Human Adenovirus Type 5 Infection Leads to Nuclear Envelope Destabilization and Membrane Permeability Independently of Adenovirus Death Protein
Abstract
:1. Introduction
2. Results
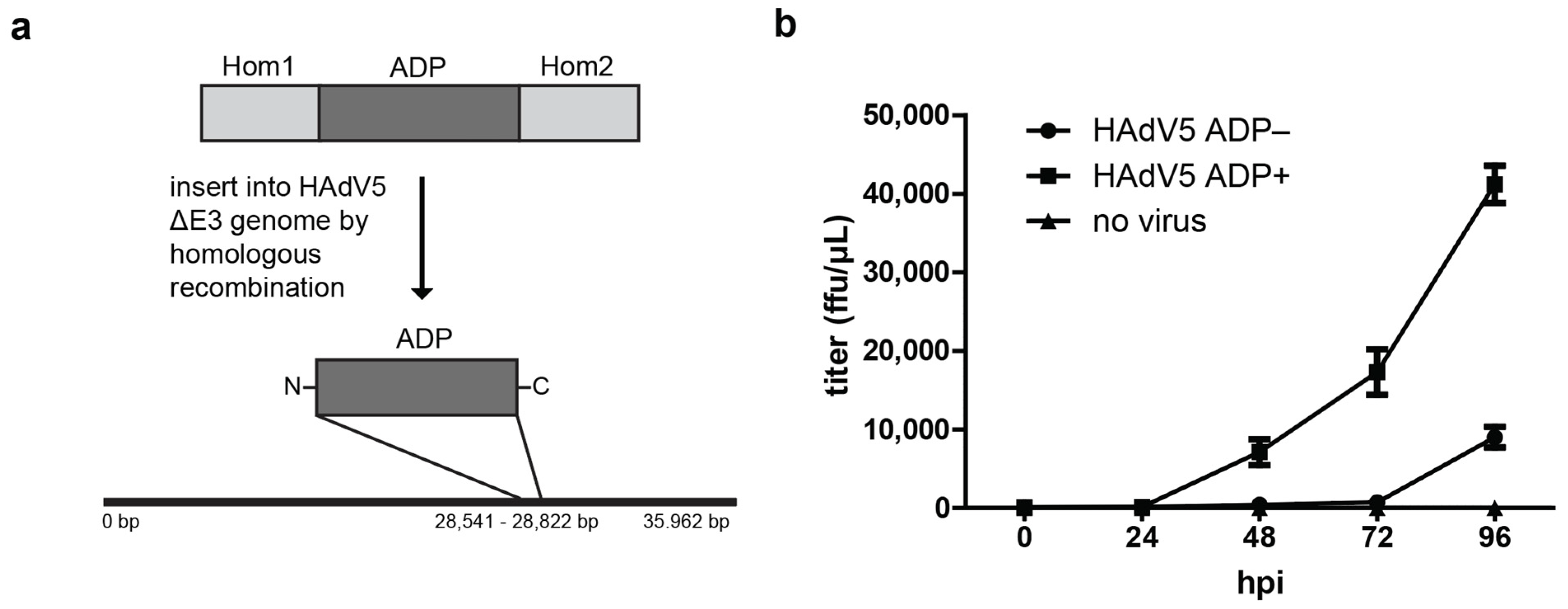
2.1. Expression of ADP Facilitates Release of Infectious Virus Particles but Is Unlikely to Result from ADP Pore Formation
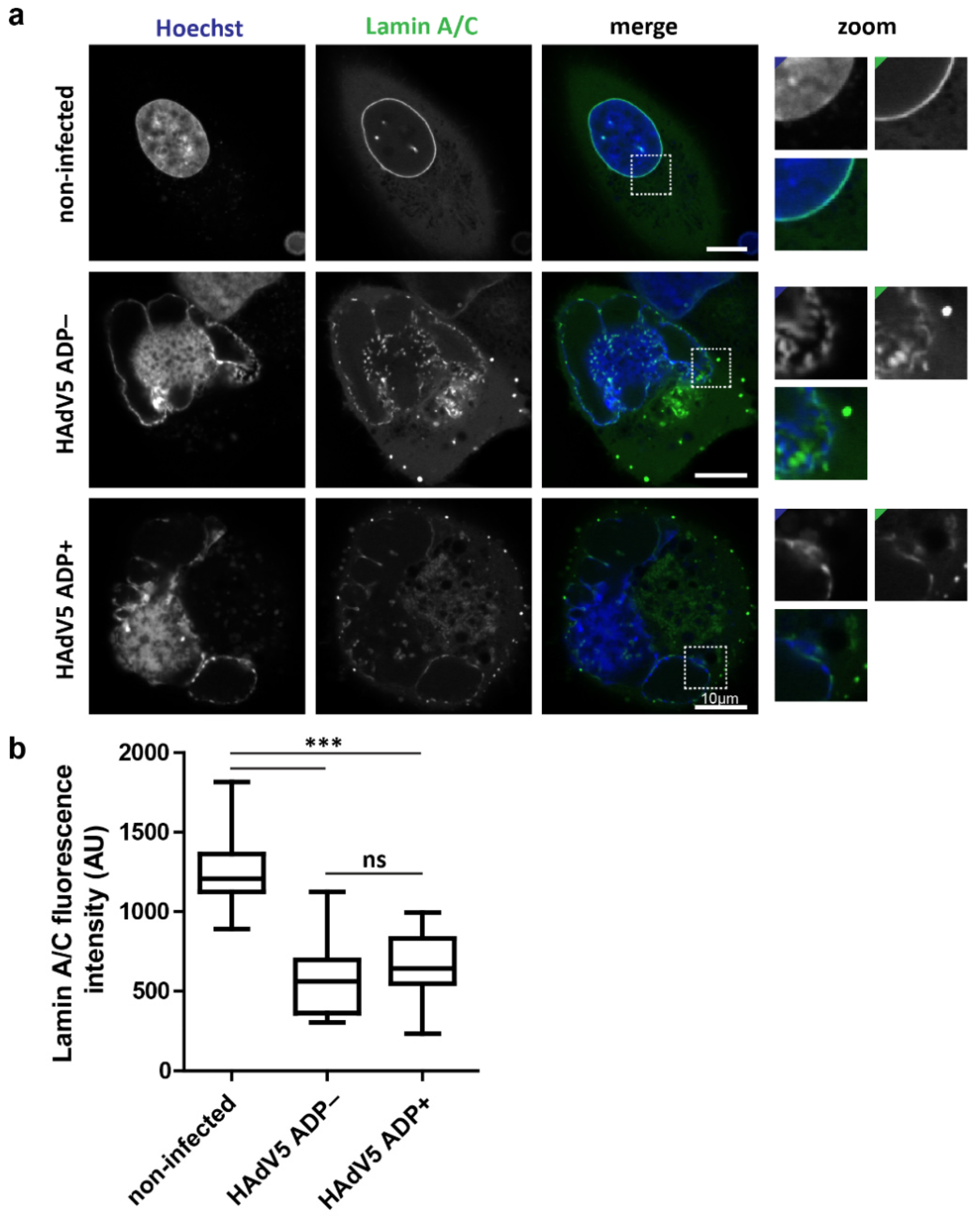
2.2. Infection Reduces Lamin A/C Levels at the Nuclear Envelope Independently of ADP Expression
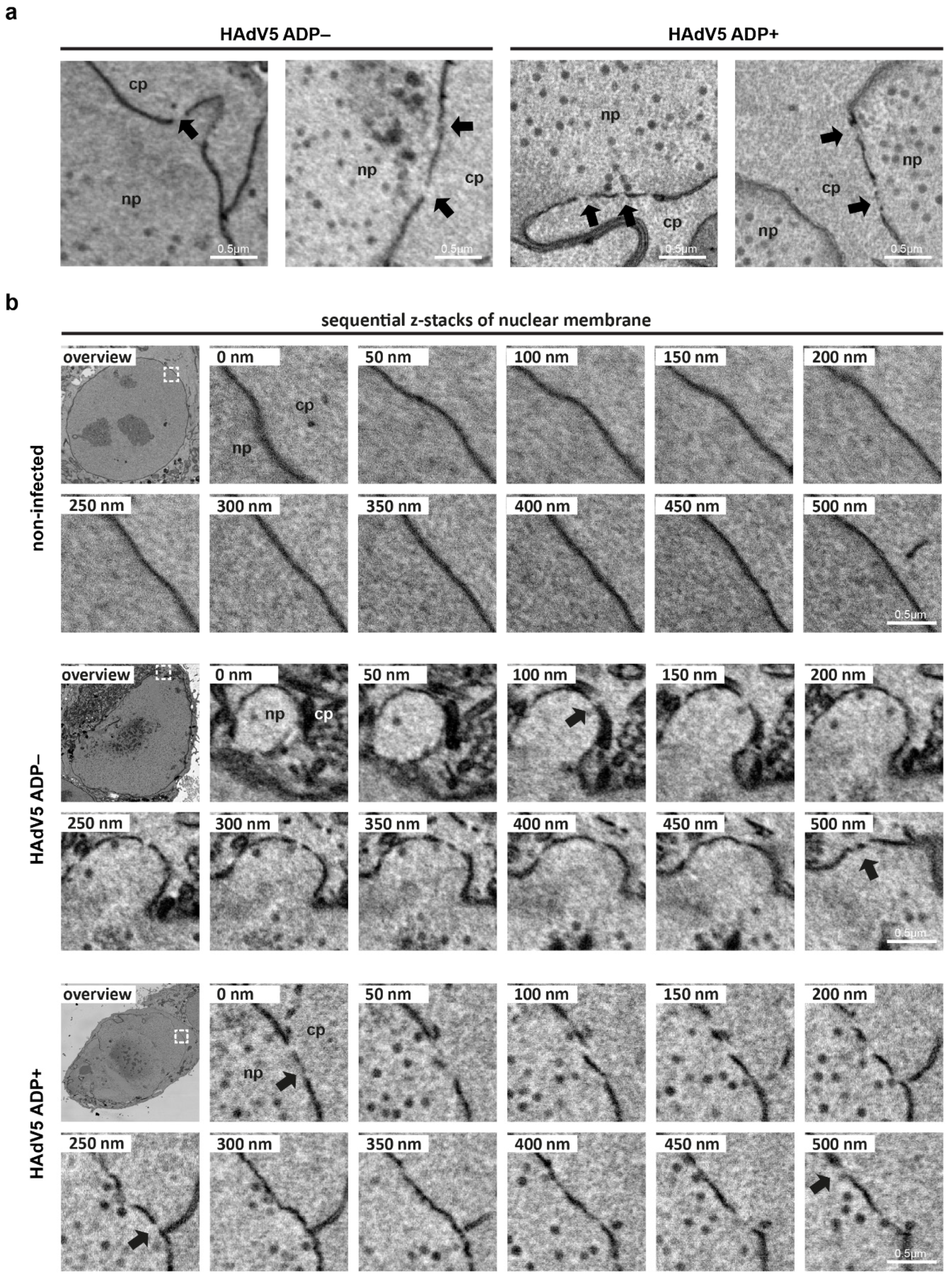
2.3. Serial Block-Face SEM and Electron Cryotomography Reveal Details of HAdV5-Induced Nuclear Membrane Deformation
2.4. Adenovirus Infection Leads to Damage of the Nuclear Envelope and an Increased Nuclear Permeability Independent of ADP
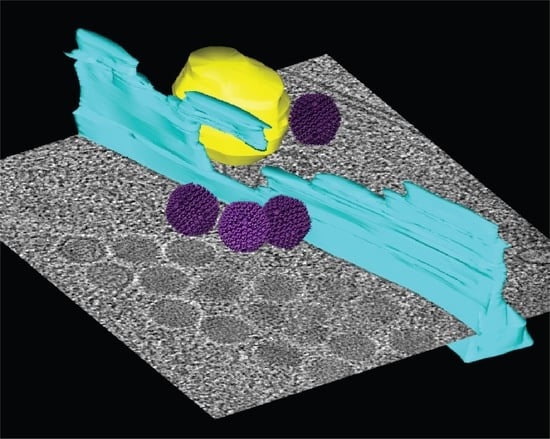
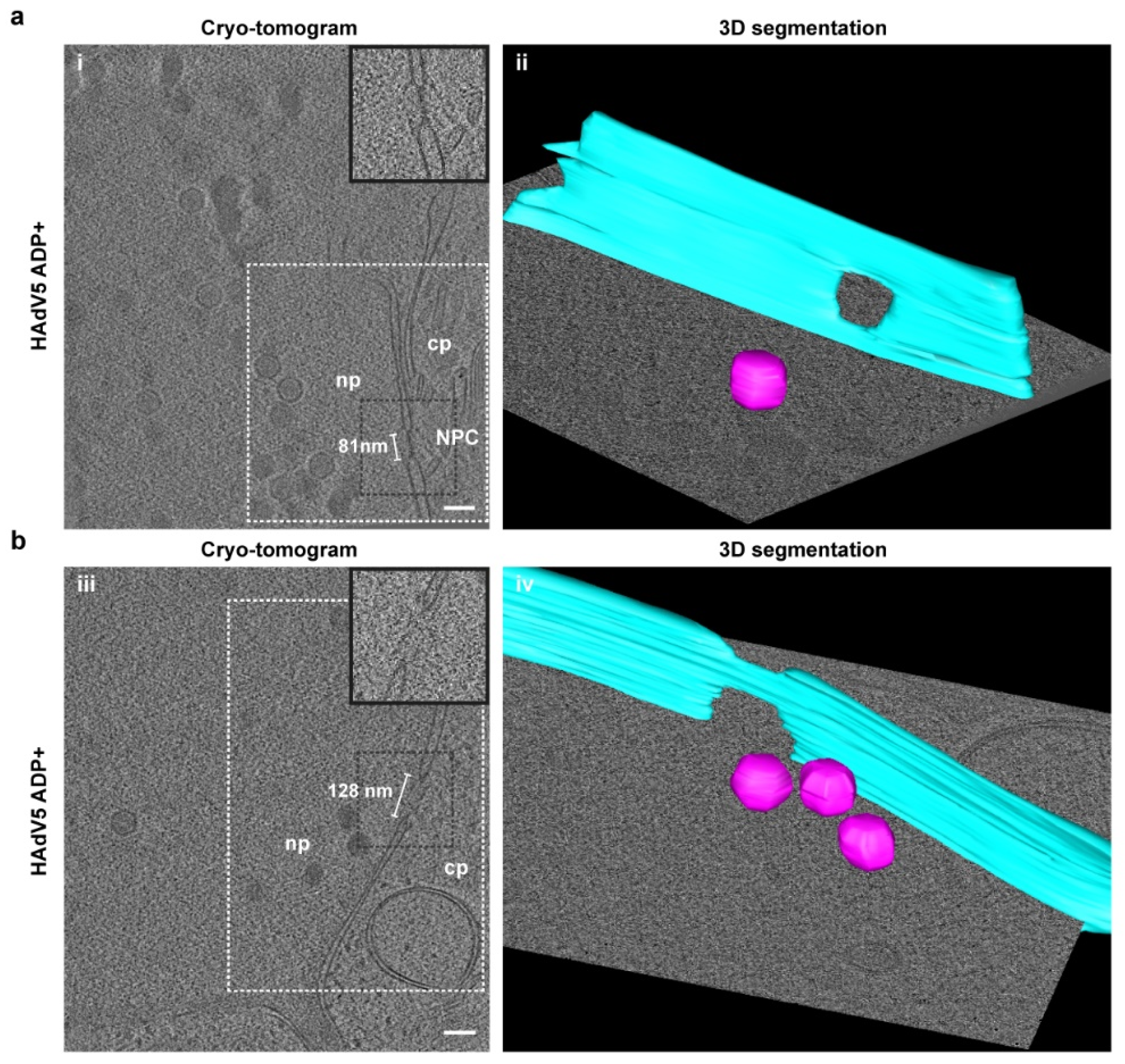
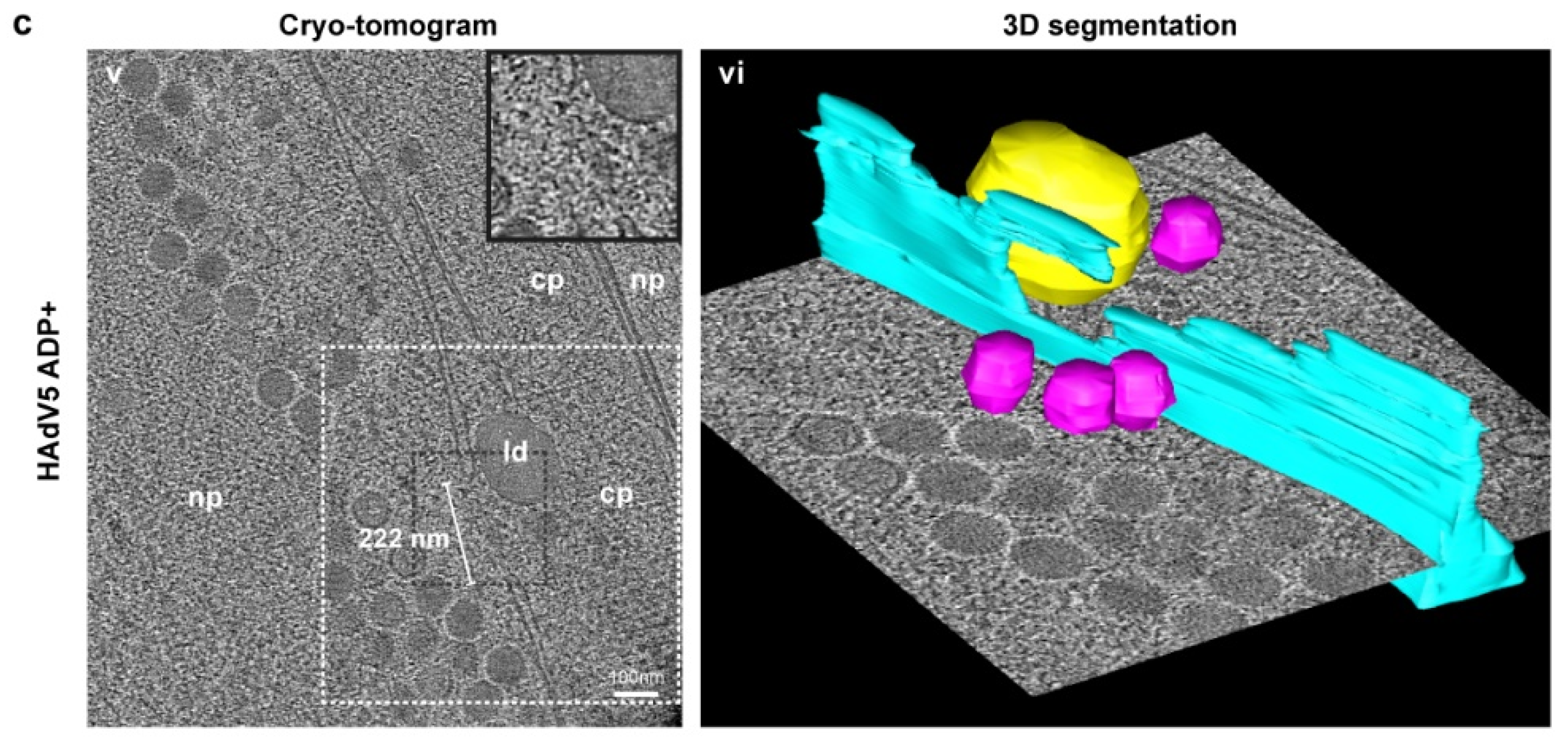
2.5. Nuclear Envelope Lesions Are also Detectable under Cryocondition in Electron Cryotomograms of FIB-Milled Infected Cells
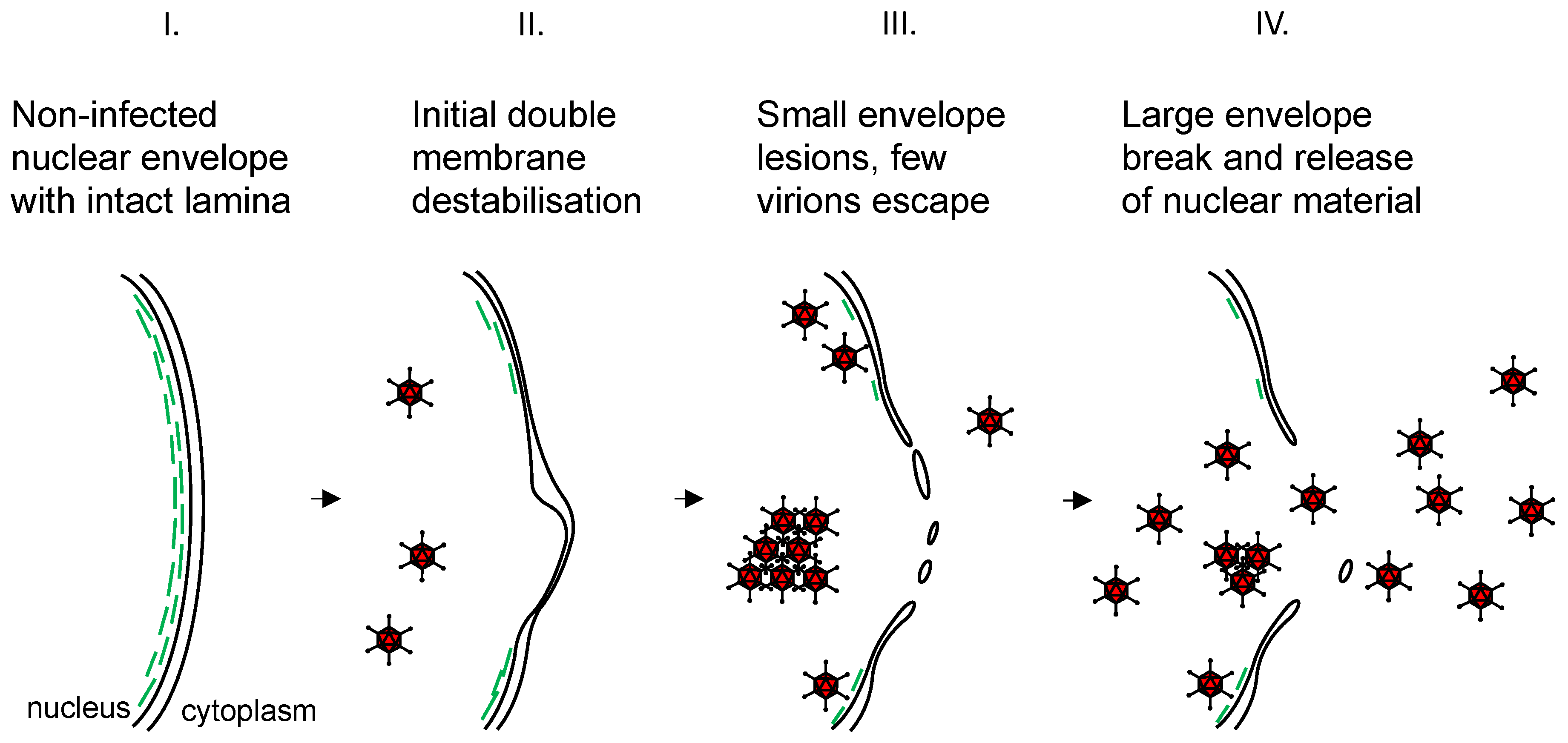
3. Discussion
4. Materials and Methods
