Abstract
Lignin biosynthesis enzymes form complexes for metabolic channelling during lignification and these enzymes also play an essential role in biotic and abiotic stress response. Cinnamyl alcohol dehydrogenase (CAD) is a vital enzyme that catalyses the reduction of aldehydes to alcohols, which is the final step in the lignin biosynthesis pathway. In the present study, we identified 49 CAD enzymes in five Bambusoideae species and analysed their phylogenetic relationships and conserved domains. Expression analysis of Moso bamboo PheCAD genes in several developmental tissues and stages revealed that among the PheCAD genes, PheCAD2 has the highest expression level and is expressed in many tissues and PheCAD1, PheCAD6, PheCAD8 and PheCAD12 were also expressed in most of the tissues studied. Co-expression analysis identified that the PheCAD2 positively correlates with most lignin biosynthesis enzymes, indicating that PheCAD2 might be the key enzyme involved in lignin biosynthesis. Further, more than 35% of the co-expressed genes with PheCADs were involved in biotic or abiotic stress responses. Abiotic stress transcriptomic data (SA, ABA, drought, and salt) analysis identified that PheCAD2, PheCAD3 and PheCAD5 genes were highly upregulated, confirming their involvement in abiotic stress response. Through yeast two-hybrid analysis, we found that PheCAD1, PheCAD2 and PheCAD8 form homo-dimers. Interestingly, BiFC and pull-down experiments identified that these enzymes form both homo- and hetero- dimers. These data suggest that PheCAD genes are involved in abiotic stress response and PheCAD2 might be a key lignin biosynthesis pathway enzyme. Moreover, this is the first report to show that three PheCAD enzymes form complexes and that the formation of PheCAD homo- and hetero- dimers might be tissue specific.
1. Introduction
Lignin is the major component of the secondary cell wall and is the most abundant aromatic compound produced by plants. Lignin is more hydrophobic than cellulose, thereby playing a critical role in water transport in the xylem vessels [1,2]. Due to its rigidity, lignin also plays a crucial role in plant defence [3]. Thus, abiotic and biotic stresses trigger the induction of the lignin polymer synthesis [3]. Lignin is derived from the polymerisation of three phenylpropanoid monomers p-coumaryl, coniferyl and sinapyl alcohols also called p-hydroxyphenyl (H), guaiacyl (G) and syringyl (S) monolignols, respectively [4]. A sequence of eleven phenylpropanoid pathway enzyme families is involved in the biosynthesis of monolignols, starting from the deamination of phenylalanine to monolignol precursors in the cytoplasm (Figure S1) [4]. The composition of monolignols is highly diverse across plant taxa, cell types, developmental stages and cell wall layers [5]. For example, lignin is polymerised from S and G monolignols in angiosperms, whereas in gymnosperms, lignin is composed of purely G lignin [5].
Cinnamyl alcohol dehydrogenase (CAD) is the last enzyme in the lignin biosynthesis pathway, and it reduces sinapaldehyde, coniferaldehyde and p-coumaraldehyde to sinapyl, coniferyl and p-coumaryl alcohols, respectively [6]. CAD enzymes contain catalytic and structural Zn-binding sites and the NADP(H) binding sites [7]. Due to the economic importance and involvement in various biological roles, CAD enzyme has been well studied in several plant species. Earlier studies showed a significant reduction in the lignin content in the natural pine and maize CAD mutants [8,9]. Further, downregulation of CAD genes expression in tobacco, pine and poplar results in the incorporation of sinapaldehyde and coniferaldehyde in place of sinapyl and coniferyl alcohols in the lignin polymer [8,10,11,12]. The incorporation of aldehyde molecules imparts the reddish-brown colour to the stems of both natural and induced CAD mutants in Zea mays, Oryza sativa, pine and poplar [8,9,13]. Similarly, in Arabidopsis double mutant (cad-c cad-d), coniferyl and sinapyl aldehydes were incorporated in place of coniferyl and sinapyl alcohols resulting in a limp floral stem [14]. Moreover, a significant reduction in the stem lignin content was also observed in these natural or induced mutants [8,9,14].
AtCAD-D gene acts as a crucial component for resistance to Pseudomonas syringae pv. Tomato infection [15]. Expression of CAD1 gene in Ipomoea batatas is highly induced by cold, reactive oxygen species and wounding [16]. Overexpression of I. batatas CAD1 and Sedum alfredii CAD gene in Arabidopsis increases oxidative stress tolerance and cadmium stress tolerance, respectively [17,18]. Ginkgo biloba CAD1 gene expression is increased after SA, UV, ethephon and wounding treatment [19]. Similarly, CAD1 and CAD2 genes of Plagiochasma appendiculatum are induced by MeJA treatment [20]. Deng et al. (2013) [21] identified the CsCAD3 gene was highly upregulated after wounding and Ectropis oblique attack. Further, CsCAD1 and CsCAD2 had elevated expression after MeJA and SA treatment. Likewise, the Cucumis melo CmCAD2 gene becomes strongly upregulated after salt, drought and wounding treatments [22].
Bamboo is the fastest-growing plant (7.5–100 cm per day) and is used for human food, animal diet, timber, textile, musical instruments, fuel and paper production [23,24,25]. The industry related to bamboo was estimated to be USD 72.1 billion in 2019 (www.globenewswire.com, accessed on 1 November 2021). The fast growth of bamboo forests promotes more than 30% more high-carbon sequestration than the tropical mountain rainforest and fast-growing Chinese fir, Cunninghamia lanceolata [26,27]. Moso bamboo (Phyllostachys edulis) is the most crucial bamboo in China, covering around 3 million ha, and is the main species for edible shoot, timber and paper production. Moso bamboo is a tetraploid woody plant containing 48 chromosomes (2n = 4X = 48) [28]. Peng et al. (2013) [29] published the first scaffold draft genome for Moso bamboo. Zhao et al. (2018) [30] recently released the chromosome-level reference genome and also provided the transcriptomic data for multiple developmental tissues belonging to different heights of Moso bamboo. Recently, the draft genome sequences of diploid herbaceous bamboo (Raddia guianensis and Olyra latifolia), tetraploid neotropical woody bamboo (Guadua angustifolia) and hexaploid palaeotropical woody bamboo (Bonia amplexicaulis) were published [31].
Lignin biosynthesis enzymes interact with each other and forms complexes for metabolic channelling [32]. Recently Yan et al. (2019) [33] identified that Populus trichocarpa PtrCAD1 forms homo-dimers through BiFC experiments, but it needs more studies in different plant species to characterise the CAD enzyme complexes. Further, the CAD genes response to abiotic or biotic stress data is also lacking in Moso bamboo. Therefore, the availability of whole-genome or draft genome sequences of different bamboo species, different kinds of Moso bamboo tissues and developmental stages transcriptomic data and our laboratory’s unpublished abiotic stress treatment transcriptomic data provided an opportunity to study the CAD gene family in Bambusoideae. In this study, we identified CAD genes belonging to five bamboo species and their phylogenetic relationship and conserved domains. Further, we analysed the PheCAD genes expression in different Moso bamboo tissues and identified the most expressed PheCAD genes, PheCAD1, PheCAD2 and PheCAD8, forms both homo- and hetero- dimers in planta. Then we analysed the response of the PheCAD genes to abiotic stress and the co-expression network and identified the lignin biosynthesis genes co-expressed with PheCAD genes.
2. Results
2.1. Identification of Bambusoideae CAD Enzymes
Our yeast two-hybrid (Y2H) one-to-one interaction experiments to identify the enzyme–enzyme interactions of lignin biosynthesis enzymes identified the PheCAD2 enzyme forms homo-dimers in Moso bamboo. To characterise the Moso bamboo CAD enzymes, we identified them from a P. edulis genome database using the rice CAD protein sequences (downloaded from the Oryzabase) as a reference. The enzymes containing the catalytic Zn-binding site “GHE(X)2G(X)5V”, structural Zn-binding site “GD(X)10C(X)2C(X)7C” and the NADP(H) binding site “G(X)3G(X)2GLGG(X)GH(X)2VK(X)2K(X)2G-(X)VTV(X)S(X)S(X)2K” were considered as CAD enzymes [6]. A total of 14 PheCAD enzymes were identified in the Moso bamboo genome database (Table S1). Further, to study the phylogenesis and domains, we also isolated the CAD enzymes from available bamboo draft genome sequences R. guianensis (2n), O. latifolia (2n), G. angustifolia (4n), and B. amplexicaulis (6n) [31]. A total of 11 RguCAD, seven OlaCAD, eight GanCAD and 16 BamCAD enzymes were identified, respectively (Table S1). Most of the identified CAD enzymes contain intact catalytic and structural Zn-binding sites and the NADP(H) binding sites with minor mutations. Unlike other Bambusoideae species, six of the 11 RguCAD enzymes (RguCAD6-11) have mutations in the NADP(H) binding site (File S1). Further, in angiosperms, polyploidy is considered a vital mechanism for speciation and family expansion [34]. However, the number of CAD enzymes in Bambusoideae is poorly correlated with the polyploidisation, which may be due to the lower sequence and assembly quality of the draft genomes [31].
2.2. Evolution of Bambusoideae CAD Enzymes
To understand the evolution of CAD enzymes, a phylogenetic tree containing 87 CAD enzymes (AtCADs, BamCADs, GanCADs, OlaCADs, OsCADs, PheCADs, PtrCADs and RguCADs) was constructed (Figure 1). The aligned amino acid sequences of CAD enzymes were used to build the phylogenetic tree using both a maximum likelihood and neighbor-joining method with 1000 bootstrap replicates [35]. The phylogenetic tree is divided into Clade I, Clade II, Clade III and Clade IV. Clade I contains more CAD enzymes than other Clades. The evolutionary topologies of the subclades are divided between monocot and dicot CAD enzymes. The OsCAD and diploid herbaceous bamboo OlaCAD, RguCAD enzymes are separated in the subclades compared to tetraploid and other woody bamboo CAD enzymes. Interestingly, the RguCAD6-11 enzymes which have mutations in the NADP(H) binding site are entirely separated in the phylogenetic tree (Figure 1 and Figure S2). These results support that CAD enzymes were present before the divergence of dicot and monocot plants, and also, they evolved separately in the Bambusoideae after polyploidisation.
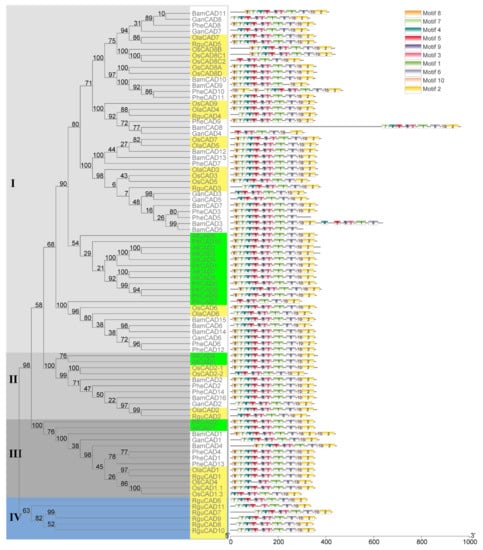
Figure 1.
