein 1 | 8314 | Regulation of cell cycle and growth | Chronic phase Blast crisis | [87] |
| BCL2 (BCL2) | BCL2 apoptosis regulator | 596 | Regulation of apoptosis | Chronic phase Blast crisis | [47] |
| BTK (BTK) | Bruton tyrosine kinase | 695 | B-cell development | TKIs1 resistance | [88] |
| CDKN2A | Cyclin dependent kinase inhibitor 2A | 1029 | Regulation of cell cycle progression | Blast crisis | [44] |
| CLU (CLU) | Clusterin | 1191 | Regulation of apoptosis and cell proliferation | Chronic phase Blast crisis | [44] |
| C-MYC (c-MYC) | MYC proto-oncogene, bHLH transcription factor | 4609 | Regulation of cell cycle progression, apoptosis, and cellular transformation | Good response to TKIs 1 | [44] |
| CTNNB1 (CTNNB1) | Catenin beta 1 | 1499 | Cell adhesion | Chronic phase Blast crisis TKIs 1 resistance | [44,47] |
| CXCR4 (CXCR4) | C-X-C motif chemokine receptor 4 | 7852 | Regulator of apoptosis, calcium-mediated signaling, response to cytokine stimulus | Chronic phase Blast crisis TKIs 1 resistance | [44] |
| E2F (E2F) | E2F transcription factor 1 | 1869 | Cell cycle regulation | Good response to TKIs 1 | [44] |
| EGFR (EGFR) | Epidermal growth factor receptor | 1956 | Cell proliferation | Chronic phase Blast crisis | [47] |
| FCER1A (FCER1A) | Fc fragment of IgE receptor 1a | 2205 | Alpha subunit of immunoglobulin E involved in allergic response | Chronic phase Blast crisis | [44] |
| FOS (FOS) | Fos proto-oncogene, AP-1 transcription factor | 2353 | Regulation of cell proliferation, differentiation, and transformation | TKIs 1 resistance | [44] |
| FYN (Fyn) | FYN proto-oncogene, Src family tyrosine | 2534 | Regulation of cell growth | Chronic phase Blast crisis | [47] |
| GAS-2 (GAS-2) | Growth arrest specific 2 | 2620 | Apoptosis | Chronic phase Blast crisis TKIs 1 resistance | [44] |
| HIF1A (HIF1A) | Hypoxia inducible factor 1 subunit alpha | 3091 | Response to hypoxia | TKIs 1resistance | [44] |
| IL1B (ILB1) | Interleukin 1 beta | 3553 | Cytokine involved in inflammatory response, cell proliferation, differentiation, and apoptosis | Chronic phase Blast crisis | [47] |
| JAK2 (JAK2) | Janus kinase 2 | 3717 | Regulation of cell growth, development, and differentiation | Chronic phase Blast crisis | [47] |
| KDR (VEGFR) | Vascular endothelial growth factor receptor | 3791 | Proliferation and migration of vascular endothelial cells | Mutations correlate to poor prognosis | [45] |
| MAPK1 (MAPK1) | Mitogen-activated protein kinase 1 | 5594 | Regulation of proliferation, differentiation, transcription, and development | Chronic phase Blast crisis | [47] |
| MAPK3 (MAPK3) | Mitogen-activated protein kinase 3 | 5595 | Regulation of proliferation, differentiation, and cell cycle progression | Chronic phase Blast crisis | [47] |
| MZB1 (MZB1) | Marginal zone B and B1 cell specific protein | 51237 | Regulation of apoptosis | Chronic phase Blast crisis | [44] |
| NFKB1 (NFKB1) | Nuclear factor kappa B subunit 1 | 4790 | Regulator of NF-kB pathway | TKIs 1 resistance | [44] |
| PDGFR-β (PDGFRB) | Platelet-derived growth factor receptor, beta polypeptide | 100487523 | Regulation of cell proliferation, survival, differentiation, chemotaxis, and migration | Chronic phase Blast crisis | [47] |
| PTGS1 (PTGS1) | Prostaglandin-endoperoxide synthase 1 | 5742 | Drug metabolism; Regulation of cell proliferation | TKIs 1 resistance | [88] |
| PTK2 (PTK2) | Protein tyrosine kinase 2 | 5747 | Regulation of cell growth and cell adhesion | Chronic phase Blast crisis | [47] |
| PTKB2 (PTKB2) | Protein tyrosine kinase 2 beta | 2185 | Calcium-induced regulation of ion channels | Chronic phase Blast crisis | [47] |
| PTPN22 (PTPN22) | Protein tyrosine phosphatase non-receptor type 22 | 26191 | CBL function in the T-cell receptor signaling pathway | TKIs 1 resistance | [88] |
| RGS2 (RGS2) | Regulator of G protein signaling 2 | 5997 | Regulator of myeloid differentiation | Chronic phase Blast crisis | [44] |
| SKIL (SKIL) | SKI like proto-oncogene | 6498 | TGF-β pathway–regulation of cell growth and differentiation | TKIs 1 resistance | [44] |
| SORBS3 (SORBS3) | Sorbin and SH3 domain containing 3 | 10174 | Cell adhesion | TKIs 1 resistance | [88] |
| SQSTM1 (SQSTM1) | Sequestosome 1 | 8878 | Regulator of NF-kB pathway | TKIs 1 resistance | [44] |
