3D Structures of IgA, IgM, and Components
Abstract
:1. Introduction
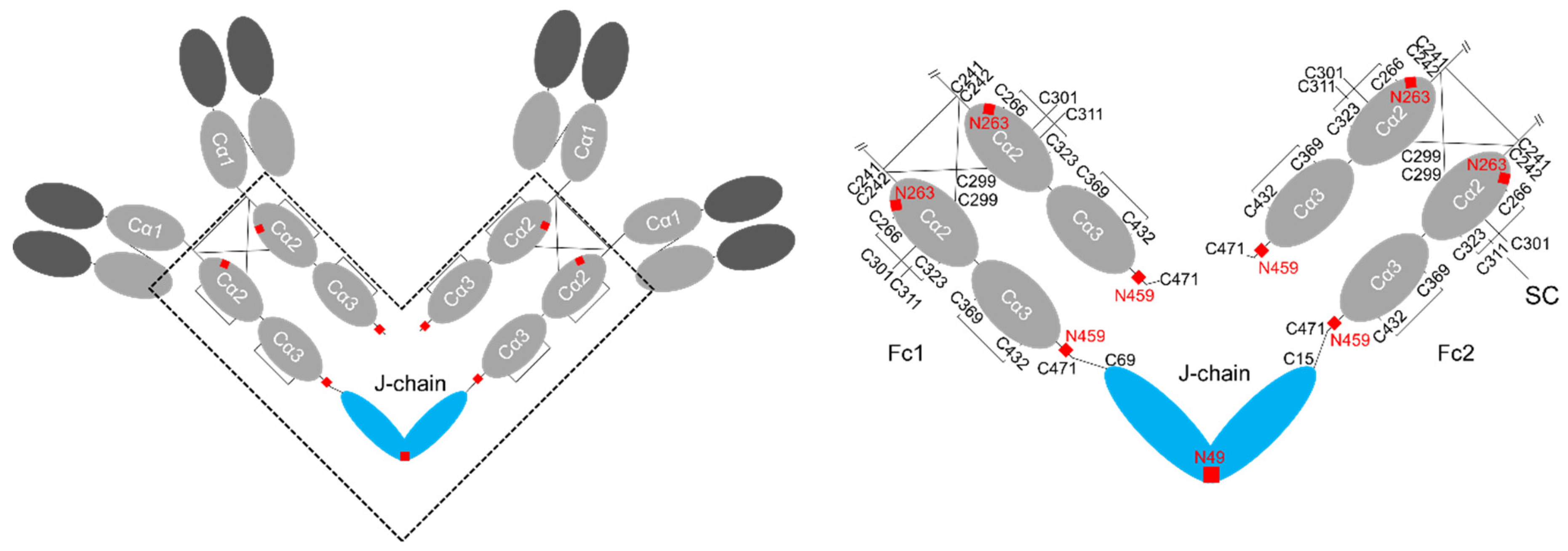
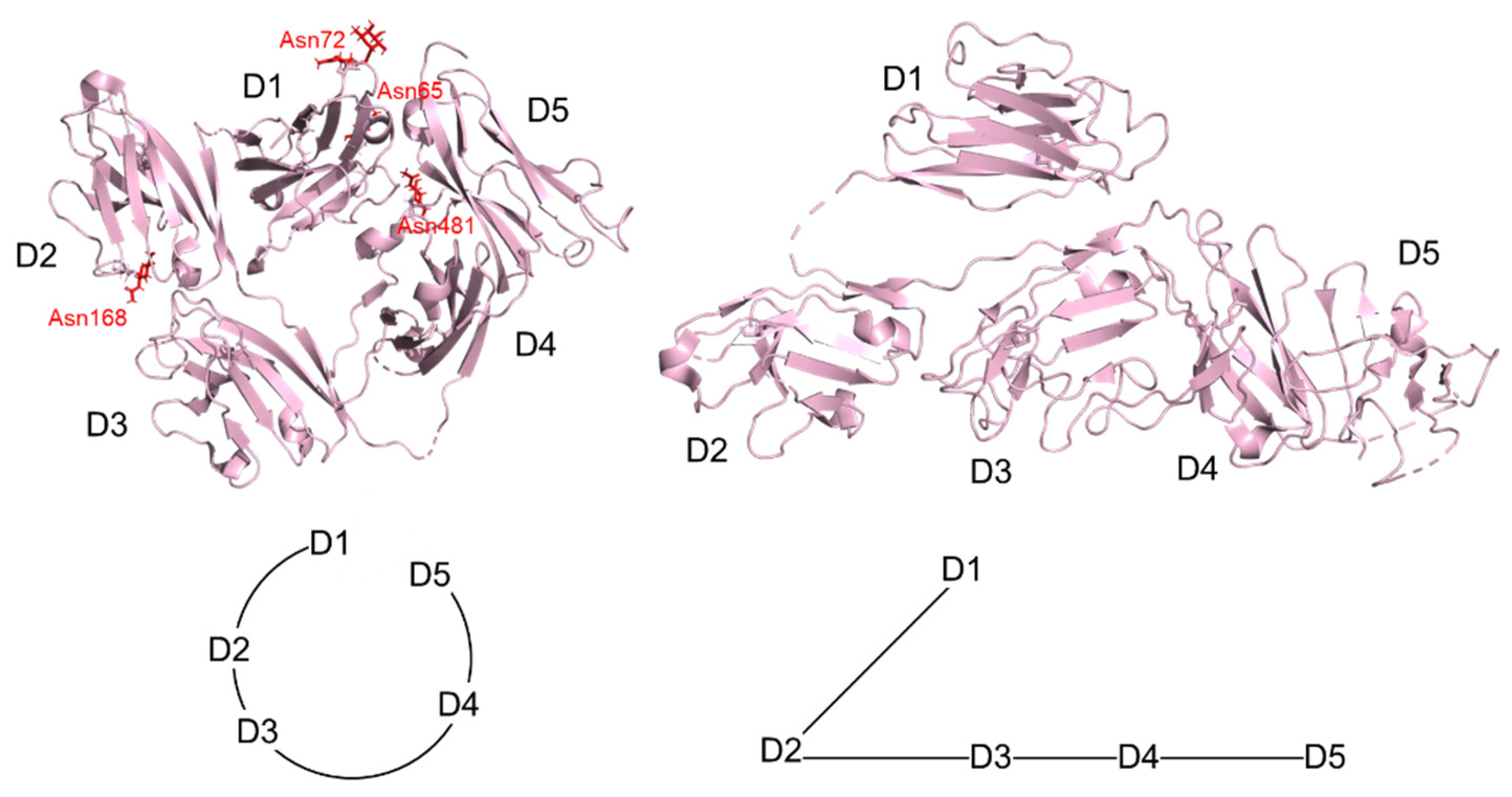
2. Structure of IgA
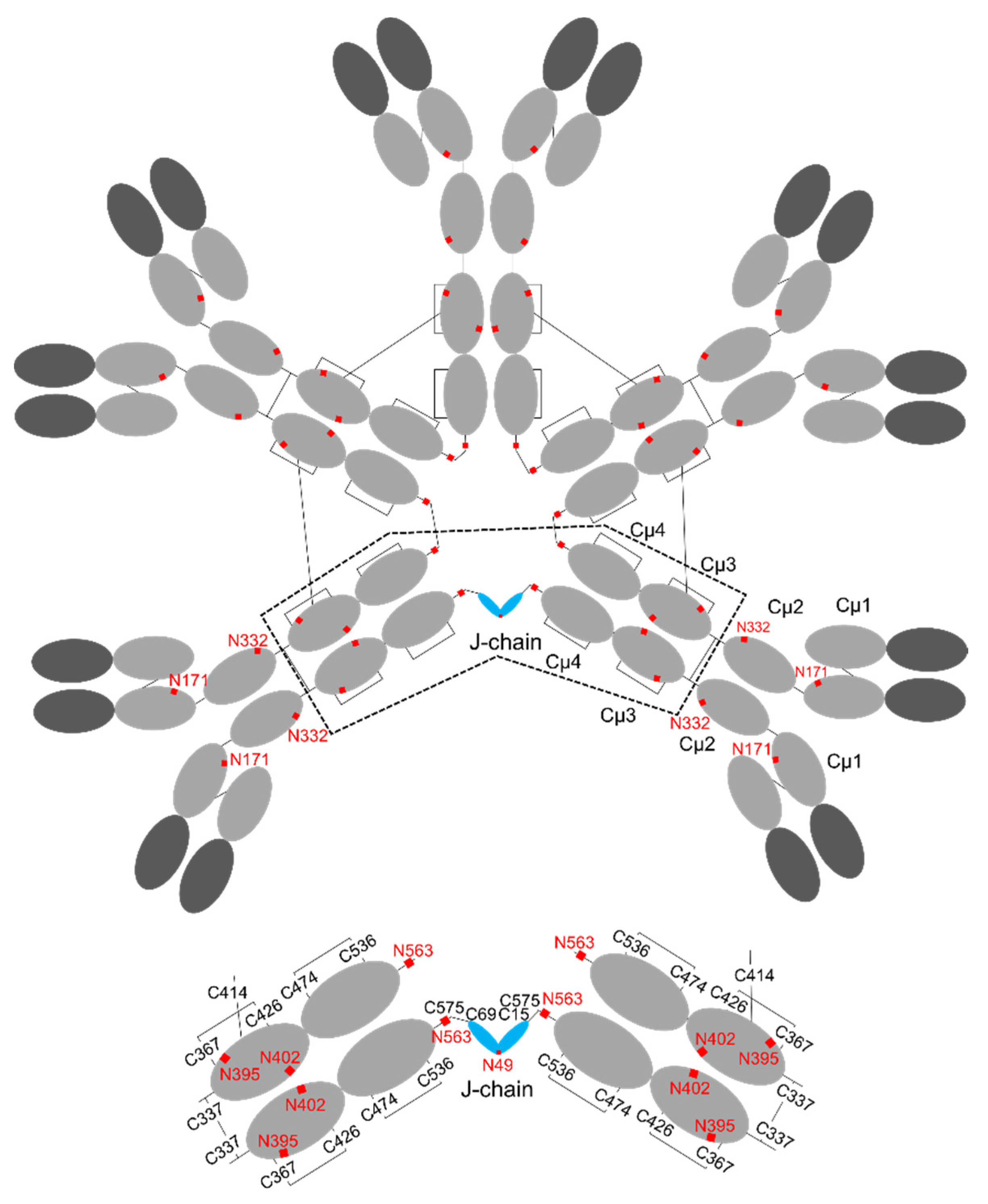
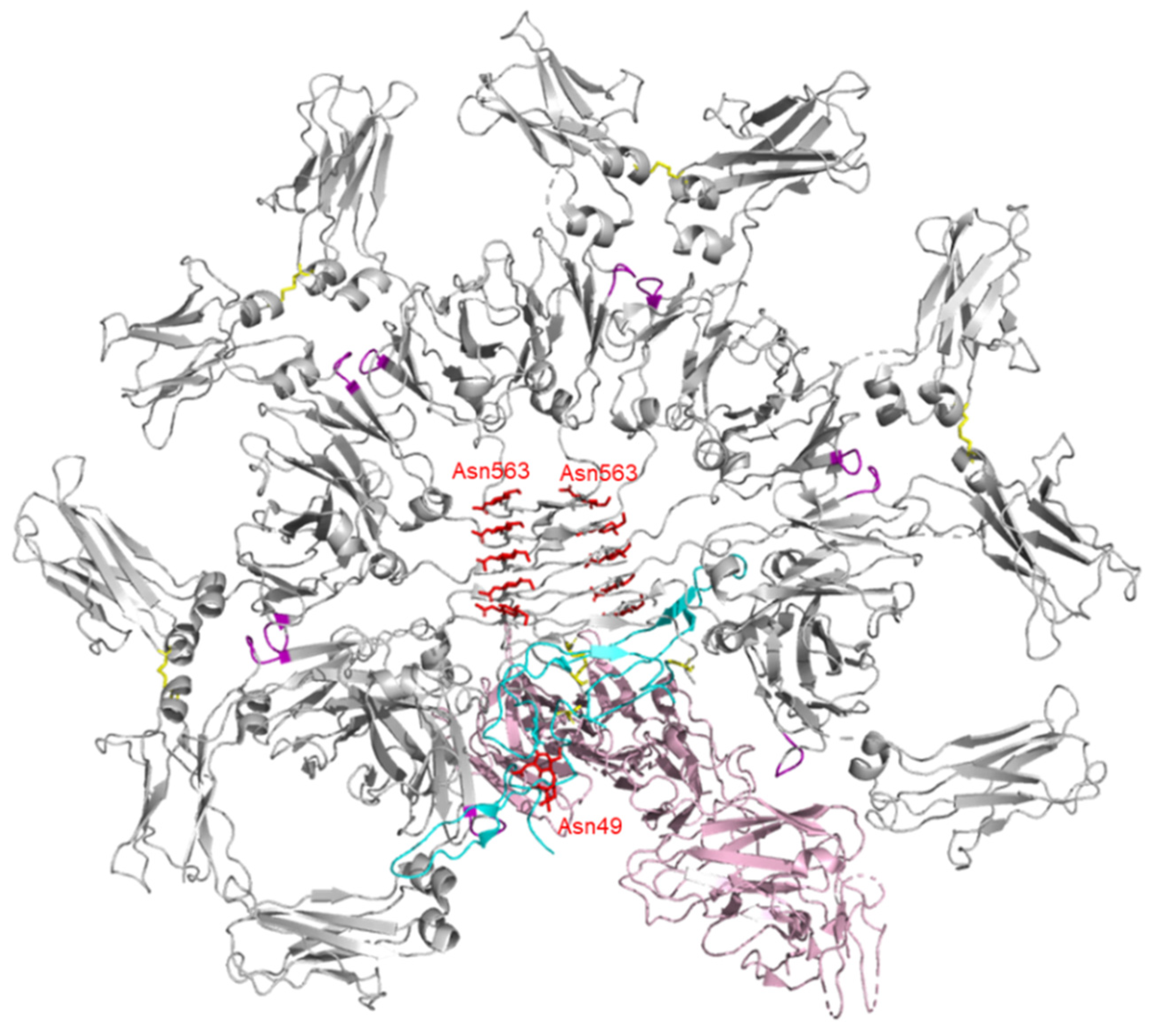
3. Structure of IgM
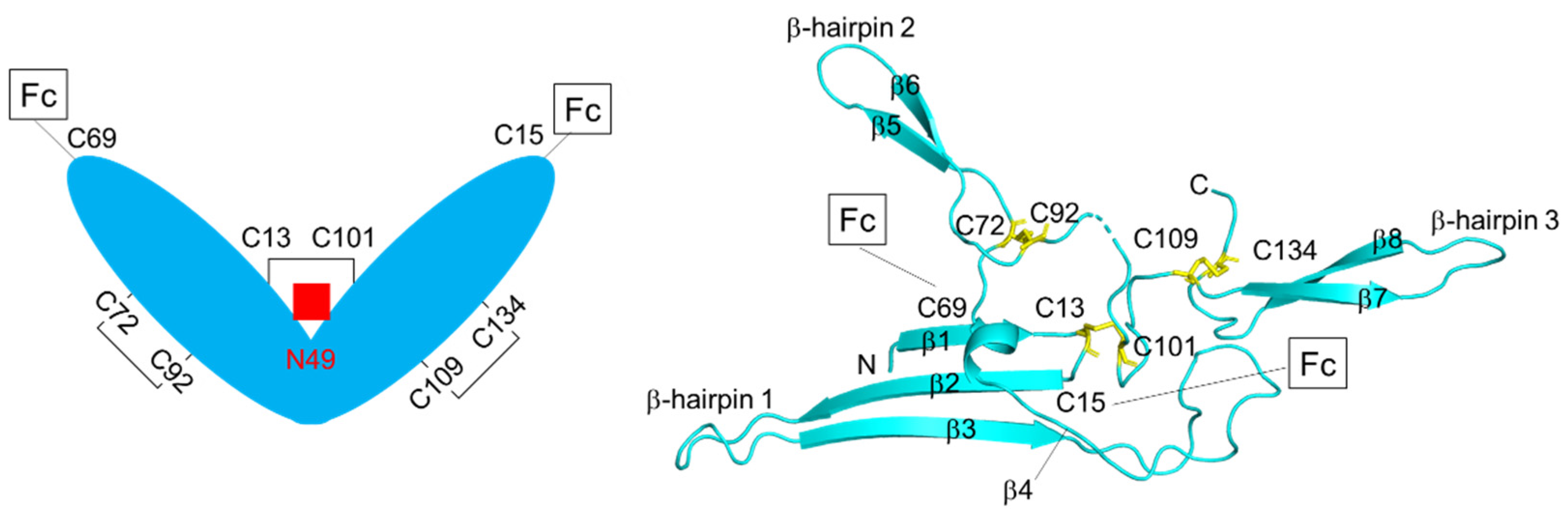
4. Structure of J-chain
5. Structure of Secretory Component
6. Interaction with Fc Receptors and Others
7. Therapeutic Potential of IgA and IgM
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Macpherson, A.J.; Gatto, D.; Sainsbury, E.; Harriman, G.R.; Hengartner, H.; Zinkernagel, R.M. A Primitive T Cell-Independent Mechanism of Intestinal Mucosal IgA Responses to Commensal Bacteria. Science 2000, 288, 2222–2226. [Google Scholar] [CrossRef]
- Bakema, J.E.; Van Egmond, M. Immunoglobulin A: A next generation of therapeutic antibodies? In mAbs; Taylor & Francis: Abingdon, UK, 2011; Volume 3, pp. 352–361. [Google Scholar]
- De Sousa-Pereira, P.; Woof, J.M. IgA: Structure, Function, and Developability. Antibodies 2019, 8, 57. [Google Scholar] [CrossRef] [Green Version]
- Michaud, E.; Mastrandrea, C.; Rochereau, N.; Paul, S. Human Secretory IgM: An Elusive Player in Mucosal Immunity. Trends Immunol. 2020, 41, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Kaetzel, C.S. The polymeric immunoglobulin receptor: Bridging innate and adaptive immune responses at mucosal surfaces. Immunol. Rev. 2005, 206, 83–99. [Google Scholar] [CrossRef]
- Mestecky, J.; Russell, M.; Jackson, S.; Brown, T.A. The human IgA system a reassessment. Clin. Immunol. Immunopathol. 1986, 41, 105–114. [Google Scholar] [CrossRef]
- Yoo, E.M.; Morrison, S.L. IgA: An immune glycoprotein. Clin. Immunol. 2005, 116, 3–10. [Google Scholar] [CrossRef]
- Leusen, J.H. IgA as therapeutic antibody. Mol. Immunol. 2015, 68, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Brandtzaeg, P. Role of J chain and secretory component in receptor-mediated glandular and hepatic transport of immunoglobulins in man. Scand. J. Immunol. 2010, 22, 111–146. [Google Scholar] [CrossRef]
- Putnam, F.W.; Liu, Y.S.; Low, T.L. Primary structure of a human IgA1 immunoglobulin. IV. Streptococcal IgA1 protease, digestion, Fab and Fc fragments, and the complete amino acid sequence of the alpha 1 heavy chain. J. Biol. Chem. 1979, 254, 2865–2874. [Google Scholar] [CrossRef]
- Sørensen, V.; Sundvold, V.; Michaelsen, T.E.; Sandlie, I. Polymerization of IgA and IgM: Roles of Cys309/Cys414 and the secretory tailpiece. J. Immunol. 1999, 162, 3448–3455. [Google Scholar] [PubMed]
- Woof, J.M.; Russell, M.W. Structure and function relationships in IgA. Mucosal Immunol. 2011, 4, 590–597. [Google Scholar] [CrossRef] [Green Version]
- Chintalacharuvu, K.R.; Raines, M.; Morrison, S.L. Divergence of human alpha-chain constant region gene sequences. A novel recombinant alpha 2 gene. J. Immunol. 1994, 152, 5299–5304. [Google Scholar]
- Chintalacharuvu, K.R.; Morrison, S.L. Residues critical for H-L disulfide bond formation in human IgA1 and IgA2. J. Immunol. 1996, 157, 3443–3449. [Google Scholar] [PubMed]