The phylogenetic relationship among CAD genes of Bambusoideae. The phylogenetic tree was constructed using CAD protein sequences of P. trichocarpa (Ptr), Arabidopsis thaliana (At), Oryza sativa (Os), O. latifolia (Ola), P. edulis (Phe), G. angustifolia (Gan), R. guianensis (Rgu) and B. amplexicaulis (Bam). Clade I, II and III are indicated in different ash shades and Clade IV in blue. The dicot plant CAD genes were indicated in the green boxes and the diploid rice and bamboo species are shown in light yellow boxes. The conserved motifs (1–10) are indicated in ten different colour boxes.
2.3. Identification of Conserved Domains of the CAD Enzymes
To better understand the protein structures of CAD enzymes, we analysed the conserved motifs through MEME software. A total of ten conserved motifs were identified in the CAD enzymes in both monocot and dicot plants analysed in this study. Except for some, all ten motifs were present in the same order for most CAD enzymes (Figure 1). Interestingly, the starting three motifs 8, 7 and 4 are absent in the OsCAD8C2, GanCAD4 and PtrCAD7. The first motif, 8, is missing in 14 CAD enzymes and the starting two motifs 8 and 7 in six CAD enzymes. Further, the last two motifs 10 and 2, are absent in the BamCAD5 and PheCAD5 and duplicated in the PheCAD10 and BamCAD1, whereas motif 6 and 5 are lacking in the BamCAD9 and OsCAD4. Moreover, motifs 4, 5, 9, 3, 1 and 6 are duplicated in the BamCAD3 enzyme (Figure 1 and Figure S2).
2.4. Cis-Acting Elements Responsive to Abiotic Stress
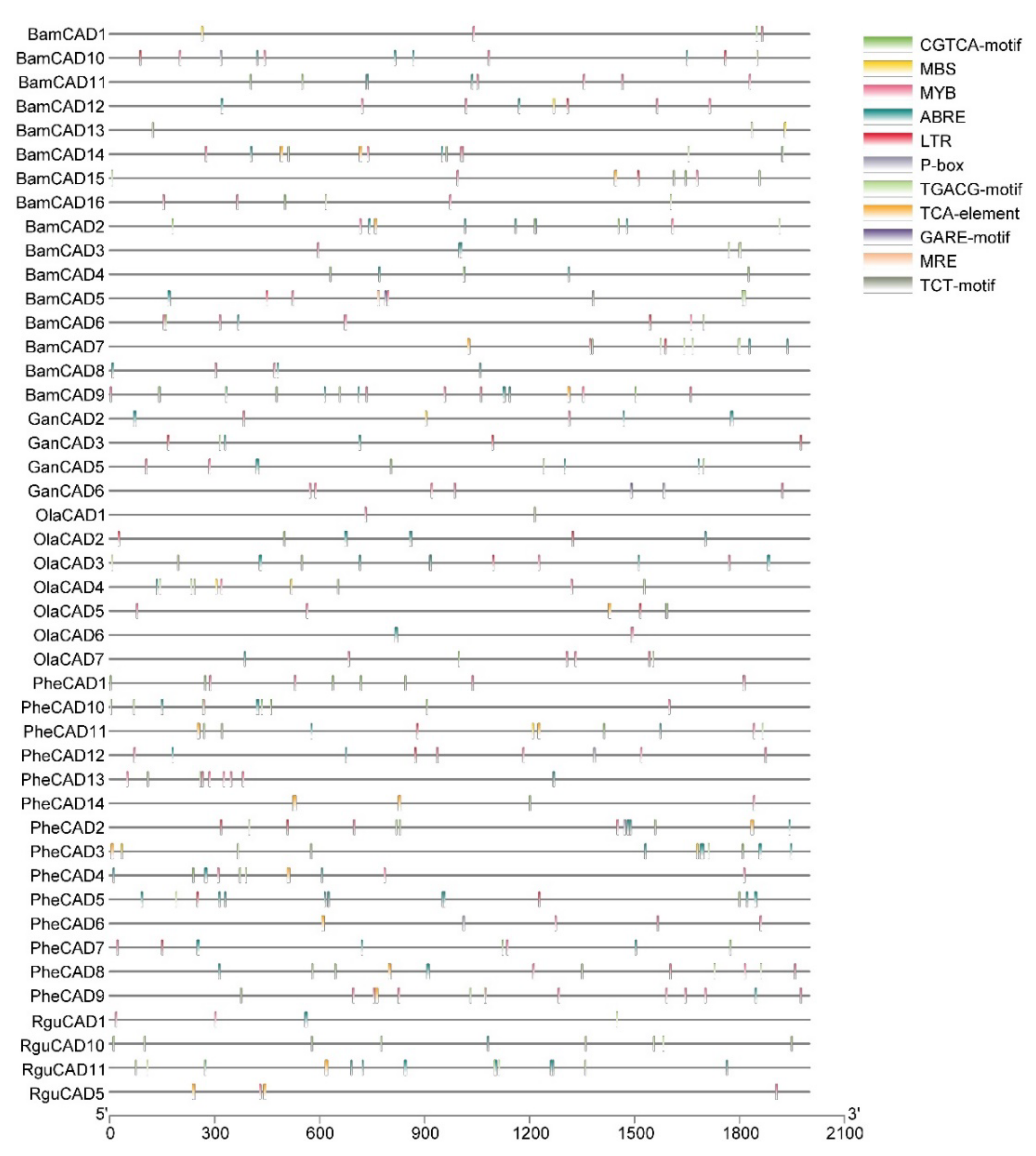
Cis-acting elements trigger the genes involved in abiotic stress. Therefore, studying cis-acting elements in the promotor regions of CAD genes will help to understand their role in abiotic stress response. Hence, we analysed the 2kb upstream region to the PheCAD, BamCAD, GanCAD, OlaCAD and RguCAD genes. Due to the lack of proper genome assembly, we were unable to retrieve all the upstream 2kb regions of GanCAD and RguCAD genes. Further, among the cis-acting elements analysed we focused on ABA, SA and GA-responsive and other important cis-elements such as ABRE, CGTCA-motif, GARE-motif, LTR, MBS, MRE, MYB, P-box, TCA-element, TCT-motif and TGACG-motif (Figure 2 and Table S2).
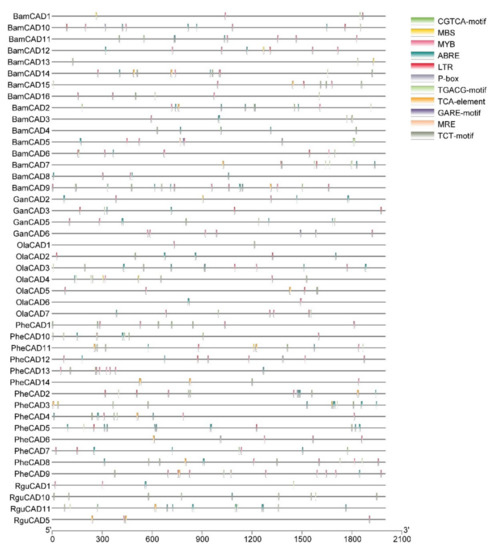
Figure 2.
The conserved cis-elements analysis of CAD genes in the promoter regions of Bambusoideae (ABRE, CGTCA-motif, GARE-motif, LTR, MBS, MRE, MYB, P-box, TCA-element, TCT-motif and TGACG-motif).
2.5. Expression of Moso Bamboo CAD Genes during Plant Development
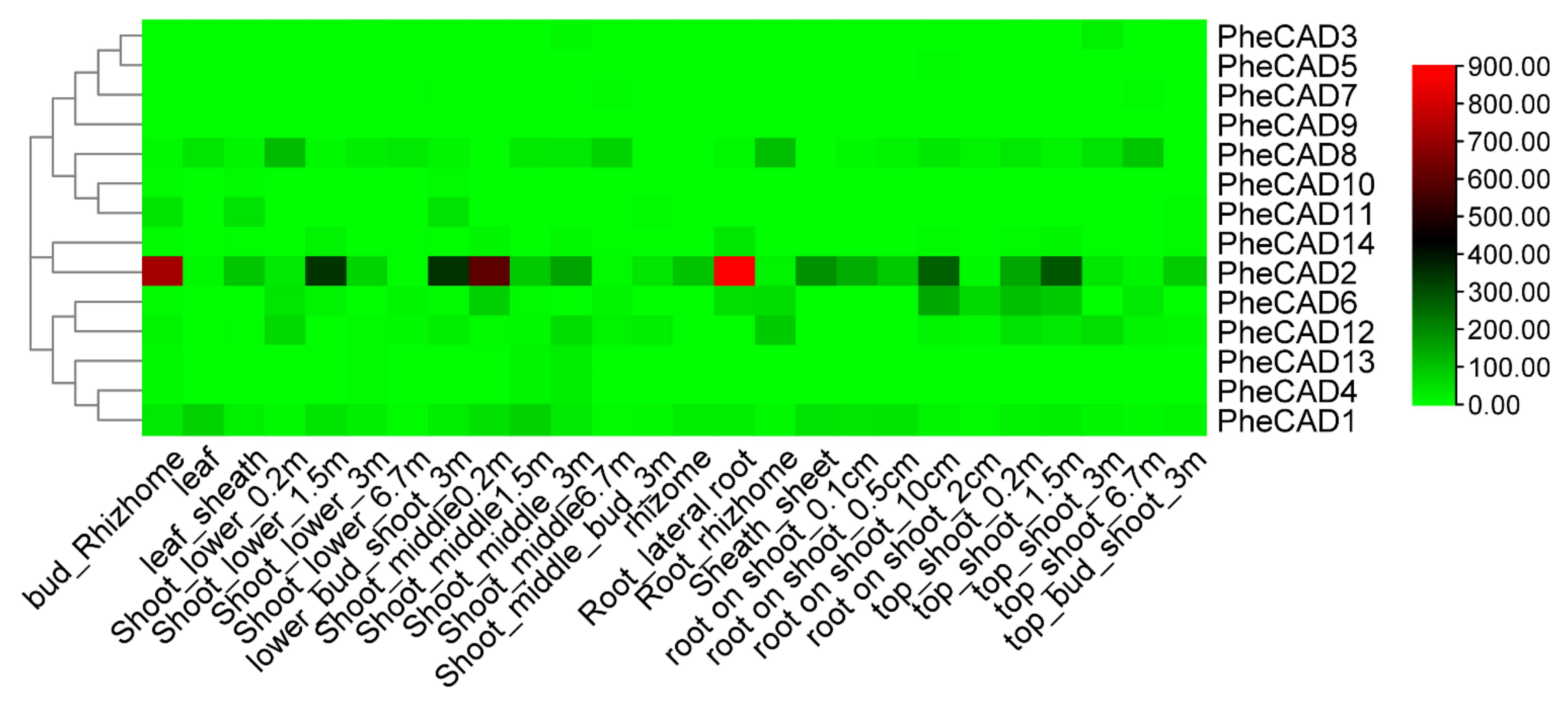
We analysed the transcriptomic data of twenty-six bamboo tissues published by Zhao et al., (2018) [30] and used the FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values of 14 PheCAD genes (Table S3). A heatmap was constructed using the FPKM values to identify the most-expressed genes (Figure 3). The five Moso bamboo CAD genes PheCAD1, PheCAD2, PheCAD6, PheCAD8, and PheCAD12 are expressed in most of the analysed tissues. In these, the PheCAD2 (PH02Gene39617) gene is highly expressed but not in the leaf and 6.7 m height shoot. At the same time, PheCAD8 (PH02Gene13789) is also expressed in most tissues, including leaf and shoot 6.7 m. Further, PheCAD1 (PH02Gene46268) is also expressed in different kinds of tissues, including leaves.