4.1. Mammalian Cell Line Culture
4.2. Construction of Virus Strains and Expression Plasmids
4.3. Virus Production, Titration, Infection and Growth Curve
4.4. Live-Cell Fluorescence Microscopy
4.5. Immunofluorescence Microscopy
4.6. Nuclear Permeability Assay
4.7. Serial Block-Face Scanning Electron Microscopy
4.8. Electron Cryotomography
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tollefson, A.E.; Ryerse, J.S.; Scaria, A.; Hermiston, T.W.; Wold, W.S.M. The E3-11.6-kDa adenovirus death protein (ADP) is required for efficient cell death: Characterization of cells infected with adp mutants. Virology 1996, 220, 152–162. [Google Scholar] [CrossRef] [Green Version]
- Abou El Hassan, M.A.I.; van der Meulen-Muileman, I.; Abbas, S.; Kruyt, F.A.E. Conditionally Replicating Adenoviruses Kill Tumor Cells via a Basic Apoptotic Machinery-Independent Mechanism That Resembles Necrosis-Like Programmed Cell Death. J. Virol. 2004, 78, 12243–12251. [Google Scholar] [CrossRef] [Green Version]
- Weigert, M.; Binks, A.; Dowson, S.; Leung, E.Y.L.; Athineos, D.; Yu, X.; Mullin, M.; Walton, J.B.; Orange, C.; Ennis, D.; et al. RIPK3 promotes adenovirus type 5 activity. Cell Death Dis. 2017, 8, 3206. [Google Scholar] [CrossRef] [PubMed]
- Raghava, S.; Giorda, K.M.; Romano, F.B.; Heuck, A.P.; Hebert, D.N. The SV40 late protein VP4 is a viroporin that forms pores to disrupt membranes for viral release. PLoS Pathog. 2011, 7, e1002116. [Google Scholar] [CrossRef] [Green Version]
- Klupp, B.G.; Granzow, H.; Mettenleiter, T.C. Nuclear Envelope Breakdown Can Substitute for Primary Envelopment-Mediated Nuclear Egress of Herpesviruses. J. Virol. 2011, 85, 8285–8292. [Google Scholar] [CrossRef] [Green Version]
- Engelsma, D.; Valle, N.; Fish, A.; Salome, N.; Almendral, J.M.; Fornerod, M. A Supraphysiological Nuclear Export Signal Is Required for Parvovirus Nuclear Export. Mol. Biol. Cell 2008, 219, 2544–2552. [Google Scholar] [CrossRef] [Green Version]
- Skepper, J.N.; Whiteley, A.; Browne, H.; Minson, A. Herpes Simplex Virus Nucleocapsids Mature to Progeny Virions by an Envelopment → Deenvelopment → Reenvelopment Pathway. J. Virol. 2001, 75, 5697–5702. [Google Scholar] [CrossRef] [Green Version]