- Toraño, A.; Putnam, F.W. Complete amino acid sequence of the α2 heavy chain of a human IgA2 immunoglobulin of the A2m(2) allotype. Proc. Natl. Acad. Sci. USA 1978, 75, 966–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toraño, A.; Tsuzukida, Y.; Liu, Y.; Putnam, F.W. Location and Structural Significance of the Oligosaccharides in Human IgA1 and IgA2 Immunoglobulins. Proc. Natl. Acad. Sci. USA 1977, 74, 2301–2305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chintalacharuvu, K.R.; Yu, L.J.; Bhola, N.; Kobayashi, K.; Fernandez, C.Z.; Morrison, S.L. Cysteine Residues Required for the Attachment of the Light Chain in Human IgA2. J. Immunol. 2002, 169, 5072–5077. [Google Scholar] [CrossRef] [Green Version]
- Norderhaug, I.N.; Johansen, F.E.; Schjerven, H.; Brandtzaeg, P. Regulation of the formation and external transport of secretory immunoglobulins. Crit. Rev. Immunol. 1999, 19, 481–508. [Google Scholar]
- Stadtmueller, B.M.; Huey-Tubman, K.E.; Lopez, C.J.; Yang, Z.; Hubbell, W.L.; Bjorkman, P.J. The structure and dynamics of secretory component and its interactions with polymeric immunoglobulins. eLife 2016, 5, e10640. [Google Scholar] [CrossRef]
- Dourmashkin, R.R.; Virella, G.; Parkhouse, R.M. Electron microscopy of human and mouse myeloma serum IgA. J. Mol. Biol. 1971, 56, 207–208. [Google Scholar] [CrossRef]
- Kumar, N.; Arthur, C.; Ciferri, C.; Matsumoto, M.L. Structure of the secretory immunoglobulin A core. Science 2020, 368, 1008–1014. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, G.; Li, Y.; Zhu, Q.; Shen, H.; Gao, N.; Xiao, J. Structural insights into secretory immunoglobulin A and its interaction with a pneumococcal adhesin. Cell Res. 2020, 30, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Herr, A.B.; Ballister, E.; Bjorkman, P.J. Insights into IgA-mediated immune responses from the crystal structures of human FcαRI and its complex with IgA1-Fc. Nature 2003, 423, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Ramsland, P.A.; Willoughby, N.; Trist, H.M.; Farrugia, W.; Hogarth, P.M.; Fraser, J.D.; Wines, B.D. Structural basis for evasion of IgA immunity by Staphylococcus aureus revealed in the complex of SSL7 with Fc of human IgA1. Proc. Natl. Acad. Sci. USA 2007, 104, 15051–15056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar Bharathkar, S.; Parker, B.W.; Malyutin, A.G.; Haloi, N.; Huey-Tubman, K.E.; Tajkhorshid, E.; Stadtmueller, B.M. The structures of secretory and dimeric immunoglobulin A. eLife 2020, 9, e56098. [Google Scholar] [CrossRef]
- Kawamura, S.; Saitou, N.; Ueda, S. Concerted evolution of the primate immunoglobulin alpha-gene through gene conversion. J. Biol. Chem. 1992, 267, 7359–7367. [Google Scholar] [CrossRef]
- Royle, L.; Roos, A.; Harvey, D.J.; Wormald, M.R.; van Gijlswijk-Janssen, D.; Redwan, E.R.M.; Wilson, I.A.; Daha, M.R.; Dwek, R.A.; Rudd, P.M. Secretory IgA N- and O-glycans provide a link between the innate and adaptive immune systems. J. Biol. Chem. 2003, 278, 20140–20153. [Google Scholar] [CrossRef]