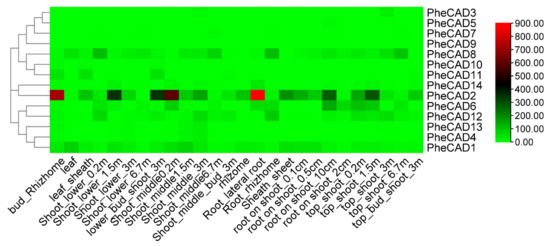
Figure 3.
Expression heatmap of PheCAD genes in 26 different kinds of tissues and stages of bamboo development. The log2 expression values are depicted in colour shades from red (highest) to green (lowest).
2.6. Co-Expression Analysis of PheCAD Genes in Moso Bamboo
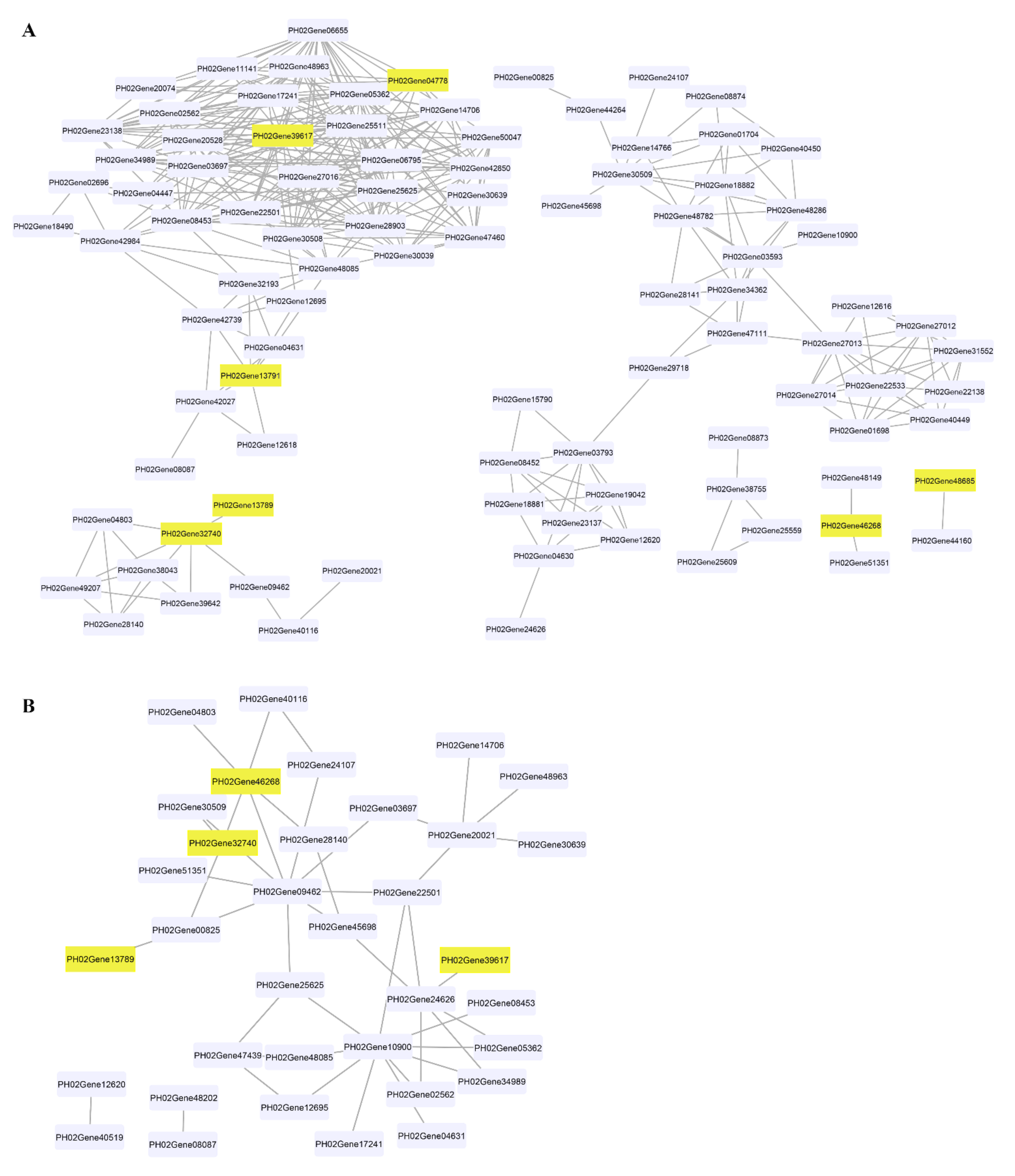
We analysed the gene co-expression network for lignin biosynthesis genes to identify the lignin biosynthesis genes co-expressed with PheCAD genes. A total of 160 lignin biosynthesis genes’ FPKM values in 26 different tissues were used for co-expression analysis (Table S3). The Pearson correlation coefficient (PCC) was calculated between two genes as the correlation coefficient to measure the co-expression relationship. The highest PCC values > 0.7 and lowest PCC values < −0.5 were considered thresholds for positive and negative correlation, respectively (Table S4). Interestingly, the PheCAD2 gene has positive correlation with most of the lignin biosynthesis genes PhePAL (PH02Gene08453, PH02Gene23138, PH02Gene30508 and PH02Gene42984), PheC4H (PH02Gene03697), Phe4CL (PH02Gene34989), PheHCT (PH02Gene06655, PH02Gene25625), PheCCoAOMT (PH02Gene02562), PheCCR (PH02Gene06795, PH02Gene11141, PH02Gene14706, PH02Gene25511, PH02Gene28903, PH02Gene42850, PH02Gene47460, PH02Gene48085 and PH02Gene48963) and PheCOMT (PH02Gene05362, PH02Gene17241) (Figure 4A). These co-expression results indicate that except for the PheC3H, PheCSE and PheF5H genes, all other lignin biosynthesis genes involved in producing H, S and G lignin units were co-expressed with PheCAD2.
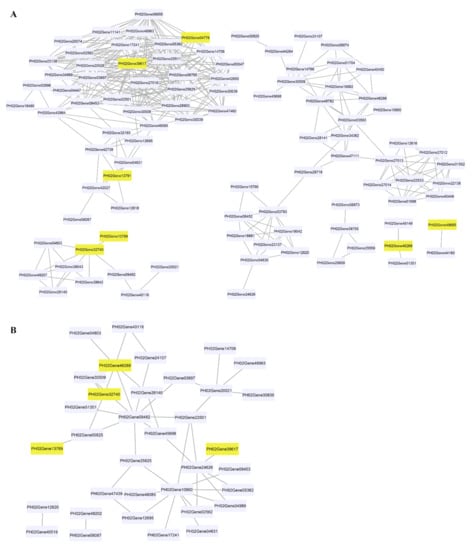
Figure 4.
Co-expression network of PheCAD genes with lignin biosynthesis pathway genes. (A) Positive correlation of PheCAD genes with lignin biosynthesis pathway genes; (B) negative correlation of PheCAD genes with lignin biosynthesis pathway genes. Pink-coloured boxes indicate the PheCAD genes and violet boxes indicate the other lignin biosynthesis genes.
Similarly, PheCAD2 also has a negative correlation with Phe4CL (PH02Gene24626) (Figure 4B), whereas PheCAD1 has a positive correlation with PheCCR (PH02Gene51351, PH02Gene48149) and negative correlation with Phe4CL (PH02Gene09462) and PheCCR (PH02Gene04803, PH02Gene28140, PH02Gene41539). Further, PheCAD8 has a positive correlation with PheCAD7 (PH02Gene32740) and a negative correlation with Phe4CL (PH02Gene00825). At the same time, PheCAD7 has a positive correlation with Phe4CL (PH02Gene09462), PheCSE (PH02Gene39642, PH02Gene49207), PheCCR (PH02Gene04803, PH02Gene28140). Whereas negative correlation with PheCAD1, Phe4CL (PH02Gene00825) and PhePAL (PH02Gene30509). Further, we also analysed the co-expression analysis of PheCAD genes to identify the transcription factors or regulators and other proteins involved in abiotic or biotic stress. Interestingly, more than 35% of the annotated co-expressed genes with PheCAD genes were involved in biotic or abiotic stress response (Table S5).
