Molecular Determinants and Specificity of mRNA with Alternatively-Spliced UPF1 Isoforms, Influenced by an Insertion in the ‘Regulatory Loop’
Abstract
:1. Introduction
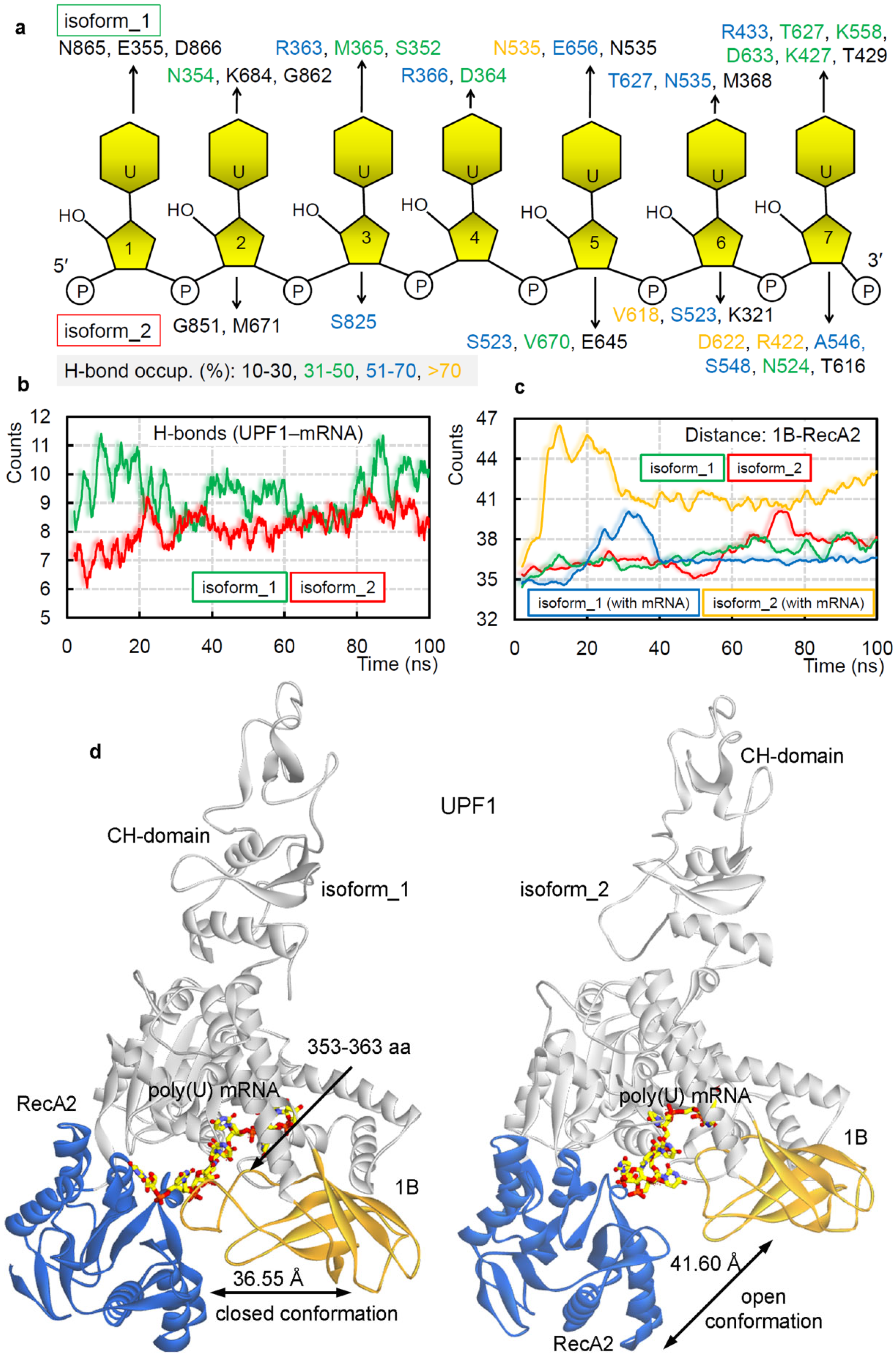
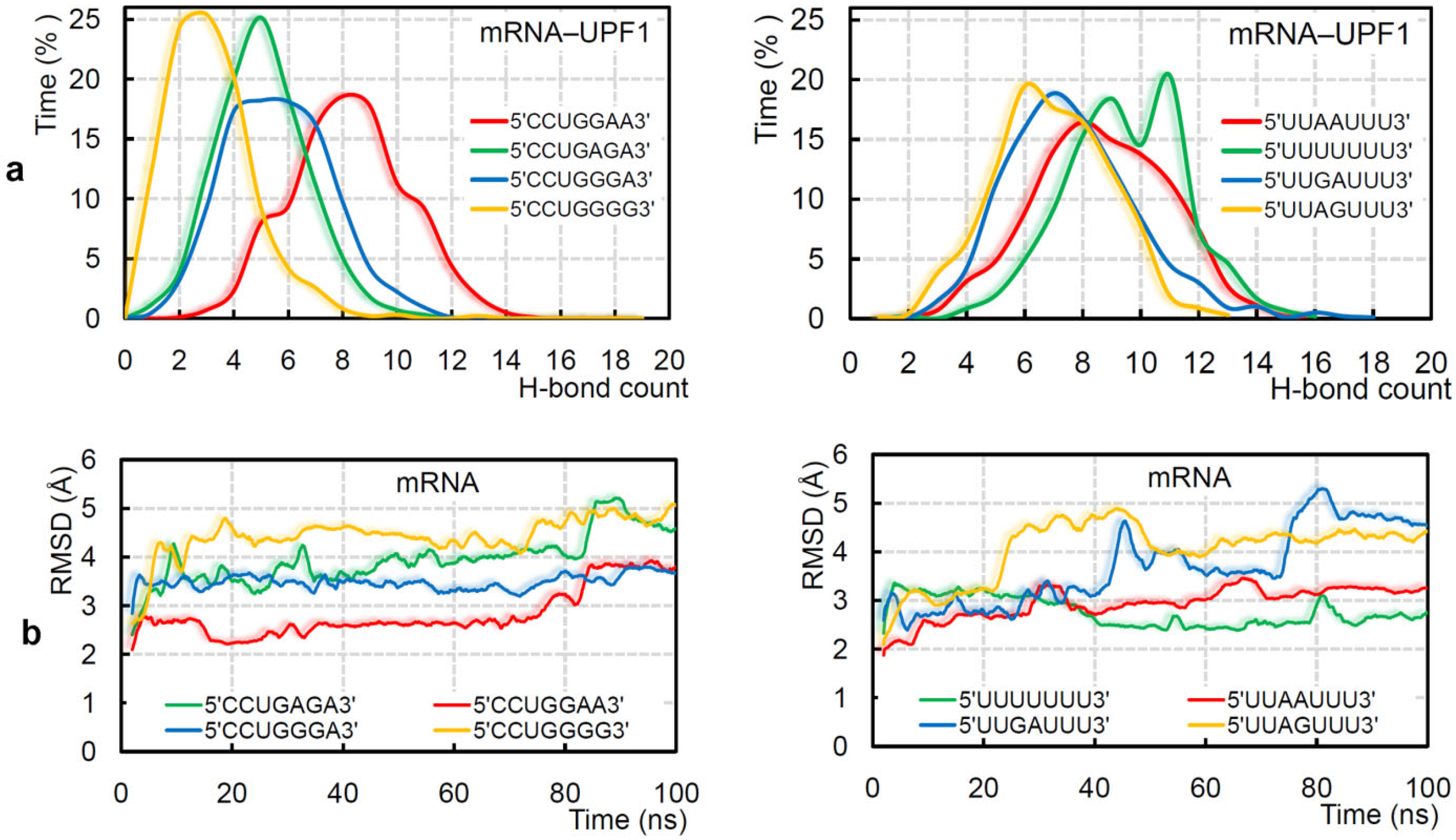
2. Results and Discussion
2.1. Different UPF1 Isoforms and Their Selectivity towards mRNA Motifs
2.2. GC- and AU-Rich mRNA Motifs Binding Pattern with UPF1 Isoforms
2.3. UPF1A839T and UPF1P533L/T Cancer Mutants Influencing the UPF1-mRNA Binding
3. Materials and Methods
Systems Build-Up for Different mRNA Motifs with UPF1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| aa | amino acids |
| CLIP | cross-linking immunoprecipitation |
| CH domain | cysteine-histidine-rich domain |
| CHARMm | Chemistry at Harvard Macromolecular Mechanics |
| DECID | decay-inducing complex |
| EJC | exon–exon junction complex |
| eRF1 | eukaryotic translation termination factor 1 |
| H-bond | hydrogen bond |
| LINCS | linear constraint solver |
| mRNP | messenger ribonucleoprotein |
| MDS | molecular dynamics simulation |
| MOE | Molecular Operating Environment |
| NGD | no-go decay |
| NMD | nonsense-mediated mRNA decay |
| NSD | non-stop decay |
| ns | nanosecond |
| PAB1 | poly(A)-binding protein cytoplasmic 1 |
| pdb | protein data bank |
| PME | particle mesh Ewald |
| PTC | premature termination codon |
| ps | picosecond |
| RMSD | root-mean-square deviation |
| RMSF | root-mean-square fluctuations |
| SMG | suppressors with morphogenetic effect on genitalia |
| SPC | simple point charge |
| uORF | upstream open reading frames |
| UPF1 | UP-frameshift 1 |
| UTR | untranslated region |
| VMD | Visual Molecular Dynamics |
References
- Stefani, M.; Dobson, C.M. Protein aggregation and aggregate toxicity: New insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. 2003, 81, 678–699. [Google Scholar] [CrossRef]
- Gregersen, N.; Bross, P.; Vang, S.; Christensen, J.H. Protein Misfolding and Human Disease. Annu. Rev. Genom. Hum. Genet. 2006, 7, 103–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, R.; Müller, L.; Wullich, B. Protein transport into the endoplasmic reticulum: Mechanisms and pathologies. Trends Mol. Med. 2006, 12, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Hebert, D.N.; Molinari, M. In and Out of the ER: Protein Folding, Quality Control, Degradation, and Related Human Diseases. Physiol. Rev. 2007, 87, 1377–1408. [Google Scholar] [CrossRef]
- Jarjanazi, H.; Savas, S.; Pabalan, N.; Dennis, J.W.; Ozcelik, H. Biological implications of SNPs in signal peptide domains of human proteins. Proteins Struct. Funct. Bioinform. 2007, 70, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Hipp, M.S.; Park, S.-H.; Hartl, F.U. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014, 24, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Karamyshev, A.L.; Karamysheva, Z.N. Lost in Translation: Ribosome-Associated mRNA and Protein Quality Controls. Front. Genet. 2018, 9, 431. [Google Scholar] [CrossRef] [Green Version]