| SRC (SRC) | SRC proto-oncogene, non-receptor tyrosine kinase | 6714 | Regulation of cell growth | Chronic phase Blast crisis | [47] |
| TGFB1 (TGFB1) | Transforming growth factor beta 1 | 7040 | Regulation of cell proliferation, differentiation, and growth | Chronic phase Blast crisis | [47] |
| TGFB2 (TGFB2) | Transforming growth factor beta 2 | 7042 | Regulation of cell proliferation, differentiation, and growth | Chronic phase Blast crisis | [47] |
| TNC (TNC) | Tenascin C | 3371 | Regulation of cell adhesion | TKIs 1 resistance | [88] |
| TNFA (TNF) | Tumor necrosis factor | 7124 | Cytokine involved in inflammatory response. Cell proliferation and differentiation | Chronic phase Blast crisis | [47] |
| TP53 (p53) | Tumor protein p53 | 7157 | Regulation of cell-cycle, apoptosis, and autophagy | Chronic phase Blast crisis Mutations correlated to poor prognosis | [46,47,48,49,50] |
| VEGFA (VEGFA) | Vascular endothelial growth factor A | 7422 | Proliferation and migration of vascular endothelial cells | Chronic phase Blast crisis | [47] |
| WTAP (WTAP) | Wilms tumor (WT1) associated protein | 9589 | Cell cycle regulation | TKIs 1 resistance | [44] |
| miRNA | Expression in CML | Process Involved/Biological Relevance | References |
|---|---|---|---|
| miRNA-21 | Upregulated | One of the most upregulated miRNAs in cancer, Implicated in drug resistance. Low levels at diagnosis are associated with an optimal response to TKI therapy. Biomarker of disease progression: expression increases from CP to BP. Inhibition leads to G1-phase arrest, growth inhibition, and apoptosis. | [149,150,151,152] |
| miRNA-29b | Downregulated | Expression level is negatively correlated with MCL-1 and BCL-2 expression. Downregulation involved in apoptosis evasion. | [153] |
| miRNA-30a | Downregulated | Inhibits autophagy, thus promoting IM cytotoxicity: upregulated in IM-responders. Expression levels are negatively correlated with Sokal score and BCR-ABL1 following treatment. | [154,155] |
| miRNA-138 | Downregulated | Associated with downregulation of BCR-ABL1, inhibition of proliferation, and IM-induced apoptosis. | [156] |
| miRNA-203 | Downregulated | Frequently silenced in CML (monoallelic loss and promotor hypermethylation). IM inducemiRNA-203 promoter demethylation: miRNA-203 upregulation decreases BCR-ABL1 expression and reduces CML cells proliferation rate. | [157,158] |
| miRNA-451 | Downregulated | Biomarker of disease progression: expression decreases from CP to BP. Reciprocal regulatory loop between miRNA-451 and BCR-ABL1 Biomarker of prognosis and treatment response: involved in IM resistance, expression levels at diagnosis predict TKI response. | [149,159,160,161,162] |
| miRNA-144/451 | Downregulated | Regulatory pathway between miRNA-144/451 and c-MYC Downregulated in IM resistant patients (c-MYC upregulated); Restoration of miRNA-144/451 expression levels sensitize CML cells to IM. | [163] |
| miRNA-486-5p | Downregulated | Valuable prognosis biomarker: higher miRNA-486-5p expression levels at diagnosis are associated with better prognosis and faster achievement of complete hematologic response to IM treatment. | [164] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdulmawjood, B.; Costa, B.; Roma-Rodrigues, C.; Baptista, P.V.; Fernandes, A.R. Genetic Biomarkers in Chronic Myeloid Leukemia: What Have We Learned So Far? Int. J. Mol. Sci. 2021, 22, 12516. https://doi.org/10.3390/ijms222212516
Abdulmawjood B, Costa B, Roma-Rodrigues C, Baptista PV, Fernandes AR. Genetic Biomarkers in Chronic Myeloid Leukemia: What Have We Learned So Far? International Journal of Molecular Sciences. 2021; 22(22):12516. https://doi.org/10.3390/ijms222212516
Chicago/Turabian StyleAbdulmawjood, Bilal, Beatriz Costa, Catarina Roma-Rodrigues, Pedro V. Baptista, and Alexandra R. Fernandes. 2021. "Genetic Biomarkers in Chronic Myeloid Leukemia: What Have We Learned So Far?" International Journal of Molecular Sciences 22, no. 22: 12516. https://doi.org/10.3390/ijms222212516
APA StyleAbdulmawjood, B., Costa, B., Roma-Rodrigues, C., Baptista, P. V., & Fernandes, A. R. (2021). Genetic Biomarkers in Chronic Myeloid Leukemia: What Have We Learned So Far? International Journal of Molecular Sciences, 22(22), 12516. https://doi.org/10.3390/ijms222212516