2.7. PheCAD Genes Response to Abiotic Stress Treatment
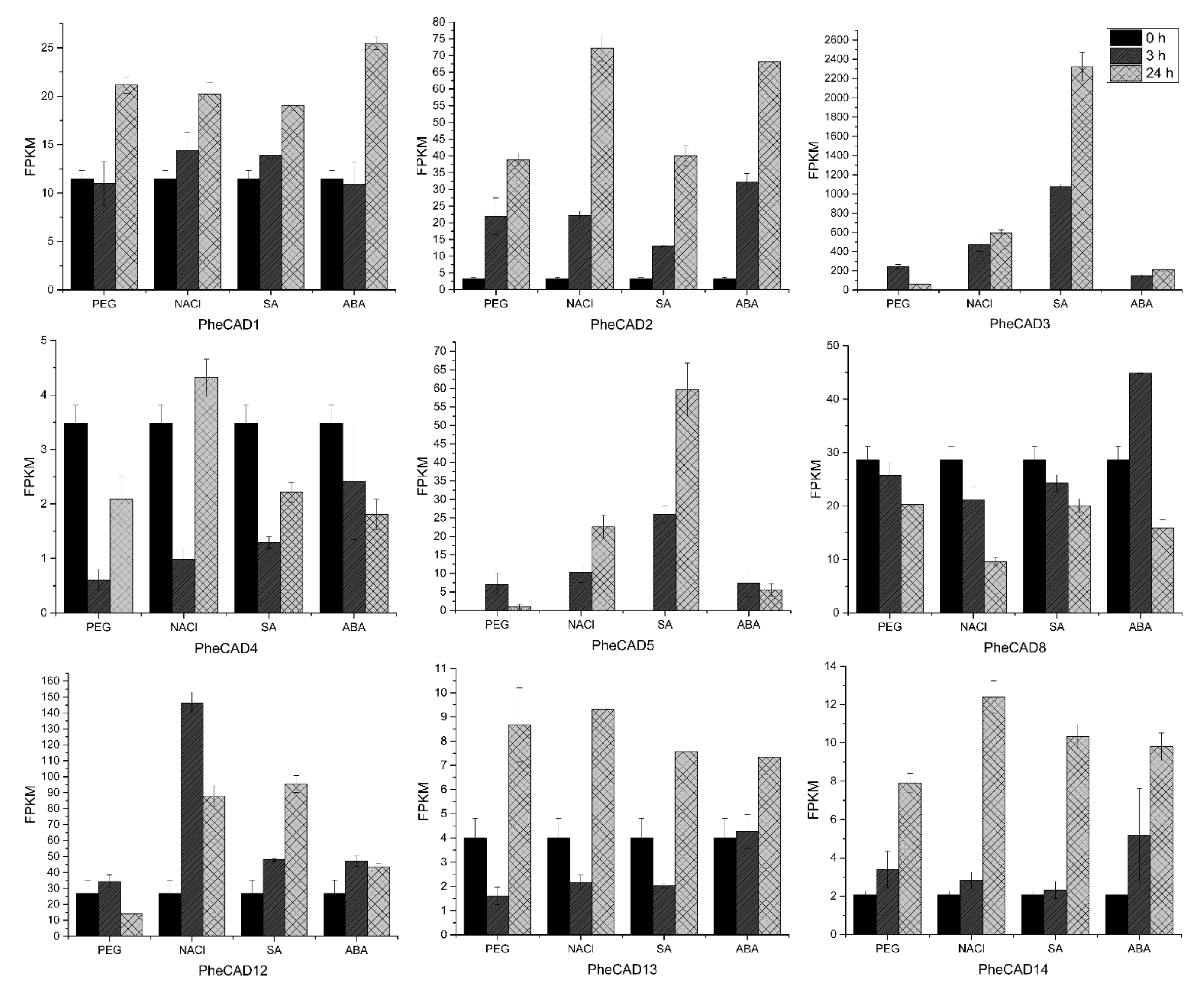
Plant cells respond to abiotic stress by increasing the cell wall thickness, i.e., accumulation of hemicellulose and lignin polymer [36]. Our co-expression analysis studies also indicate that PheCAD genes are involved in biotic or abiotic stress responses. To identify the PheCAD genes involved in response to the abiotic stress, we analysed the transcriptomic data of Moso bamboo seedlings treated with SA, ABA, drought and salt (unpublished data). In this study, the genes with a two-fold change in expression compared to the control after abiotic stress treatment were considered differentially expressed. Among PheCAD1, PheCAD2 and PheCAD8 genes, the PheCAD2 gene was significantly upregulated after all abiotic stress treatments, especially after 24 h of NaCl and ABA treatment (Figure 5).
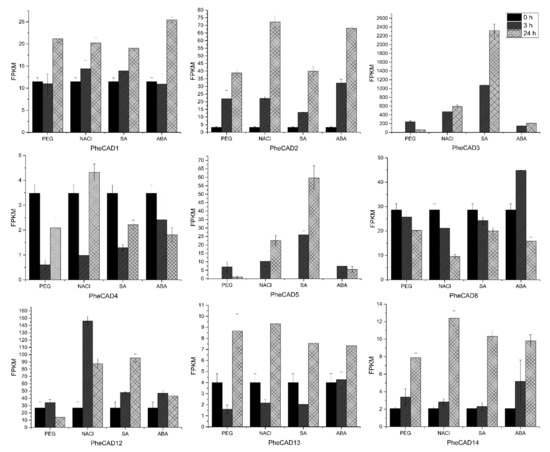
Figure 5.
Expression analysis of PheCAD genes in response to PEG, NaCl, ABA and SA. The FPKM values of transcriptomic data (Moso bamboo seedlings treated with PEG (25%), NaCl (200mM), ABA (1uM), SA (1mM) for 3 and 24 h) were used to develop graphs.
Of other Moso bamboo CAD genes, PheCAD3 and PheCAD5 were highly upregulated after all abiotic stress treatments. After 24 h of NaCl, SA and ABA treatment, the relative expression of PheCAD3 was upregulated to 274, 1069 and 97 times to the control, respectively. After 3 h of PEG treatment, PheCAD3 expression was upregulated to about 111 times that of the control and decreased 29 times after 24 h PEG treatment. Further, the PheCAD5 gene expression was found to be nil in control, but after NaCl and SA treatment, the relative expression was upregulated to about 100 and 250 after 3 h and 226 and 597 times after 24 h, respectively. Whereas after PEG and ABA treatment, the relative expression was upregulated to 70 and 74 times that of control after 3 h but decreased to 10 and 55 times after 24 h, respectively. Interestingly, both PheCAD3 and PheCAD5 are also very closely related in the phylogenetic tree (Figure 1). After 3 h of PEG, NaCl and SA treatment, the PheCAD4 gene expression was downregulated 0.1–0.3 times to control. At the same time, the PheCAD12 gene expression was upregulated 5.5 to 3.3 times of control after 3 and 24 h of NaCl treatment, respectively. In contrast, the expression was upregulated 3.6 times of control after 24 h of SA treatment. Similarly, after 3 h of PEG treatment, the relative expression of PheCAD13 was downregulated 0.4 times that of control, where the expression was upregulated two times after 24 h of PEG and NaCl treatment. The relative expression of PheCAD14 was upregulated 3.8–6 times of control after 24 h of all abiotic treatments (Figure 5). The expression levels of PheCAD6, PheCAD7, PheCAD9, PheCAD10 and PheCAD11 genes were too low for analysis.
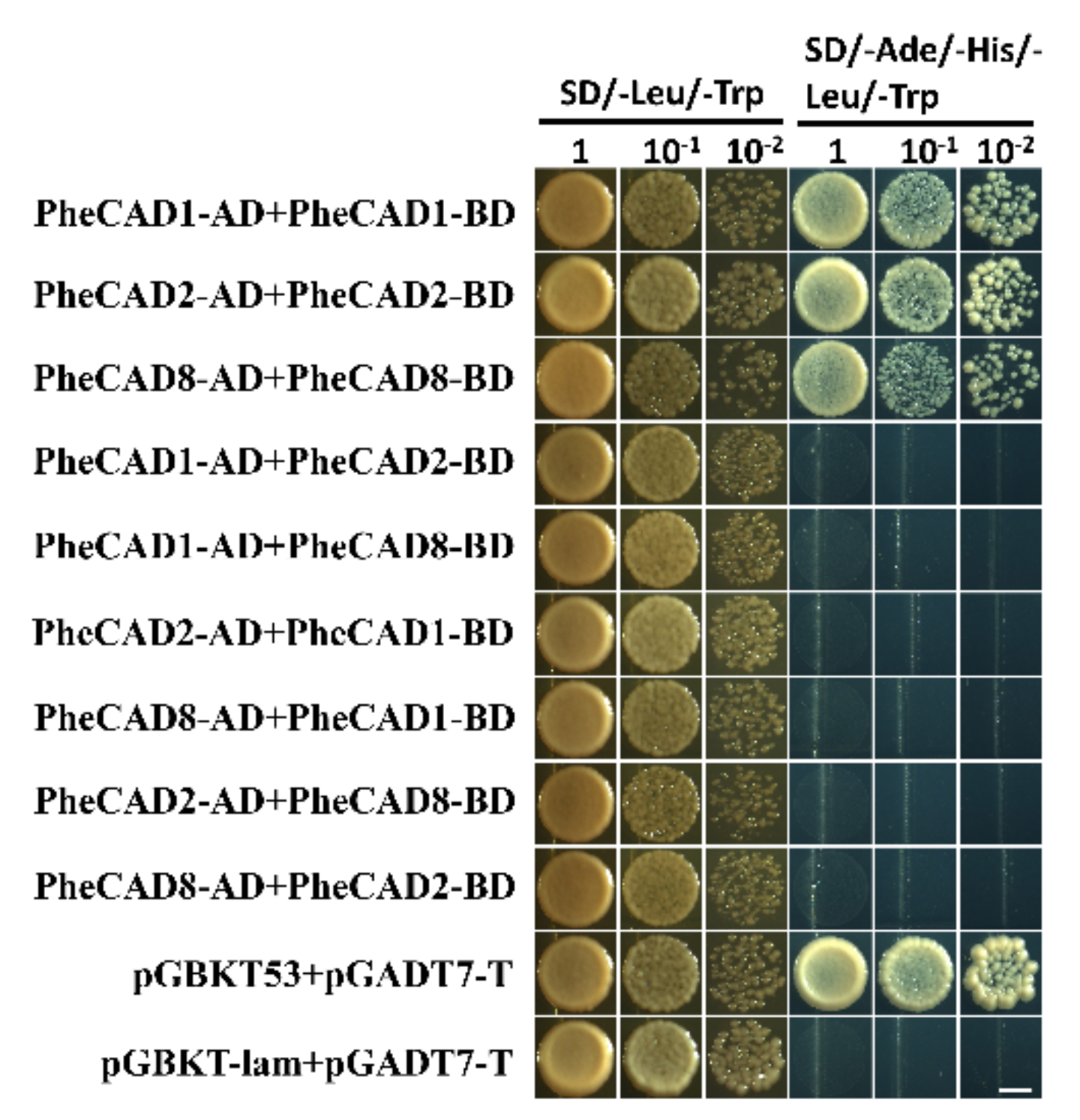
2.8. The PheCAD Enzymes Form Only Homo-Dimers in Yeast
Lignin biosynthesis pathway enzymes form homo- and hetero-dimers and also interact with each other for metabolic channelling [32]. Further, the formation of homo- and hetero-dimers increases the catalytic efficiency compared with individual enzymes [37]. Recently Yan et al., (2019) identified PtrCAD1 forms homo-dimers in P. trichocarpa [33]. Moreover, it was reported that Ptr4CL, PtrC4H and PtrC3H also form homo- and hetero-dimers in P. trichocarpa [37,38]. Therefore, based on the tissue expression analysis, we selected the highly expressed CAD enzymes PheCAD1, PheCAD2 and PheCAD8 for Y2H experiments to confirm whether Moso bamboo CAD genes can also form the homo- or hetero-dimers. The yeast cell cotransformed with pGBKT-7-PheCAD1 and pGADT7-PheCAD1, pGBKT-7-PheCAD2 and pGADT7-PheCAD2, pGBKT-7-PheCAD8 and pGADT7-PheCAD8 and positive control grew normally on SD/–Ade/–His/–Leu/–Trp medium. The yeast cells cotransformed with other combinations, and the negative control failed to grow on SD/–Ade/–His/–Leu/–Trp medium (Figure 6). These results suggest that PheCAD1, PheCAD2 and PheCAD8 enzymes form only homo-dimers and fail to form hetero-dimers in the yeast cells.