- Field, M.C.; Amatayakul-Chantler, S.; Rademacher, T.W.; Rudd, P.M.; Dwek, R.A. Structural analysis of the N-glycans from human immunoglobulin A1: Comparison of normal human serum immunoglobulin A1 with that isolated from patients with rheumatoid arthritis. Biochem. J. 1994, 299 Pt 1, 261–275. [Google Scholar] [CrossRef] [Green Version]
- Hui, G.K.; Wright, D.W.; Vennard, O.L.; Rayner, L.E.; Pang, M.; Yeo, S.C.; Gor, J.; Molyneux, K.; Barratt, J.; Perkins, S.J. The solution structures of native and patient monomeric human IgA1 reveal asymmetric extended structures: Implications for function and IgAN disease. Biochem. J. 2015, 471, 167–185. [Google Scholar] [CrossRef] [Green Version]
- Maurer, M.A.; Meyer, L.; Bianchi, M.; Turner, H.L.; Le, N.P.L.; Steck, M.; Wyrzucki, A.; Orlowski, V.; Ward, A.B.; Crispin, M.; et al. Glycosylation of Human IgA Directly Inhibits Influenza A and Other Sialic-Acid-Binding Viruses. Cell Rep. 2018, 23, 90–99. [Google Scholar] [CrossRef] [Green Version]
- Mattu, T.S.; Pleass, R.; Willis, A.C.; Kilian, M.; Wormald, M.R.; Lellouch, A.C.; Rudd, P.M.; Woof, J.M.; Dwek, R.A. The Glycosylation and Structure of Human Serum IgA1, Fab, and Fc Regions and the Role of N-Glycosylation on Fcα Receptor Interactions. J. Biol. Chem. 1998, 273, 2260–2272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woof, J.M.; Burton, D.R. Human antibody-Fc receptor interactions illuminated by crystal structures. Nat. Rev. Immunol. 2004, 4, 89–99. [Google Scholar] [CrossRef]
- Lombana, T.N.; Rajan, S.; Zorn, J.A.; Mandikian, D.; Chen, E.C.; Estevez, A.; Yip, V.; Bravo, D.D.; Phung, W.; Farahi, F.; et al. Production, characterization, and in vivo half-life extension of polymeric IgA molecules in mice. mAbs 2019, 11, 1122–1138. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Cao, X.; Liu, C.; Li, W.; Lu, H. N-glycopeptide Signatures of IgA2 in Serum from Patients with Hepatitis B Virus-related Liver Diseases. Mol. Cell. Proteom. 2019, 18, 2262–2272. [Google Scholar] [CrossRef] [Green Version]
- Yoo, E.M.; Li, J.Y.; Wims, L.A.; Goldberg, D.; Morrison, S.L. Differences in N-glycan structures found on recombinant IgA1 and IgA2 produced in murine myeloma and CHO cell lines. mAbs 2010, 2, 320–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steffen, U.; Koeleman, C.A.; Sokolova, M.V.; Bang, H.; Kleyer, A.; Rech, J.; Unterweger, H.; Schicht, M.; Garreis, F.; Hahn, J. IgA subclasses have different effector functions associated with distinct glycosylation profiles. Nat. Commun. 2020, 11, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grönwall, C.; Vas, J.; Silverman, G.J. Protective Roles of Natural IgM Antibodies. Front. Immunol. 2012, 3, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eskeland, T.; Christensen, T. IgM Molecules with and without J Chain in Serum and after Purification, Studied by Ultra-centrifugation, Electrophoresis, and Electron Microscopy. Scand. J. Immunol. 1975, 4, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Dolder, F. Occurrence, isolation and interchain bridges of natural 7-S immunoglobulin M in human serum. Biochim. Biophys. Acta 1971, 236, 675–685. [Google Scholar]