- Karamyshev, A.L.; Tikhonova, E.B.; Karamysheva, Z.N. Translational Control of Secretory Proteins in Health and Disease. Int. J. Mol. Sci. 2020, 21, 2538. [Google Scholar] [CrossRef] [Green Version]
- Welch, E.M.; Jacobson, A. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 1999, 18, 6134–6145. [Google Scholar] [CrossRef] [Green Version]
- Doma, M.K.; Parker, R. RNA Quality Control in Eukaryotes. Cell 2007, 131, 660–668. [Google Scholar] [CrossRef] [Green Version]
- Shoemaker, C.J.; Green, R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012, 19, 594–601. [Google Scholar] [CrossRef]
- Popp, M.W.-L.; Maquat, L.E. Organizing Principles of Mammalian Nonsense-Mediated mRNA Decay. Annu. Rev. Genet. 2013, 47, 139–165. [Google Scholar] [CrossRef] [Green Version]
- Lewis, B.P.; Green, R.; Brenner, S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA 2002, 100, 189–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, P.; Mühlemann, O. Cutting the nonsense: The degradation of PTC-containing mRNAs. Biochem. Soc. Trans. 2010, 38, 1615–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frischmeyer, P.A.; Dietz, H.C. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999, 8, 1893–1900. [Google Scholar] [CrossRef] [Green Version]
- Mort, M.; Ivanov, D.; Cooper, D.N.; Chuzhanova, N.A. A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat. 2008, 29, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Hurt, J.A.; Robertson, A.D.; Burge, C.B. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013, 23, 1636–1650. [Google Scholar] [CrossRef] [Green Version]
- Colombo, M.; Karousis, E.; Bourquin, J.; Bruggmann, R.; Mühlemann, O. Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA 2016, 23, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Imamachi, N.; Salam, K.A.; Suzuki, Y.; Akimitsu, N. A GC-rich sequence feature in the 3′ UTR directs UPF1-dependent mRNA decay in mammalian cells. Genome Res. 2016, 27, 407–418. [Google Scholar] [CrossRef] [Green Version]
- Karousis, E.D.; Gurzeler, L.A.; Annibaldis, G.; Dreos, R.; Mühlemann, O. Human NMD ensues independently of stable ribosome stalling. Nat. Commun. 2020, 11, 4134. [Google Scholar] [CrossRef]
- Yepiskoposyan, H.; Aeschimann, F.; Nilsson, D.; Okoniewski, M.; Mühlemann, O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 2011, 17, 2108–2118. [Google Scholar] [CrossRef] [Green Version]