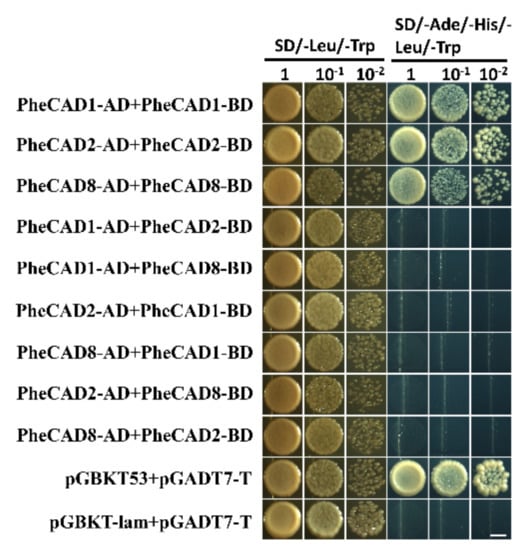
Figure 6.
PheCAD1, PheCAD2 and PheCAD8 enzyme homo-dimers were detected with Y2H experiments. All the combinations of PheCAD1, PheCAD2 and PheCAD8 enzymes were used as both prey and bait. Yeast carrying pGBKT53, pGADT7-T used as positive control and pGBKT-lam, pGADT7-T used as a negative control. The transformed yeast cells grew on growth (SD/-Trp/-His) and selective (SD/-Ade/-His/-Trp/-His) media. Images were taken after 5 days of incubation at 30 °C. Scale bar, 3 mm.
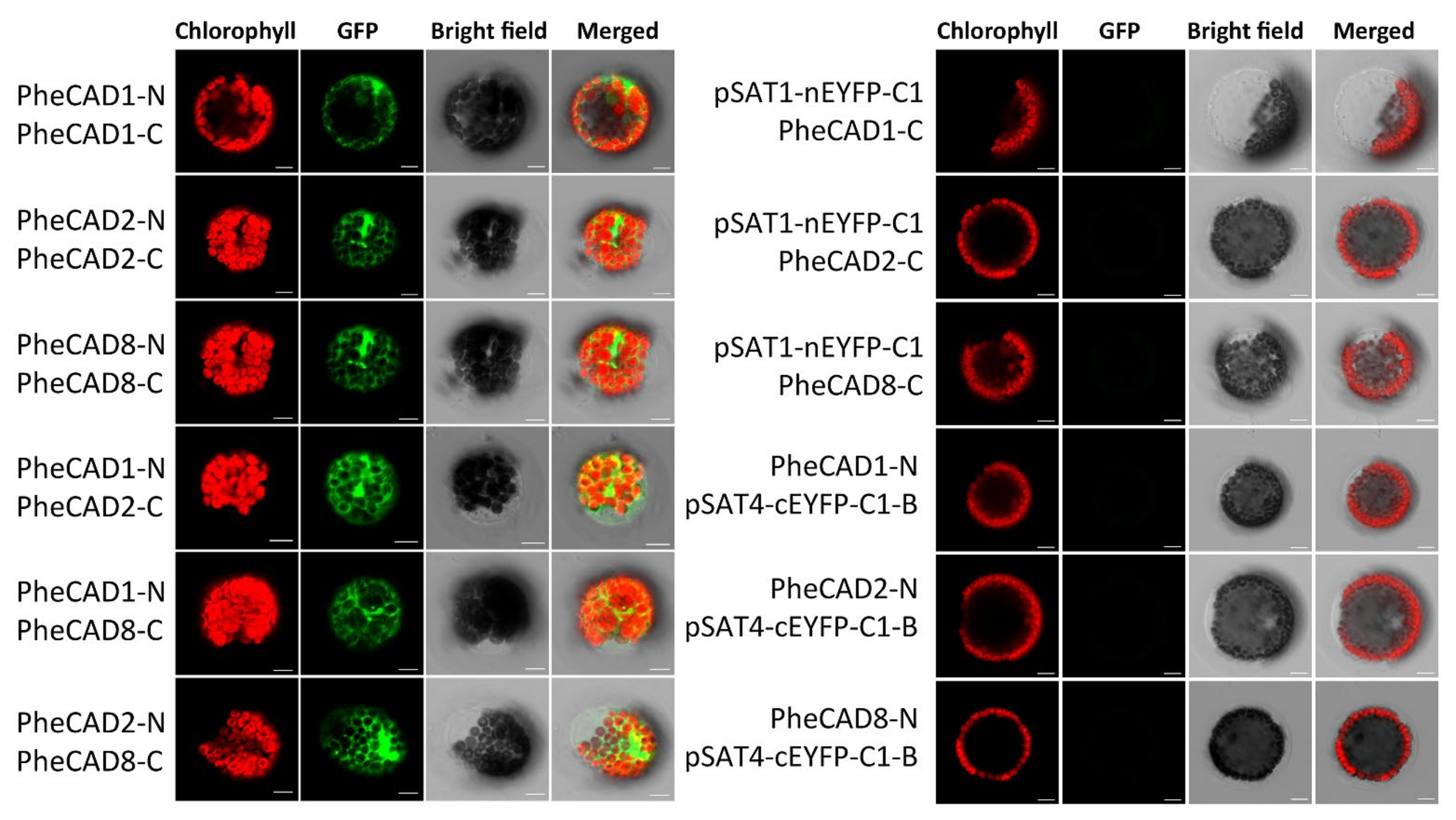
2.9. Formation of Both PheCAD Homo- and Hetero-Dimers in Planta
Bimolecular fluorescence complementation assay (BiFC) in Arabidopsis protoplast cells was performed to confirm the Y2H results. We generated constructs containing PheCAD1, PheCAD2 and PheCAD8 genes fused with both N-terminal and C-terminal fragments of EYFP in the pSAT1-nEYFP-C1 and pSAT4-cEYFP-C1-B vectors, respectively. A strong fluorescence signal in the Arabidopsis protoplast cells was observed when pairwise cotransformation of 35S-PheCAD1:pSAT1-nEYFP-C1 and 35S-PheCAD1:pSAT4-cEYFP-C1-B; 35S-PheCAD2: pSAT1-nEYFP-C1 and 35S-PheCAD2:pSAT4-cEYFP-C1-B and 35S-PheCAD8:pSAT1-nEYFP-C1 and 35S-PheCAD8:pSAT4-cEYFP-C1-B vectors (Figure 7). These results confirmed the Y2H results of the formation of homo-dimers. At the same time, in contrast to Y2H results, a strong signal was also observed in the Arabidopsis protoplast cells when pairwise cotransformation of 35S-PheCAD1:pSAT1-nEYFP-C1 and 35S-PheCAD2:pSAT4-cEYFP-C1-B; 35S-PheCAD1:pSAT1-nEYFP-C1 and 35S-PheCAD8:pSAT4-cEYFP-C1-B; 35S-PheCAD2:pSAT1-nEYFP-C1 and 35S-PheCAD8:pSAT4-cEYFP-C1-B vectors into Arabidopsis protoplast cells (Figure 7). These results indicate that PheCAD1, PheCAD2 and PheCAD8 enzymes interact with each other and can form hetero-dimers in planta.
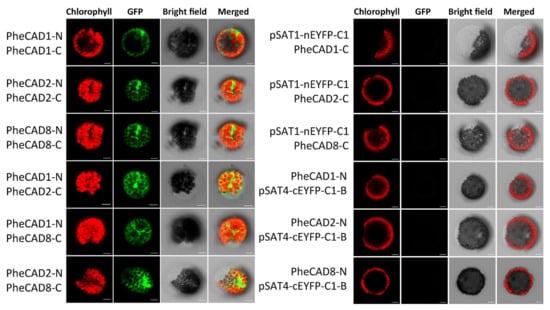
Figure 7.
Pairwise bimolecular fluorescence complementation assay of PheCAD1, PheCAD2 and PheCAD8 enzymes in Arabidopsis protoplast cells. Empty vector and corresponding C-terminal or N-terminal PheCAD genes were used as the negative control. Scale bar, 10 μm.