- Davis, A.C.; Roux, K.H.; Pursey, J.; Shulman, M.J. Intermolecular disulfide bonding in IgM: Effects of replacing cysteine residues in the μ heavy chain. EMBO J. 1989, 8, 2519–2526. [Google Scholar] [CrossRef]
- Pasalic, D.; Weber, B.; Giannone, C.; Anelli, T.; Müller, R.; Fagioli, C.; Felkl, M.; John, C.; Mossuto, M.F.; Becker, C.F.W.; et al. A peptide extension dictates IgM assembly. Proc. Natl. Acad. Sci. USA 2017, 114, E8575–E8584. [Google Scholar] [CrossRef] [Green Version]
- Keyt, B.A.; Baliga, R.; Sinclair, A.M.; Carroll, S.F.; Peterson, M.S. Structure, Function, and Therapeutic Use of IgM Antibodies. Antibodies 2020, 9, 53. [Google Scholar] [CrossRef]
- Gall, W.E.; Edelman, G.M. The covalent structure of a human γG-immunoglobulin. X. Intrachain disulfide bonds. Biochemistry 1970, 9, 3188–3196. [Google Scholar] [CrossRef] [PubMed]
- Feinstein, A.; Munn, E. Conformation of the free and antigen-bound IgM antibody molecules. Nature 1969, 224, 1307–1309. [Google Scholar] [CrossRef]
- Davis, A.C.; Roux, K.H.; Shulman, M.J. On the structure of polymeric IgM. Eur. J. Immunol. 1988, 18, 1001–1008. [Google Scholar] [CrossRef]
- Li, Y.; Wang, G.; Li, N.; Wang, Y.; Zhu, Q.; Chu, H.; Wu, W.; Tan, Y.; Yu, F.; Su, X.D.; et al. Structural insights into immunoglobulin M. Science 2020, 367, 1014–1017. [Google Scholar] [CrossRef]
- Hiramoto, E.; Tsutsumi, A.; Suzuki, R.; Matsuoka, S.; Arai, S.; Kikkawa, M.; Miyazaki, T. The IgM pentamer is an asymmetric pentagon with an open groove that binds the AIM protein. Sci. Adv. 2018, 4, eaau1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colucci, M.; Stöckmann, H.; Butera, A.; Masotti, A.; Baldassarre, A.; Giorda, E.; Petrini, S.; Rudd, P.M.; Sitia, R.; Emma, F. Sialylation of N-linked glycans influences the immunomodulatory effects of IgM on T cells. J. Immunol. 2015, 194, 151–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muraoka, S.; Shulman, M.J. Structural requirements for IgM assembly and cytolytic activity. Effects of mutations in the oligosaccharide acceptor site at Asn402. J. Immunol. 1989, 142, 695–701. [Google Scholar]
- Moh, E.S.; Lin, C.; Thaysen-Andersen, M.; Packer, N.H. Site-Specific N-Glycosylation of Recombinant Pentameric and Hexameric Human IgM. Am. Soc. Mass Spectrom. 2016, 27, 1443–1455. [Google Scholar] [CrossRef]
- Mestecky, J.Z.J.; Zikan, J.; Butler, W.T. Immunoglobulin M and Secretory Immunoglobulin A: Presence of a Common Polypeptide Chain Different from Light Chains. Science 1971, 171, 1163–1165. [Google Scholar] [CrossRef]
- Weinheimer, P.F.; Mestecky, J.; Acton, R.T. Species distribution of J chain. J. Immunol. 1971, 107, 1211. [Google Scholar] [PubMed]