- Thoren, L.A.; Nørgaard, G.A.; Weischenfeldt, J.; Waage, J.; Jakobsen, J.S.; Damgaard, I.; Bergström, F.C.; Blom, A.M.; Borup, R.; Bisgaard, H.C.; et al. UPF2 is a critical regulator of liver development, function and regeneration. PLoS ONE 2010, 5, e11650. [Google Scholar] [CrossRef] [PubMed]
- Kuzmiak, H.A.; Maquat, L.E. Applying nonsense-mediated mRNA decay research to the clinic: Progress and challenges. Trends Mol. Med. 2006, 12, 306–316. [Google Scholar] [CrossRef]
- Khajavi, M.; Inoue, K.; Lupski, J.R. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur. J. Hum. Genet. 2006, 14, 1074–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- A Holbrook, J.; Neu-Yilik, G.; Hentze, M.; E Kulozik, A. Nonsense-mediated decay approaches the clinic. Nat. Genet. 2004, 36, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Czaplinski, K.; Weng, Y.; Hagan, K.W.; Peltz, S.W. Purification and characterization of the Upf1 protein: A factor involved in translation and mRNA degradation. RNA 1995, 1, 610–623. [Google Scholar] [PubMed]
- Atkin, A.L.; Altamura, N.; Leeds, P.; Culbertson, M.R. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol. Biol. Cell 1995, 6, 611–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, Y.; Czaplinski, K.; Peltz, S.W. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell. Biol. 1996, 16, 5491–5506. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, A.; Czaplinski, K.; Trifillis, P.; He, F.; Jacobson, A.; Peltz, S.W. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 2000, 6, 1226–1235. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Perlick, H.A.; Dietz, H.C.; Maquat, L.E. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 1998, 95. [Google Scholar] [CrossRef] [Green Version]
- Lasalde, C.; Rivera, A.V.; León, A.J.; González-Feliciano, J.A.; Estrella, L.A.; Rodríguez-Cruz, E.N.; Correa, M.E.; Cajigas, I.J.; Bracho, D.P.; Vega, I.E.; et al. Identification and functional analysis of novel phosphorylation sites in the RNA surveillance protein Upf1. Nucleic Acids Res. 2013, 42, 1916–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, Y.; Czaplinski, K.; Peltz, S.W. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol. 1996, 16, 5477–5490. [Google Scholar] [CrossRef] [Green Version]
- Atkin, A.L.; Schenkman, L.R.; Eastham, M.; Dahlseid, J.N.; Lelivelt, M.J.; Culbertson, M.R. Relationship between Yeast Polyribosomes and Upf Proteins Required for Nonsense mRNA Decay. J. Biol. Chem. 1997, 272, 22163–22172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, Y.; Czaplinski, K.; Peltz, S.W. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA 1998, 4, 205–214. [Google Scholar]
- Kadlec, J.; Guilligay, D.; Ravelli, R.B.; Cusack, S. Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA 2006, 12, 1817–1824. [Google Scholar] [CrossRef] [Green Version]