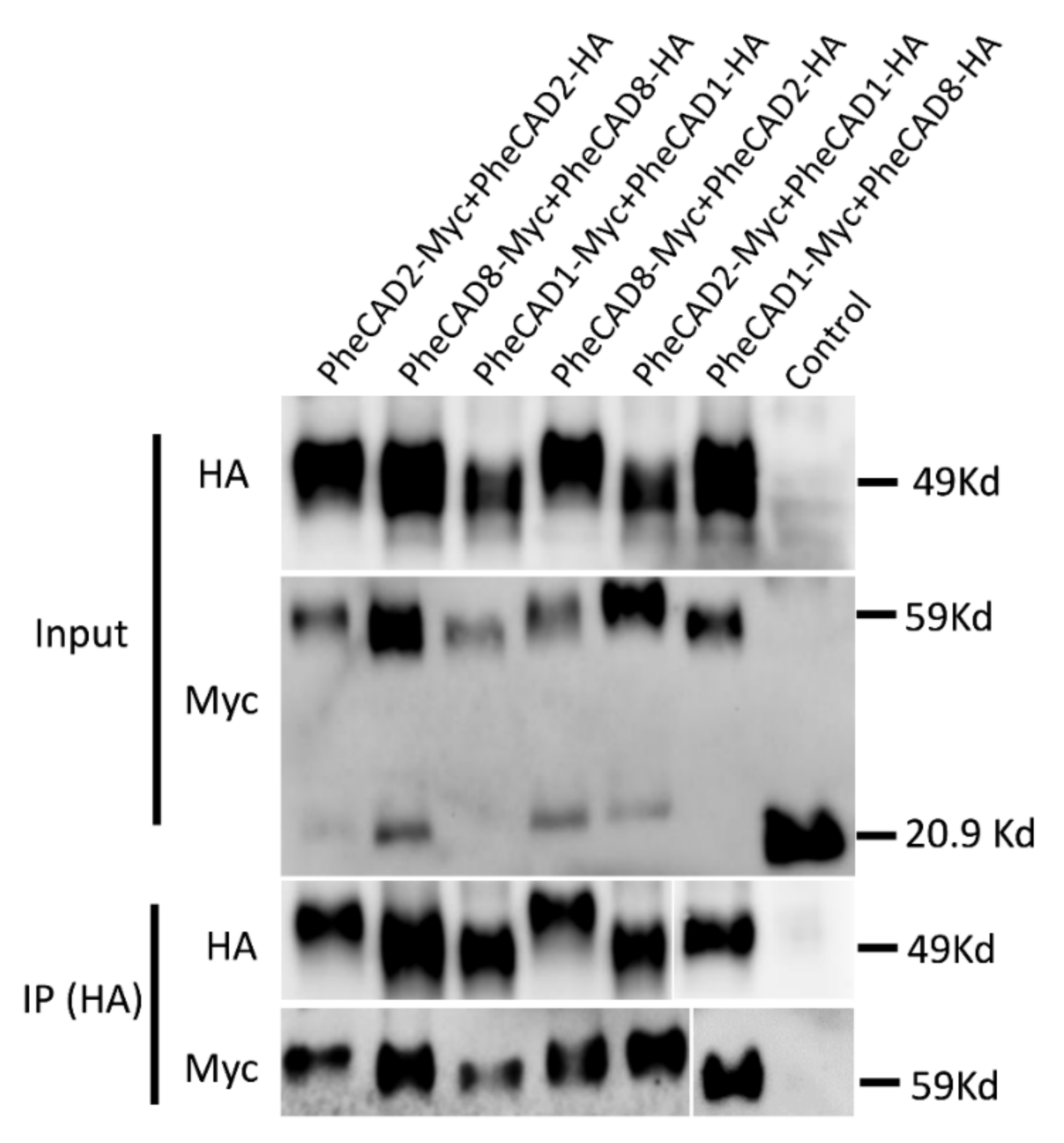
2.10. Confirmation of PheCAD Homo-Dimers and Hetero-Dimers through Co-Immunoprecipitation Experiments
To further confirm the PheCAD homo- and hetero-dimers through co-immunoprecipitation coupled with immunoblotting, SPYNE173 (Myc-tag) and SPYCE(M) (HA-tag) constructs were used. Different combinations of PheCAD genes (PheCAD1-Myc + PheCAD1-HA, PheCAD2-Myc + PheCAD2-HA and PheCAD8-Myc + PheCAD8-HA, PheCAD2-Myc + PheCAD1-HA, PheCAD1-Myc + PheCAD8-HA and PheCAD8-Myc + PheCAD2-HA) were injected into tobacco leaves. The total protein was isolated from the injected leaves, and immunoprecipitation experiments were conducted with anti-HA-antibodies to pull down the HA fusion proteins. If HA tag protein interacts with Myc tag protein, the HA beads should also pull down the Myc-tag protein. Both total protein and the purified protein was subjected to Western blotting using anti-HA and anti-Myc antibodies. The detection of bands with anti-Myc antibodies in pull-down assay confirmed that the PheCAD1, PheCAD2, and PheCAD3 enzymes form homo-dimers, and these proteins interact with each other and form hetero-dimers in planta (Figure 8). The uncropped Western blot figures were provided as a Supplementary Materials.
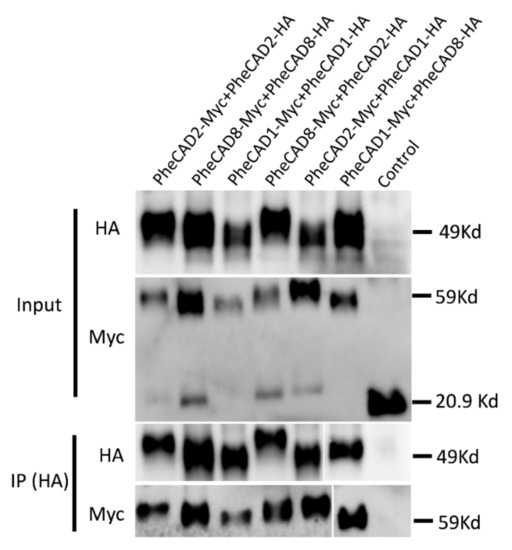
Figure 8.
CO-IP assay of PheCAD1, PheCAD2 and PheCAD8 enzymes using proteins expressed in N. benthamiana leaf epidermal cells BiFC assay. The proteins were immunoprecipitated with anti-HA antibodies and detected with both anti-HA and anti-Myc antibodies.
3. Discussion
Lignin is the major structural component of the secondary cell wall that provides mechanical strength for the upright growth of vascular plants and provides rigidity to the cell wall [39]. It interacts with cellulose and hemicellulose and helps in effective water transport by creating a hydrophobic environment in the vascular bundle. However, lignin renders the cell wall processability and is the primary barrier for biofuel and paper production [40]. Bamboos are the fastest-growing plants that are useful for food, timber and paper production. Thus, studying lignin biosynthesis in bamboo is not only crucial for understanding plant biology but also for genetic engineering strategies to improve pulp quality for paper production. CAD is the key rate-limiting enzyme in the lignin biosynthesis involved in the catalysis of coniferaldehyde, p-coumaraldehyde, and sinapaldehyde to corresponding alcohols and also for biotic and abiotic stress response [41]. The CAD gene family is investigated in so many plant species’ Arabidopsis [7,42]: rice [43], wheat [44], sorghum [45], P. trichocarpa [46], sweet potato [17], Brachypodium distachyon [6], tea [21], melon [41] and pear [47], etc. The present study aims to identify CAD genes in available Bambusoideae genome sequences and functional characterisation of Moso bamboo CAD genes.
In Angiosperms, CAD genes have distinct roles and are expressed in different stages and tissues during plant growth and development [17,41]. In Arabidopsis, AtCAD4, AtCAD5, AtCAD7 and AtCAD8 genes were involved in lignin biosynthesis. Among them, AtCAD4 was strongly expressed in roots and flowers, whereas AtCAD5 was in lignified roots [7,15]. Similarly, BdCAD5 in B. distachyon was expressed in most tissues and highest in stem and root. At the same time, the BdCAD3 expression was confined to stem and spikes [6]. In melon, CmCAD1 was expressed from 25–33 days after anthesis, whereas CmCAD2 and CmCAD5 genes were strongly expressed during fruit development. The CmCAD5 is also expressed in most vegetative tissues, with the highest in flower and negligible in mature leaves [41]. Similarly, this study identified that PheCAD2 had intense expression in most tissues except leaves, root rhizome, root on shoot—2 cm and 6.7m height shoot, especially PheCAD8 expressed in the tissues where the PheCAD2 gene expression was absent. PheCAD1, PheCAD6 and PheCAD12 were also expressed in most tissues, indicating that these five PheCAD genes might be involved in lignification (Figure 3).
It has been demonstrated that some of the lignin biosynthesis enzymes are loosely or transiently associated with each other to form metabolic channelling or metabolon and also form homo- or hetero- dimers. The early steps of lignin biosynthesis enzymes such as PAL isoforms in tobacco cells associate with C4H and substrate channel for better catalytic efficiency and regulation of metabolic flux [48,49,50]. In Arabidopsis, the cytochrome P450 enzymes of lignin biosynthesis (AtC4H, AtC3H and AtF5H) were found to associate with each other to form an enzyme complex or a metabolon with the help of membrane steroid-binding proteins (AtMSBPs) [51,52]. Further, in P. trichocarpa, the isozymes of PtrC4H, PtrC3H form hetero-dimeric (PtrC4H1/C4H2, PtrC4H1/C3H3, and PtrC4H2/C3H3) and heterotrimeric (PtrC4H1/C4H2/C3H3) enzyme complexes and increase the catalytic efficiency of the complex by 70–6500 fold as compared with individual enzymes [37]. Moreover, Chen et al., (2013, 2014) [38,53] have demonstrated that Ptr4CL3 and Ptr4CL5 isoforms interact to form a tetramer both in vivo and in vitro and proposed a regulatory role for it in P. trichocarpa. Additionally, PtrCAD1 interacts with PtrCCR2, and both enzymes were found to form homo-dimers in P. trichocarpa [33]. This study identified that PheCAD1, PheCAD2 and PheCAD8 enzymes interact and form homo-dimers in yeast (Figure 6) and both homo- and hetero- dimers in planta (Figure 7 and Figure 8). The failure to form hetero- dimers in yeast is due to post-translation modifications and the unavailability of other components, which might be needed in the formation of hetero-dimers. These results and the above literature indicate that PheCCR might interact with the PheCAD homo- or hetero- dimers. Therefore, studying the CAD homo- and hetero- dimers and PheCCR ratio within the plants and the catalytic efficiency would be interesting.
Besides lignification, CAD genes are also involved in biotic and abiotic stress. Previous studies reported that CAD1 gene expression is strongly induced by both biotic and abiotic stress in sweet potatoes and Ginkgo biloba [17,19]. Similarly, the oriental melon CmCAD2 gene was significantly induced after salt and drought stress [22]. Further, in Arabidopsis, among the most expressed CAD genes AtCAD-C and AtCAD-D, only AtCAD-D was strongly induced after biotic stress. Whereas among the least active CAD genes, AtCAD-B1 was induced in response to biotic stress [15]. Similarly, our investigation of moso bamboo PheCAD genes in response to abiotic stress identified that PheCAD2 was significantly induced after all the abiotic stress treatments among the highly expressed CAD genes. These results indicate involvement in both lignification and also in defence of the abiotic stress response. Among least-expressed PheCAD genes (PheCAD3, PheCAD4, PheCAD5, PheCAD7, PheCAD9, PheCAD10, PheCAD11, PheCAD13 and PheCAD14), PheCAD3 and PheCAD5 genes were induced tremendously after all abiotic stress treatments (Figure 5). These results indicate the involvement of PheCAD3 and PheCAD5 genes only in response to abotic stress. In conclusion, our results identified the major PheCAD genes involved in the lignin biosynthesis pathway and abiotic stress response. Further, identifying homo- and hetero-dimers of the PheCAD enzymes provides new evidence that PheCAD enzymes might act as a complex in a tissue-specific manner.