- Frutiger, S.; Hughes, G.J.; Paquet, N.; Lüthy, R.; Jaton, J.C. Disulfide bond assignment in human J chain and its covalent pairing with immunoglobulin M. Biochemistry 1992, 31, 12643–12647. [Google Scholar] [CrossRef]
- Kumar, N.; Arthur, C.P.; Ciferri, C.; Matsumoto, M.L. Structure of the human secretory immunoglobulin M core. Structure 2021, 29, 564–571. [Google Scholar] [CrossRef]
- Baenziger, J.U. Structure of the oligosaccharide of human J chain. J. Biol. Chem. 1979, 254, 4063–4071. [Google Scholar] [CrossRef]
- Krugmann, S.; Pleass, R.J.; Atkin, J.D.; Woof, J.M. Structural requirements for assembly of dimeric IgA probed by site-directed mutagenesis of J chain and a cysteine residue of the α-chain CH2 domain. J. Immunol. 1997, 159, 244–249. [Google Scholar]
- Brandtzaeg, P.; Prydz, H. Direct evidence for an integrated function of J chain and secretory component in epithelial transport of immunoglobulins. Nature 1984, 311, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Mostov, K.E.; Friedlander, M.; Blobel, G. The receptor for transepithelial transport of IgA and IgM contains multiple immunoglobulin-like domains. Nature 1984, 308, 37–43. [Google Scholar] [CrossRef]
- Frutiger, S.; Hughes, G.J.; Hanly, W.C.; Kingzette, M.; Jaton, J.C. The amino-terminal domain of rabbit secretory component is responsible for noncovalent binding to immunoglobulin A dimers. J. Biol. Chem. 1986, 261, 16673–16681. [Google Scholar] [CrossRef]
- Hamburger, A.E.; West, A.P.; Bjorkman, P.J. Crystal Structure of a Polymeric Immunoglobulin Binding Fragment of the Human Polymeric Immunoglobulin Receptor. Structure 2004, 12, 1925–1935. [Google Scholar] [CrossRef] [Green Version]
- Hughes, G.J.; Reason, A.J.; Savoy, L.A.; Jaton, J.C.; Frutiger-Hughes, S. Carbohydrate moieties in human secretory component. Biochim. Biophys. Acta-Protein Struct. Mol. Enzymol. 1999, 1434, 86–93. [Google Scholar] [CrossRef]
- Coyne, R.S.; Siebrecht, M.; Peitsch, M.C.; Casanova, J.E. Mutational analysis of polymeric immunoglobulin receptor/ligand interactions. Evidence for the involvement of multiple complementarity determining region (CDR)-like loops in receptor domain I. J. Biol. Chem. 1994, 269, 31620–31625. [Google Scholar] [CrossRef]
- Longet, S.; Miled, S.; Lötscher, M.; Miescher, S.M.; Zuercher, A.W.; Corthésy, B. Human plasma-derived polymeric IgA and IgM antibodies associate with secretory component to yield biologically active secretory-like antibodies. J. Biol. Chem. 2013, 288, 4085–4094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteiro, R.; Kubagawa, H.; Cooper, M.D. Cellular distribution, regulation, and biochemical nature of an Fcα receptor in humans. J. Exp. Med. 1990, 171, 597–613. [Google Scholar] [CrossRef] [PubMed]
- Aleyd, E.; Heineke, M.H.; Egmond, M.V. The era of the immunoglobulin A Fc receptor FcRI; its function and potential as target in disease. Immunol. Rev. 2015, 268, 123–138. [Google Scholar] [CrossRef]