- Hagen, C.; Dent, K.C.; Zeev-Ben-Mordehai, T.; Grange, M.; Bosse, J.B.; Whittle, C.; Klupp, B.G.; Siebert, C.A.; Vasishtan, D.; Bäuerlein, F.J.B.; et al. Structural Basis of Vesicle Formation at the Inner Nuclear Membrane. Cell 2015, 163, 1692–1701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tollefson, A.E.; Scaria, A.; Hermiston, T.W.; Ryerse, J.S.; Wold, L.J.; Wold, W.S.M. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 1996, 70, 2296–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scaria, A.; Tollefson, A.E.; Saha, S.K.; Wold, W.S.M. The E3-11. 6K Protein of adenovirus is an Asn-glycosylated integral membrane protein that localizes to the nuclear membrane. Virology 1992, 753, 743–753. [Google Scholar] [CrossRef]
- Hausmann, J.; Ortmann, D.; Witt, E.; Veit, M.; Seidel, W. Adenovirus death protein, a transmembrane protein encoded in the E3 region, is palmitoylated at the cytoplasmic tail. Virology 1998, 244, 343–351. [Google Scholar] [CrossRef] [Green Version]
- Doronin, K.; Toth, K.; Kuppuswamy, M.; Krajcsi, P.; Tollefson, A.E.; Wold, W.S.M. Overexpression of the ADP (E3-11.6K) protein increases cell lysis and spread of Adenovirus. Virology 2003, 305, 378–387. [Google Scholar] [CrossRef]