4. Materials and Methods
4.1. Identification of CAD Enzymes from Bambusoideae Genome Database
The CAD enzymes amino acid sequences of rice were downloaded from Oryzabase (https://shigen.nig.ac.jp/rice/oryzabase/, acessed on 1 November 2021), and these sequences were used to identify CAD enzymes from the Bambusoideae genome database. The Moso bamboo CAD enzymes were identified from the P. edulis genome database through local BLASTP [30]. Similarly, for herbaceous diploid bamboo species R. guianensis and O. latifolia, the tetraploid and hexaploid woody species G. angustifolia and B. amplexicaulis CAD enzymes were identified from available draft genomes [31]. The sequences containing the catalytic and structural Zn-binding and NADP(H) binding sites were considered CAD enzymes.
4.2. Phylogenetic Tree and Motif Analysis of CAD Enzymes
The phylogenetic tree was constructed in the MEGA-X software using both a maximum likelihood and a neighbour-joining method [35]. The aligned peptide sequences of A. thaliana, P. trichocarpa, Oryza sativa, O. latifolia, R. guianensis, G. angustifolia, P. edulis, and B. amplexicaulis were used to generate a phylogenetic tree (Table S1). To access the statistical significance of the clades in the phylogenetic tree, a bootstrap value of 1000 replicates was used. The conserved motifs were identified through the MEME server and visualised in TBtools [54].
4.3. Expression Analysis
We downloaded the transcriptomic data of 26 different kinds of tissues published by Zhou et al. (2018) [30] through the NCBI Short Read Archive database (SRX2408703). We analysed these data, and the FPKM values of the Moso bamboo CAD genes were used to develop a heatmap by TBtools [54]. For abiotic stress treatment, 30-day-old Moso bamboo seedlings with similar height were used. The seedlings were treated individually with 200 mM sodium chloride (NaCl), 25% polyethylene glycol (PEG), 1mM salicylic acid (SA) and 1uM abscisic acid (ABA) nutrient solution for 3 h and 24 h. Total RNA was isolated from young leaves, and transcriptomic data were generated in three biological replicates (NCBI ID: GSE169067). The graphs were developed using FPKM values of the PheCAD genes transcriptomic data.
4.4. Co-Expression Analysis
The FPKM values of 26 different kinds of transcriptomic tissue data developed by Zhou et al., (2018) [30] were used for the co-expression analysis. A minimum threshold of 0.05 FPKM value in more than six tissues out of 26 tissues was chosen as a cutoff to identify lignin biosynthesis genes. The Pearson correlation coefficient (PCC) was calculated, and the gene pairs with PCC values 0.7 to 1 and −0.5 to −1were considered to have positive and negative correlation, respectively. These gene pairs were visualised using Cytoscape. Further to identify total genes co-expressed with PheCAD genes, we submitted the PheCAD genes to the BambooNET (http://bioinformatics.cau.edu.cn/bamboo/index.html, accessed on 1 November 2021) and developed the co-expression network.
4.5. Yeast Two-Hybrid (Y2H) Assay
The coding sequences of PheCAD genes were amplified by using specific primers designed from 5′ UTR and 3′ UTR using leaf and stem cDNA as a template. This purified full-length PCR product was used to amplify adapter containing gene-specific primers (Table S6) and cloned into the pGBKT-7 and pGADT7 vectors (Clontech, Beijing, China). The recombinant vectors pGBKT-7 and pGADT7 containing PheCAD1, PheCAD2 and PheCAD3 genes were cotransformed into AH109 yeast strain following the manufacturer’s instructions (MatchmakerTM GAL4 two-hybrid system, Clontech). The vectors pGBKT53+pGADT-Lam and pGBKT-53+pGADT7-T were used as negative and positive controls, respectively. The transformed yeast cells were cultured on SD/-Leu/-Trp with Agar medium (Coolaber, Beijing, China) at 30 °C for three days. The colonies grown in this medium were selected, dissolved into ddH2o and plated onto the selection plates containing SD/-Ade/-His/-Leu/-Trp medium (Coolaber, Beijing, China) as serial dilutions; the plates were incubated at 30 °C for 5 days. The pictures were taken on the 5th day under a light microscope.
4.6. Bimolecular Fluorescence Complementation Assay in Arabidopsis Protoplast
The three PheCAD genes were cloned into pSAT1- nEYFP-C1 and pSAT4-cEYFP-C1-B vectors. The recombinant YFPN and YFPC fusion vectors transiently co-expressed in mesophyll protoplasts of Arabidopsis, according to Yoo et al., (2007) [55]. The transformed cells images were captured in a confocal microscope at wavelength 488 and 594 nm argon laser.
4.7. Co-Immunoprecipitation and Immunoblot Analysis
To generate pull-down constructs, the three PheCAD genes were cloned under the 35S promoter of SPYNE173 and SPYCE(M) vectors (contain Myc- and HA- tag, respectively). The recombinant vectors and empty vectors were transformed into Agrobacterium strain GV3101. Overnight cultures were collected and resuspended in buffer containing 10 mM MgCl2, 10 mM MES-K (pH 5.6) and 100 μM acetosyringone to OD600 nm at 0.4. The resuspended agrobacterium solutions carrying Myc- and HA- tag vectors combined in 1:1 ration and then co-infiltrated into the abaxial side of N. benthamiana 4-week-old leaves. On the 3rd day after the infiltration, the total protein from tobacco leaves was extracted using Plant Protein Extraction Reagent (CWBIO, Beijing, China). An equal amount of total protein was taken as input for CO-IP experiments. Pierce Anti-HA magnetic beads (Thermo Scientific, Waltham, MA, USA) were used to immunoprecipitate HA-tagged proteins as per the manufacturer’s instructions. The proteins were separated on SDS-PAGE gels (TGX Stain-Free FastCast Acrylamide Kit, 12% [Biorad, Hercules, CA, USA]) and blotted onto PVDF membrane (0.45 μm, Merck Millipore, Burlington, VT, USA). The HA and Myc fusion proteins were detected using mouse anti-HA (Affinity Biosciences, Cincinnati, OH, USA) and rabbit anti-Myc (Hangzhou HuaAn Biotechnology Co., Ltd., Hangzhou, China) antibodies. The dilutions of antibodies were used as per the instructions of the manufacture. The bands were visualised in ChemiDocTM MP Imaging System (Biorad, Hercules, CA, USA) by chemiluminescence assay (The ECLTM Prime Western Blotting Detection Reagent kit, AmershamTM, Amersham, UK).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222312917/s1.
Author Contributions
Conceptualisation, N.V., X.L.; methodology, N.V., D.H., H.W., R.M.S.; software, N.V., D.H., L.-H.Z.; validation, N.V.; formal analysis, N.V., H.W., R.M.S.; investigation, N.V., X.L.; resources, X.L., A.W.; data curation, N.V., D.H., H.W., L.-H.Z.; writing—original draft preparation, N.V.; writing—review and editing, N.V., R.M.S., K.Y., X.L.; visualisation, N.V., L.-H.Z.; supervision, X.L., A.W.; project administration, X.L., A.W.; funding acquisition, X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research work was supported by National Key Research & Development Program of China (2021YFD220000303).
Institutional Review Board Statement
Not applicable as no animal and human trials were needed for this study.
Informed Consent Statement
Not applicable.
Data Availability Statement
SRX2408703, GSE169067. All the data pertaining to this study can be accessed from the lead authors on reasonable request.
Acknowledgments
We thank Cheng Jiang, State Key Laboratory of Subtropical Silviculture, Zhejiang A & F University, for providing the BiFC vectors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Holbrook, N.M.; Zwieniecki, M.A. Embolism repair and xylem tension: Do We need a miracle? Plant Physiol. 1999, 120, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.Y.; Walker, A.M.; Kim, H.; Ralph, J.; Vermerris, W.; Sattler, S.E.; Kang, C. The Enzyme Activity and Substrate Specificity of Two Major Cinnamyl Alcohol Dehydrogenases in Sorghum (Sorghum bicolor), SbCAD2 and SbCAD4. Plant Physiol. 2017, 174, 2128–2145. [Google Scholar] [CrossRef] [PubMed]
- Park, H.L.; Kim, T.L.; Bhoo, S.H.; Lee, T.H.; Lee, S.W.; Cho, M.H. Biochemical Characterization of the Rice Cinnamyl Alcohol Dehydrogenase Gene Family. Molecules 2018, 23, 2659. [Google Scholar] [CrossRef] [PubMed]
- Bonawitz, N.D.; Chapple, C. The genetics of lignin biosynthesis: Connecting genotype to phenotype. Annu. Rev. Genet. 2010, 44, 337–363. [Google Scholar] [CrossRef]
- Mottiar, Y.; Vanholme, R.; Boerjan, W.; Ralph, J.; Mansfield, S.D. Designer lignins: Harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 2016, 37, 190–200. [Google Scholar] [CrossRef]
- Bukh, C.; Nord-Larsen, P.H.; Rasmussen, S.K. Phylogeny and structure of the cinnamyl alcohol dehydrogenase gene family in Brachypodium distachyon. J. Exp. Bot. 2012, 63, 6223–6236. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Kim, M.-R.; Bedgar, D.L.; Moinuddin, S.G.; Cardenas, C.L.; Davin, L.B.; Kang, C.; Lewis, N.G. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 1455–1460. [Google Scholar] [CrossRef]
- MacKay, J.J.; O’Malley, D.M.; Presnell, T.; Booker, F.L.; Campbell, M.M.; Whetten, R.W.; Sederoff, R.R. Inheritance, gene expression, and lignin characterization in a mutant pine deficient in cinnamyl alcohol dehydrogenase. Proc. Natl. Acad. Sci. USA 1997, 94, 8255–8260. [Google Scholar] [CrossRef] [PubMed]
- Guillaumie, S.; Pichon, M.; Martinant, J.P.; Bosio, M.; Goffner, D.; Barriere, Y. Differential expression of phenylpropanoid and related genes in brown-midrib bm1, bm2, bm3, and bm4 young near-isogenic maize plants. Planta 2007, 226, 235–250. [Google Scholar] [CrossRef]