- Egmond, M.V.; Bakema, J.E.; Woof, J.M. Fc Receptors in Mucosal Immunology; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Xue, J.; Zhu, L.; Zhang, W.; Zhao, Q. Deglycosylation of FcαR at N58 increases its binding to IgA. Glycobiology 2010, 20, 905–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wines, B.D.; Sardjono, C.T.; Trist, H.M.; Lay, C.S.; Hogarth, P.M. The interaction of FcαRI with IgA and its implications for ligand binding by immunoreceptors of the leukocyte receptor cluster. J. Immunol. 2001, 166, 1781–1789. [Google Scholar] [CrossRef] [Green Version]
- Herr, A.B.; White, C.L.; Milburn, C.; Wu, C.; Bjorkman, P.J. Bivalent Binding of IgA1 to FcαRI Suggests a Mechanism for Cytokine Activation of IgA Phagocytosis. J. Mol. Biol. 2003, 327, 645–657. [Google Scholar] [CrossRef]
- Göritzer, K.; Turupcu, A.; Maresch, D.; Novak, J.; Strasser, R. Distinct Fcα receptor N-glycans modulate the binding affinity to immunoglobulin A (IgA) antibodies. J. Biol. Chem. 2019, 294, 13995–14008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, N.; Shibuya, K.; Shimizu, Y.; Yotsumoto, K.; Miyabayashi, T.; Sakano, S.; Tsuji, T.; Nakayama, E.; Nakauchi, H.; Shibuya, A. A novel Fc receptor for IgA and IgM is expressed on both hematopoietic and non-hematopoietic tissues. Eur. J. Immunol. 2015, 31, 1310–1316. [Google Scholar] [CrossRef]
- Kinet, J.-P.; Launay, P. Fcα/μR: Single member or first born in the family? Nat. Immunol. 2000, 31, 1310–1316. [Google Scholar] [CrossRef]
- Cho, Y.; Usui, K.; Honda, S.I.; Tahara-Hanaoka, S.; Shibuya, K.; Shibuya, A. Molecular characteristics of IgA and IgM Fc binding to the Fcα/μR. Biochem. Biophys. Res. Commun. 2006, 345, 474–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibuya, A.; Sakamoto, N.; Shimizu, Y.; Shibuya, K.; Osawa, M.; Hiroyama, T.; Eyre, H.J.; Sutherland, G.R.; Endo, Y.; Fujita, T. Fcα/μ receptor mediates endocytosis of IgM-coated microbes. Nat. Immunol. 2000, 1, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, Y.; Honda, S.I.; Yotsumoto, K.; Tahara-Hanaoka, S.; Eyre, H.J.; Sutherland, G.R.; Endo, Y.; Shibuya, K.; Koyama, A.; Nakauchi, H. Fcα/μ receptor is a single gene-family member closely related to polymeric immunoglobulin receptor encoded on Chromosome 1. Immunogenetics 2001, 53, 709–711. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, M.; Bjorkman, P. Fc Receptors and Their interactions with immunolobulins. Annu. Rev. Cell Dev. Biol. 1996, 12, 181. [Google Scholar] [CrossRef] [Green Version]
- Konstantin, A.; Lorenza, B.; Jürgen, K.; Torsten, S. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar]
- Yang, X.; Zhao, Q.; Zhu, L.; Zhang, W. The three complementarity-determining region-like loops in the second extracellular domain of human Fc alpha/mu receptor contribute to its binding of IgA and IgM. Immunobiology 2013, 218, 798–809. [Google Scholar] [CrossRef]
- Cho, Y.; Honda, S.; Yoshizawa, Y.; Takagaki, K.; Usui, K.; Shibuya, A. Requirement of the cytoplasmic portion for dimer formation of Fcα/μ receptor expressed on cell surface. Mol. Immunol. 2010, 47, 878–882. [Google Scholar] [CrossRef]