- He, F.; Brown, A.H.; Jacobson, A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 1997, 17, 1580–1594. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarti, S.; Jayachandran, U.; Bonneau, F.; Fiorini, F.; Basquin, C.; Domcke, S.; Le Hir, H.; Conti, E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell 2011, 41, 693–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serin, G.; Gersappe, A.; Black, J.D.; Aronoff, R.; Maquat, L.E. Identification and Characterization of Human Orthologues to Saccharomyces cerevisiae Upf2 Protein and Upf3 Protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol. 2001, 21, 209–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamieh, H.; Ballut, L.; Bonneau, F.; Le Hir, H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2007, 15, 85–93. [Google Scholar] [CrossRef]
- Kalathiya, U.; Padariya, M.; Pawlicka, K.; Verma, C.S.; Houston, D.; Hupp, T.R.; Alfaro, J.A. Hupp Insights into the Effects of Cancer Associated Mutations at the UPF2 and ATP-Binding Sites of NMD Master Regulator: UPF1. Int. J. Mol. Sci. 2019, 20, 5644. [Google Scholar] [CrossRef] [Green Version]
- Kashima, I.; Yamashita, A.; Izumi, N.; Kataoka, N.; Morishita, R.; Hoshino, S.; Ohno, M.; Dreyfuss, G.; Ohno, S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006, 20, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Echevarría, M.J.; González, C.I.; Peltz, S.W. Identifying the right stop: Determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998, 17, 575–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, J.; Sato, H.; Tang, Y.; Matsuda, D.; Maquat, L.E. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol. Cell 2010, 39, 396–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amrani, N.; Ganesan, R.; Kervestin, S.; Mangus, D.A.; Ghosh, S.; Jacobson, A. A faux 3′-UTR promotes aberrant termination and triggers nonsense- mediated mRNA decay. Nature 2004, 432, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Lejeune, F.; E Maquat, L. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 2005, 17, 309–315. [Google Scholar] [CrossRef]
- Kuroha, K.; Tatematsu, T.; Inada, T. Upf1 stimulates degradation of the product derived from aberrant messenger RNA containing a specific nonsense mutation by the proteasome. EMBO Rep. 2009, 10, 1265–1271. [Google Scholar] [CrossRef] [Green Version]
- Carter, M.S.; Li, S.; Wilkinson, M.F. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996, 15, 5965–5975. [Google Scholar] [CrossRef]
- Kervestin, S.; Li, C.; Buckingham, R.; Jacobson, A. Testing the faux-UTR model for NMD: Analysis of Upf1p and Pab1p competition for binding to eRF3/Sup35p. Biochimie 2012, 94, 1560–1571. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, T.; Yamashita, A.; Kashima, I.; Schell, T.; Anders, K.; Grimson, A.; Hachiya, T.; Hentze, M.; Anderson, P.; Ohno, S. Phosphorylation of hUPF1 Induces Formation of mRNA Surveillance Complexes Containing hSMG-5 and hSMG-7. Mol. Cell 2003, 12, 1187–1200. [Google Scholar] [CrossRef]