- Kim, H.; Ralph, J.; Lu, F.; Pilate, G.; Leple, J.C.; Pollet, B.; Lapierre, C. Identification of the structure and origin of thioacidolysis marker compounds for cinnamyl alcohol dehydrogenase deficiency in angiosperms. J. Biol. Chem. 2002, 277, 47412–47419. [Google Scholar] [CrossRef]
- Ralph, J.; Lapierre, C.; Marita, J.M.; Kim, H.; Lu, F.; Hatfield, R.D.; Ralph, S.; Chapple, C.; Franke, R.; Hemm, M.R.; et al. Elucidation of new structures in lignins of CAD-and COMT-deficient plants by NMR. Phytochemistry 2001, 57, 993–1003. [Google Scholar] [CrossRef]
- Halpin, C.; Knight, M.E.; Grima-Pettenati, J.; Goffner, D.; Boudet, A.; Schuch, W.J.P.P. Purification and characterization of cinnamyl alcohol dehydrogenase from tobacco stems. Plant Physiol. 1992, 98, 12–16. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, K.; Qian, Q.; Huang, Z.; Wang, Y.; Li, M.; Hong, L.; Zeng, D.; Gu, M.; Chu, C.; Cheng, Z. GOLD Hull and Internode2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol. 2006, 140, 972–983. [Google Scholar] [CrossRef] [PubMed]
- Sibout, R.; Eudes, A.; Mouille, G.; Pollet, B.; Lapierre, C.; Jouanin, L.; Seguin, A. Cinnamyl Alcohol Dehydrogenase-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 2005, 17, 2059–2076. [Google Scholar] [CrossRef] [PubMed]
- Tronchet, M.; Balague, C.; Kroj, T.; Jouanin, L.; Roby, D. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 2010, 11, 83–92. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Huh, G.-H. Overexpression of cinnamyl alcohol dehydrogenase gene from sweetpotato enhances oxidative stress tolerance in transgenic Arabidopsis. In Vitro Cell. Dev. Biol. Plant 2019, 55, 172–179. [Google Scholar] [CrossRef]
- Kim, Y.H.; Bae, J.M.; Huh, G.H. Transcriptional regulation of the cinnamyl alcohol dehydrogenase gene from sweet potato in response to plant developmental stage and environmental stress. Plant Cell Rep. 2010, 29, 779–791. [Google Scholar] [CrossRef]
- Qiu, W.; Song, X.; Han, X.; Liu, M.; Qiao, G.; Zhuo, R. Overexpression of Sedum alfredii cinnamyl alcohol dehydrogenase increases the tolerance and accumulation of cadmium in Arabidopsis. Environ. Exp. Bot. 2018, 155, 566–577. [Google Scholar] [CrossRef]
- Cheng, H.; Li, L.; Xu, F.; Cheng, S.; Cao, F.; Wang, Y.; Yuan, H.; Jiang, D.; Wu, C. Expression patterns of a cinnamyl alcohol dehydrogenase gene involved in lignin biosynthesis and environmental stress in Ginkgo biloba. Mol. Biol. Rep. 2013, 40, 707–721. [Google Scholar] [CrossRef]
- Sun, Y.; Wu, Y.; Zhao, Y.; Han, X.; Lou, H.; Cheng, A. Molecular cloning and biochemical characterization of two cinnamyl alcohol dehydrogenases from a liverwort Plagiochasma appendiculatum. Plant Physiol. Biochem. 2013, 70, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.W.; Zhang, M.; Wu, J.Q.; Jiang, Z.Z.; Tang, L.; Li, Y.Y.; Wei, C.L.; Jiang, C.J.; Wan, X.C. Molecular cloning, functional analysis of three cinnamyl alcohol dehydrogenase (CAD) genes in the leaves of tea plant, Camellia sinensis. J. Plant Physiol. 2013, 170, 272–282. [Google Scholar] [CrossRef]
- Liu, W.; Jin, Y.; Li, M.; Dong, L.; Guo, D.; Lu, C.; Qi, H. Analysis of CmCADs and three lignifying enzymes in oriental melon (‘CaiHong7’) seedlings in response to three abiotic stresses. Sci. Horti. 2018, 237, 257–268. [Google Scholar] [CrossRef]
- Buckingham, K.; Jepson, P.; Wu, L.; Rao, I.V.; Jiang, S.; Liese, W.; Lou, Y.; Fu, M. The potential of bamboo is constrained by outmoded policy frames. AMBIO 2011, 40, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Li, L.; Ma, T.; Pei, J.; Zhao, Z.; Lu, M.; Wu, A.; Lin, X. The SOC1-like gene BoMADS50 is associated with the flowering of Bambusa oldhamii. Hortic. Res. 2021, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Zhao, Z.; Hu, Q.; Li, L.; Vasupalli, N.; Zhuo, J.; Zeng, W.; Wu, A.; Lin, X. PeSNAC-1 a NAC transcription factor from moso bamboo (Phyllostachys edulis) confers tolerance to salinity and drought stress in transgenic rice. Tree Physiol. 2020, 40, 1792–1806. [Google Scholar] [CrossRef]
- Kuehl, Y.; Li, Y.; Henley, G. Impacts of selective harvest on the carbon sequestration potential in Moso bamboo (Phyllostachys pubescens) plantations. For. Trees Livelihoods 2013, 22, 1–18. [Google Scholar] [CrossRef]
- Yuen, J.Q.; Fung, T.; Ziegler, A.D. Carbon stocks in bamboo ecosystems worldwide: Estimates and uncertainties. For. Ecol. Manag. 2017, 393, 113–138. [Google Scholar] [CrossRef]
- Ramakrishnan, M.; Yrjälä, K.; Vinod, K.K.; Sharma, A.; Cho, J.; Satheesh, V.; Zhou, M. Genetics and genomics of moso bamboo (Phyllostachys edulis): Current status, future challenges, and biotechnological opportunities toward a sustainable bamboo industry. Food Energy Secur. 2020, 9, e229. [Google Scholar] [CrossRef]
- Peng, Z.; Lu, Y.; Li, L.; Zhao, Q.; Feng, Q.; Gao, Z.; Lu, H.; Hu, T.; Yao, N.; Liu, K.; et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 2013, 45, 456–461. [Google Scholar] [CrossRef]
- Zhao, H.; Gao, Z.; Wang, L.; Wang, J.; Wang, S.; Fei, B.; Chen, C.; Shi, C.; Liu, X.; Zhang, H.; et al. Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis). Gigascience 2018, 7, giy115. [Google Scholar] [CrossRef]
- Guo, Z.H.; Ma, P.F.; Yang, G.Q.; Hu, J.Y.; Liu, Y.L.; Xia, E.H.; Zhong, M.C.; Zhao, L.; Sun, G.L.; Xu, Y.X.; et al. Genome Sequences Provide Insights into the Reticulate Origin and Unique Traits of Woody Bamboos. Mol. Plant 2019, 12, 1353–1365. [Google Scholar] [CrossRef]
- Wang, J.P.; Liu, B.; Sun, Y.; Chiang, V.L.; Sederoff, R.R. Enzyme-Enzyme Interactions in Monolignol Biosynthesis. Front. Plant Sci. 2018, 9, 1942. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Liu, J.; Kim, H.; Liu, B.; Huang, X.; Yang, Z.; Lin, Y.J.; Chen, H.; Yang, C.; Wang, J.P.; et al. CAD1 and CCR2 protein complex formation in monolignol biosynthesis in Populus trichocarpa. New Phytol. 2019, 222, 244–260. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Marchant, D.B.; Van de Peer, Y.; Soltis, D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015, 35, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell Wall Metabolism in Response to Abiotic Stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef]
- Chen, H.C.; Li, Q.; Shuford, C.M.; Liu, J.; Muddiman, D.C.; Sederoff, R.R.; Chiang, V.L. Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, 21253–21258. [Google Scholar] [CrossRef]
- Chen, H.C.; Song, J.; Williams, C.M.; Shuford, C.M.; Liu, J.; Wang, J.P.; Li, Q.; Shi, R.; Gokce, E.; Ducoste, J.; et al. Monolignol pathway 4-coumaric acid:coenzyme A ligases in Populus trichocarpa: Novel specificity, metabolic regulation, and simulation of coenzyme A ligation fluxes. Plant Physiol. 2013, 161, 1501–1516. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Li, Q.; Song, J.; Peng, S.; Wang, J.P.; Qu, G.Z.; Sederoff, R.R.; Chiang, V.L. Plant biotechnology for lignocellulosic biofuel production. Plant Biotechnol. J. 2014, 12, 1174–1192. [Google Scholar] [CrossRef]
- Jin, Y.; Zhang, C.; Liu, W.; Qi, H.; Chen, H.; Cao, S. The cinnamyl alcohol dehydrogenase gene family in melon (Cucumis melo L.): Bioinformatic analysis and expression patterns. PLoS ONE 2014, 9, e101730. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, K.W.; Cho, M.H.; Franceschi, V.R.; Davin, L.B.; Lewis, N.G. Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development: Lessons for database annotations? Phytochemistry 2007, 68, 1957–1974. [Google Scholar] [CrossRef]
- Tobias, C.M.; Chow, E.K. Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification. Planta 2005, 220, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.H. Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. J. Exp. Bot. 2010, 61, 2735–2744. [Google Scholar] [CrossRef] [PubMed]
- Saballos, A.; Ejeta, G.; Sanchez, E.; Kang, C.; Vermerris, W. A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 2009, 181, 783–795. [Google Scholar] [CrossRef]
- Barakat, A.; Bagniewska-Zadworna, A.; Choi, A.; Plakkat, U.; DiLoreto, D.S.; Yellanki, P.; Carlson, J.E. The cinnamyl alcohol dehydrogenase gene family in Populus: Phylogeny, organization, and expression. BMC Plant Biol. 2009, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Qi, K.; Song, X.; Yuan, Y.; Bao, J.; Gong, X.; Huang, X.; Khanizadeh, S.; Zhang, S.; Tao, S. CAD Genes: Genome-Wide Identification, Evolution, and Their Contribution to Lignin Biosynthesis in Pear (Pyrus bretschneideri). Plants 2021, 10, 1444. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol. Plant. 1999, 107, 142–149. [Google Scholar] [CrossRef]
- Rasmussen, S.; Dixon, R.A. Transgene-Mediated and Elicitor-Induced Perturbation of Metabolic Channeling at the Entry Point into the Phenylpropanoid Pathway. Plant Cell 1999, 11, 1537–1551. [Google Scholar] [CrossRef] [PubMed]
- Ralston, L.; Yu, O. Metabolons involving plant cytochrome P450s. Phytochem. Rev. 2006, 5, 459–472. [Google Scholar] [CrossRef]
- Bassard, J.E.; Richert, L.; Geerinck, J.; Renault, H.; Duval, F.; Ullmann, P.; Schmitt, M.; Meyer, E.; Mutterer, J.; Boerjan, W.; et al. Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 2012, 24, 4465–4482. [Google Scholar] [CrossRef] [PubMed]
- Gou, M.; Ran, X.; Martin, D.W.; Liu, C.J. The scaffold proteins of lignin biosynthetic cytochrome P450 enzymes. Nat. Plants 2018, 4, 299–310. [Google Scholar] [CrossRef]
- Chen, H.C.; Song, J.; Wang, J.P.; Lin, Y.C.; Ducoste, J.; Shuford, C.M.; Liu, J.; Li, Q.; Shi, R.; Nepomuceno, A.; et al. Systems biology of lignin biosynthesis in Populus trichocarpa: Heteromeric 4-coumaric acid: Coenzyme A ligase protein complex formation, regulation, and numerical modeling. Plant Cell 2014, 26, 876–893. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).