- Zou, A.; Atencio, I.; Huang, W.M.; Horn, M.; Ramachandra, M. Overexpression of adenovirus E3-11.6K protein induces cell killing by both caspase-dependent and caspase-independent mechanisms. Virology 2004, 326, 240–249. [Google Scholar] [CrossRef] [Green Version]
- Wigand, R.; Kümel, G. The kinetics of adenovirus infection and spread in cell cultures infected with low multiplicity. Arch. Virol. 1977, 54, 177–187. [Google Scholar] [CrossRef]
- Yakimovich, A.; Gumpert, H.; Burckhardt, C.J.; Lutschg, V.A.; Jurgeit, A.; Sbalzarini, I.F.; Greber, U.F.; Lütschg, V.A.; Jurgeit, A.; Sbalzarini, I.F.; et al. Cell-Free Transmission of Human Adenovirus by Passive Mass Transfer in Cell Culture Simulated in a Computer Model. J. Virol. 2012, 86, 10123–10137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgi, F.; Greber, U.F. The Adenovirus Death Protein—A small membrane protein controls cell lysis and disease. FEBS Lett. 2020, 594, 1861–1878. [Google Scholar] [CrossRef] [PubMed]
- Andriasyan, V.; Yakimovich, A.; Petkidis, A.; Georgi, F.; Witte, R.; Puntener, D.; Greber, U.F. Microscopy deep learning predicts virus infections and reveals mechanics of lytic-infected cells. iScience 2021, 24, 102543. [Google Scholar] [CrossRef]
- Hoenig, E.M.; Margolis, G.; Kilham, L. Experimental adenovirus infection of the mouse adrenal gland. I. Light microscopic observation. Am. J. Pathol. 1974, 75, 363–374. [Google Scholar]
- Puvion-Dutilleul, F.; Besse, S.; Pichard, E.; Cajean-Feroldi, C. Release of viruses and viral DNA from nucleus to cytoplasm of HeLa cells at late stages of productive adenovirus infection as revealed by electron microscope in situ hybridization. Biol. Cell 1998, 90, 5–38. [Google Scholar] [CrossRef]
- Wang, H.; Bian, X.; Xia, L.; Ding, X.; Müller, R.; Zhang, Y.; Fu, J.; Stewart, A.F. Improved seamless mutagenesis by recombineering using ccdB for counterselection. Nucleic Acids Res. 2014, 42, e37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Fu, J.; Liu, J.; Wang, H.; Schiwon, M.; Janz, S.; Schaffarczyk, L.; von der Goltz, L.; Ehrke-Schulz, E.; Dörner, J.; et