- Hosoda, N.; Kim, Y.K.; Lejeune, F.; E Maquat, L. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2005, 12, 893–901. [Google Scholar] [CrossRef] [PubMed]
- González, C.I.; Ruiz-Echevarría, M.J.; Vasudevan, S.; Henry, M.F.; Peltz, S.W. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell 2000, 5, 489–499. [Google Scholar] [CrossRef]
- Lykke-Andersen, J.; Shu, M.-D.; Steitz, J.A. Human Upf Proteins Target an mRNA for Nonsense-Mediated Decay When Bound Downstream of a Termination Codon. Cell 2000, 103, 1121–1131. [Google Scholar] [CrossRef] [Green Version]
- Shibuya, T.; Tange, T.; Stroupe, M.E.; Moore, M.J. Mutational analysis of human eIF4AIII identifies regions necessary for exon junction complex formation and nonsense-mediated mRNA decay. RNA 2006, 12, 360–374. [Google Scholar] [CrossRef] [Green Version]
- Gehring, N.H.; Kunz, J.B.; Neu-Yilik, G.; Breit, S.; Viegas, M.H.; Hentze, M.W.; Kulozik, A.E. Exon-Junction Complex Components Specify Distinct Routes of Nonsense-Mediated mRNA Decay with Differential Cofactor Requirements. Mol. Cell 2005, 20, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Ohnishi, T.; Kashima, I.; Taya, Y.; Ohno, S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001, 15, 2215–2228. [Google Scholar] [CrossRef] [Green Version]
- Popp, M.W.; Maquat, L.E. Leveraging Rules of Nonsense-Mediated mRNA Decay for Genome Engineering and Personalized Medicine. Cell 2016, 165, 1319–1322. [Google Scholar] [CrossRef] [Green Version]
- Conti, E.; Izaurralde, E. Nonsense-mediated mRNA decay: Molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 2005, 17, 316–325. [Google Scholar] [CrossRef]
- Amrani, N.; Sachs, M.; Jacobson, A.J. Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell Biol. 2006, 7, 415–425. [Google Scholar] [CrossRef]
- Amrani, N.; Dong, S.; He, F.; Ganesan, R.; Ghosh, S.; Kervestin, S.; Li, C.; Mangus, D.A.; Spatrick, P.; Jacobson, A. Aberrant termination triggers nonsense-mediated mRNA decay. Biochem. Soc. Trans. 2006, 34, 39–42. [Google Scholar] [CrossRef]
- Rebbapragada, I.; Lykke-Andersen, J. Execution of nonsense-mediated mRNA decay: What defines a substrate? Curr. Opin. Cell Biol. 2019, 21, 394–402. [Google Scholar] [CrossRef]