- Yoo, E.M.; Trinh, K.R.; Lim, H.; Wims, L.A.; Morrison, S.L. Characterization of IgA and IgM binding and internalization by surface-expressed human Fcα/μ receptor. Mol. Immunol. 2011, 48, 1818–1826. [Google Scholar] [CrossRef]
- Kikuno, K.; Kang, D.W.; Tahara, K.; Torii, I.; Kubagawa, H.; Ho, K.; Baudino, L.; Nishizaki, N.; Shibuya, A.; Kubagawa, H. Unusual biochemical features and follicular dendritic cell expression of human Fcα/μ receptor. Eur. J. Immunol. 2007, 37, 3540–3550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resnick, D.; Pearson, A.; Krieger, M. The SRCR superfamily: A family reminiscent of the Ig superfamily. Trends Biochem. Sci. 1994, 19, 5–8. [Google Scholar] [CrossRef]
- Arai, S.; Maehara, N.; Iwamura, Y.; Honda, S.I.; Nakashima, K.; Kai, T.; Ogishi, M.; Morita, K.; Kurokawa, J.; Mori, M. Obesity-Associated Autoantibody Production Requires AIM to Retain the Immunoglobulin M Immune Complex on Follicular Dendritic Cells. Cell Rep. 2013, 3, 1187–1198. [Google Scholar] [CrossRef] [Green Version]
- Kai, T.; Tomoko, Y.; Satoko, A.; Toru, M.; Aguila, M.B. Stabilization and Augmentation of Circulating AIM in Mice by Synthesized IgM-Fc. PLoS ONE 2014, 9, e97037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyazaki, T.; Yamazaki, T.; Sugisawa, R.; Gershwin, M.E.; Arai, S. AIM associated with the IgM pentamer: Attackers on stand-by at aircraft carrier. Cell. Mol. Immunol. 2018, 15, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.; Ruprecht, R.M. Immunoglobulin M: An Ancient Antiviral Weapon—Rediscovered. Front. Immunol. 2020, 11, 1943. [Google Scholar] [CrossRef]
- Colombo, M.J.; Abraham, D.; Shibuya, A.; Alugupalli, K.R. B1b lymphocyte-derived antibodies control Borrelia hermsii independent of Fcα/μ receptor and in the absence of host cell contact. Immunol. Res. 2011, 51, 249–256. [Google Scholar] [CrossRef] [Green Version]
- Blandino, R.; Baumgarth, N. Secreted IgM: New tricks for an old molecule. J. Leukoc. Biol. 2019, 106, 1021–1034. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Hirokami, Y.; Matsuhashi, N.; Takatsuka, H.; Naito, M. Increased Susceptibility of Thymocytes to Apoptosis in Mice Lacking AIM, a Novel Murine Macrophage-derived Soluble Factor Belonging to the Scavenger Receptor Cysteine-rich Domain Superfamily. J. Exp. Med. 1999, 189, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Chappell, P.; Garner, L.; Yan, J.; Metcalfe, C.; Hatherley, D.; Johnson, S.; Robinson, C.; Lea, S.; Brown, M. Structures of CD6 and Its Ligand CD166 Give Insight into Their Interaction. Structure 2015, 23, 1426–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, M.; Kimura, H.; Iwamura, Y.; Arai, S.; Miyazaki, T. Modification of N-glycosylation modulates the secretion and lipolytic function of apoptosis inhibitor of macrophage (AIM). FEBS Lett. 2012, 586, 3569–3574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drugs@FDA: FDA-Approved Drugs. Available online: https://www.accessdata.fda.gov/scripts/cder/daf/ (accessed on 11 October 2020).