al. An Engineered Virus Library as a Resource for the Spectrum-wide Exploration of Virus and Vector Diversity. Cell Rep. 2017, 19, 1698–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.T.; Hung, W.C.; Chen, F.Y.; Huang, H.W. Mechanism and kinetics of pore formation in membranes by water-soluble amphipathic peptides. Proc. Natl. Acad. Sci. USA 2008, 105, 5087–5092. [Google Scholar] [CrossRef] [Green Version]
- Giorda, K.M.; Raghava, S.; Zhang, M.W.; Hebert, D.N. The viroporin activity of the minor structural proteins VP2 and VP3 is required for SV40 propagation. J. Biol. Chem. 2013, 288, 2510–2520. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Tollefson, A.E.; Scaria, A.; Ying, B.; Wold, W.S.M. Mutations within the ADP (E3-11.6K) protein alter processing and localization of ADP and the kinetics of cell lysis of adenovirus-infected cells. J. Virol. 2003, 77, 7764–7778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murali, V.K.; Ornelles, D.A.; Gooding, L.R.; Wilms, H.T.; Huang, W.; Tollefson, A.E.; Wold, W.S.; Garnett-Benson, C. Adenovirus death protein (ADP) is required for lytic infection of human lymphocytes. J. Virol. 2014, 88, 903–912. [Google Scholar] [CrossRef] [Green Version]