- Behm-Ansmant, I.; Kashima, I.; Rehwinkel, J.; Saulière, J.; Wittkopp, N.; Izaurralde, E. mRNA quality control: An ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007, 581, 2845–2853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Hir, H.; Gatfield, D.; Izaurralde, E.; Moore, M.J. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001, 20, 4987–4997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isken, O.; Kim, Y.K.; Hosoda, N.; Mayeur, G.L.; Hershey, J.W.; Maquat, L.E. Upf1 Phosphorylation Triggers Translational Repression during Nonsense-Mediated mRNA Decay. Cell 2008, 133, 314–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huntzinger, E.; Braun, J.E.; Heimstädt, S.; Zekri, L.; Izaurralde, E. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J. 2010, 29, 4146–4160. [Google Scholar] [CrossRef] [Green Version]
- Kebaara, B.W.; Atkin, A.L. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 2009, 37, 2771–2778. [Google Scholar] [CrossRef] [Green Version]
- Muhlrad, D.; Parker, R. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 1999, 5, 1299–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clerici, M.; Mourão, A.; Gutsche, I.; Gehring, N.H.; Hentze, M.; Kulozik, A.; Kadlec, J.; Sattler, M.; Cusack, S. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 2009, 28, 2293–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safaee, N.; Kozlov, G.; Noronha, A.M.; Xie, J.; Wilds, C.; Gehring, K. Interdomain Allostery Promotes Assembly of the Poly(A) mRNA Complex with PABP and eIF4G. Mol. Cell 2012, 48, 375–386. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Sternberg, M.J.E. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Roy, B.; He, F.; Jacobson, A. Nonsense suppression position effect implicates poly(A)-binding protein in the regulation of translation termination. SSRN Electron. J. 2019. [Google Scholar] [CrossRef]
- Gowravaram, M.; Bonneau, F.; Kanaan, J.; Maciej, V.D.; Fiorini, F.; Raj, S.; Croquette, V.; Le Hir, H.; Chakrabarti, S. A conserved structural element in the RNA helicase UPF1 regulates its catalytic activity in an isoform-specific manner. Nucleic Acids Res. 2018, 46, 2648–2659. [Google Scholar] [CrossRef] [Green Version]
- Kurosaki, T.; Maquat, L.E. Rules that govern UPF1 binding to mRNA 3′ UTRs. Proc. Natl. Acad. Sci. USA 2013, 110, 3357–3362. [Google Scholar] [CrossRef] [Green Version]
- Hamann, F.; Enders, M.; Ficner, R. Structural basis for RNA translocation by DEAH-box ATPases. Nucleic Acids Res. 2019, 47, 4349–4362. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Zhang, H.; Wang, L.; Zhu, C.; Sheng, K.; Du, Y.; Wang, K.; Dias, A.; Chen, S.; Whitman, M.; et al. Premature termination codons are recognized in the nucleus in a reading-frame-dependent manner. Cell Discov. 2015, 1, 15001. [Google Scholar] [CrossRef] [Green Version]
- Hogg, J.R.; Goff, S.P. Upf1 Senses 3′UTR Length to Potentiate mRNA Decay. Cell 2010, 143, 379–389. [Google Scholar] [CrossRef] [Green Version]