- Brandsma, A.M.; Bondza, S.; Evers, M.; Koutstaal, R.; Broeke, T.T. Potent Fc Receptor Signaling by IgA Leads to Superior Killing of Cancer Cells by Neutrophils Compared to IgG. Front. Immunol. 2019, 10, 704. [Google Scholar] [CrossRef] [Green Version]
- Niels, H.; Marjolein, V.E. Monoclonal antibody-mediated killing of tumor cells by neutrophils. Eur. J. Clin. Investig. 2018, 48, e12962. [Google Scholar]
- Steen, L.; Tuk, C.W.; Bakema, J.E.; Kooij, G.; Reijerkerk, A.; Vidarsson, G.; Bouma, G.; Kraal, G.; Vries, H.; Beelen, R. Immunoglobulin A: FcαRI interactions induce neutrophil migration through release of leukotriene B4. Gastroenterology 2009, 137, 2018–2029. [Google Scholar] [CrossRef]
- Otten, M.A.; Rudolph, E.; Dechant, M.; Tuk, C.W.; Reijmers, R.M.; Beelen, R.; Van, D.; Egmond, M.V. Immature Neutrophils Mediate Tumor Cell Killing via IgA but Not IgG Fc Receptors. J. Immunol. 2005, 174, 5472–5480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guettinger, Y.; Barbin, K.; Peipp, M.; Bruenke, J.; Dechant, M.; Horner, H.; Thierschmidt, D.; Valerius, T.; Repp, R.; Fey, G.H. A recombinant bispecific single-chain fragment variable specific for HLA class II and FcαRI (CD89) recruits polymorphonuclear neutrophils for efficient lysis of malignant B lymphoid cells. J. Immunol. 2010, 184, 1210–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baliga, R.; Li, K.; Manlusoc, M.; Hinton, P.; Keyt, B. High Avidity IgM-Based CD20xCD3 Bispecific Antibody (IGM-2323) for Enhanced T-Cell Dependent Killing with Minimal Cytokine Release. Blood 2019, 134 (Suppl. S1), 1574. [Google Scholar] [CrossRef]
- A Safety and Pharmacokinetic Study of IGM-2323 in Subjects with Relapsed/Refractory Non-Hodgkin Lymphoma. Available online: https://clinicaltrials.gov/ct2/show/NCT04082936 (accessed on 11 October 2020).
- Lohse, S.; Meyer, S.; Meulenbroek, L.; Jansen, J.; Nederend, M.; Kretschmer, A.; Klausz, K.; Moginger, U.; Derer, S.; Rosner, T. An Anti-EGFR IgA That Displays Improved Pharmacokinetics and Myeloid Effector Cell Engagement In Vivo. Cancer Res. 2016, 76, 403–417. [Google Scholar] [CrossRef] [Green Version]
- Rouwendal, G.J.; Van, M.M.; Meyer, S.; Reiding, K.R.; Schouten, J.; De, R.G.; Egging, D.F.; Leusen, J.H.; Boross, P.; Wuhrer, M. A comparison of anti-HER2 IgA and IgG1 in vivo efficacy is facilitated by high N-glycan sialylation of the IgA. In mAbs; Taylor & Francis: Abingdon, UK, 2016; Volume 8, pp. 74–86. [Google Scholar]
- Valasek, C.C.F.; Hensel, F.; Ye, P.; Conner, M.A.; Ultee, M.E. Production and Purification of a PER.C6-Expressed IgM Antibody Therapeutic—BioProcess—BioProcess International. BioProcess Int. 2011, 9, 28–37. [Google Scholar]
- Launay, P.; Grossetête, B.; Arcos-Fajardo, M.; Gaudin, E.; Torres, S.P.; Beaudoin, L.; Patey-Mariaud, D.; Lehuen, A.; Monteiro, R.C. Fcα Receptor (CD89) Mediates the Development of Immunoglobulin A (IgA) Nephropathy (Berger’s Disease): Evidence for Pathogenic Soluble Receptor–IgA Complexes in Patients and CD89 Transgenic Mice. J. Exp. Med. 2000, 191, 1999–2010. [Google Scholar] [CrossRef] [Green Version]
- Duchez, S.; Amin, R.; Cogné, N.; Delpy, L.; Sirac, C.; Pascal, V.; Corthésy, B.; Cogné, M. Premature replacement of μ with α immunoglobulin chains impairs lymphopoiesis and mucosal homing but promotes plasma cell maturation. Proc. Natl. Acad. Sci. USA 2010, 107, 3064–3069. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, S.; Manabe, N.; Yamaguchi, Y. 3D Structures of IgA, IgM, and Components. Int. J. Mol. Sci. 2021, 22, 12776. https://doi.org/10.3390/ijms222312776
Pan S, Manabe N, Yamaguchi Y. 3D Structures of IgA, IgM, and Components. International Journal of Molecular Sciences. 2021; 22(23):12776. https://doi.org/10.3390/ijms222312776
Chicago/Turabian StylePan, Shunli, Noriyoshi Manabe, and Yoshiki Yamaguchi. 2021. "3D Structures of IgA, IgM, and Components" International Journal of Molecular Sciences 22, no. 23: 12776. https://doi.org/10.3390/ijms222312776