- Zünd, D.; Gruber, A.R.; Zavolan, M.; Mühlemann, O. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat. Struct. Mol. Biol. 2013, 20, 936–943. [Google Scholar] [CrossRef]
- Lee, S.R.; Pratt, G.A.; Martinez, F.J.; Yeo, G.W.; Lykke-Andersen, J. Target Discrimination in Nonsense-Mediated mRNA Decay Requires Upf1 ATPase Activity. Mol. Cell 2015, 59, 413–425. [Google Scholar] [CrossRef] [Green Version]
- Hoernes, T.P.; Clementi, N.; Juen, M.A.; Shi, X.; Faserl, K.; Willi, J.; Gasser, C.; Kreutz, C.; Joseph, S.; Lindner, H.; et al. Atomic mutagenesis of stop codon nucleotides reveals the chemical prerequisites for release factor-mediated peptide release. Proc. Natl. Acad. Sci. USA 2018, 115, E382–E389. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Brooks, B.R.; Brooks, C.L., III; MacKerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef]
- Bjelkmar, P.; Larsson, P.; Cuendet, M.A.; Hess, B.; Lindahl, E. Implementation of the CHARMM Force Field in GROMACS: Analysis of Protein Stability Effects from Correction Maps, Virtual Interaction Sites, and Water Models. J. Chem. Theory Comput. 2010, 6, 459–466. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces. The Jerusalem Symposia on Quantum Chemistry and Biochemistry; Pullman, B., Ed.; Springer: Dordrecht, The Netherlands, 1981; Volume 14. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 19, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Van Gunsteren, W.F.; Berendsen, H.J.C. A Leap-frog Algorithm for Stochastic Dynamics. Mol. Simul. 1988, 1, 173–185. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padariya, M.; Fahraeus, R.; Hupp, T.; Kalathiya, U. Molecular Determinants and Specificity of mRNA with Alternatively-Spliced UPF1 Isoforms, Influenced by an Insertion in the ‘Regulatory Loop’. Int. J. Mol. Sci. 2021, 22, 12744. https://doi.org/10.3390/ijms222312744
Padariya M, Fahraeus R, Hupp T, Kalathiya U. Molecular Determinants and Specificity of mRNA with Alternatively-Spliced UPF1 Isoforms, Influenced by an Insertion in the ‘Regulatory Loop’. International Journal of Molecular Sciences. 2021; 22(23):12744. https://doi.org/10.3390/ijms222312744
Chicago/Turabian StylePadariya, Monikaben, Robin Fahraeus, Ted Hupp, and Umesh Kalathiya. 2021. "Molecular Determinants and Specificity of mRNA with Alternatively-Spliced UPF1 Isoforms, Influenced by an Insertion in the ‘Regulatory Loop’" International Journal of Molecular Sciences 22, no. 23: 12744. https://doi.org/10.3390/ijms222312744
APA StylePadariya, M., Fahraeus, R., Hupp, T., & Kalathiya, U. (2021). Molecular Determinants and Specificity of mRNA with Alternatively-Spliced UPF1 Isoforms, Influenced by an Insertion in the ‘Regulatory Loop’. International Journal of Molecular Sciences, 22(23), 12744. https://doi.org/10.3390/ijms222312744