- Pfitzner, S.; Hofmann-Sieber, H.; Bosse, J.B.; Franken, L.E.; Grünewald, K.; Dobner, T. Fluorescent protein tagging of adenoviral proteins pV and pIX reveals ‘late virion accumulation compartment’. PLoS Pathog. 2020, 16, e1008588. [Google Scholar] [CrossRef] [PubMed]
- Kremer, J.R.; Mastronarde, D.N.; McIntosh, J.R. Computer Visualization of Three-Dimensional Image Data Using IMOD. J. Struct. Biol. 1996, 116, 71–76. [Google Scholar] [CrossRef] [Green Version]
- Georgi, F.; Andriasyan, V.; Witte, R.; Murer, L.; Hemmi, S.; Yu, L.; Grove, M.; Meili, N.; Kuttler, F.; Yakimovich, A.; et al. The FDA-Approved Drug Nelfinavir Inhibits Lytic Cell-Free but not Cell-Associated Nonlytic Transmission of Human Adenovirus. Antimicrob. Agents Chemother. 2020, 64, e01002-20. [Google Scholar] [CrossRef]
- Brandenburg, B.; Lee, L.Y.; Lakadamyali, M.; Rust, M.J.; Zhuang, X.; Hogle, J.M. Imaging poliovirus entry in live cells. PLoS Biol. 2007, 5, e183. [Google Scholar] [CrossRef] [PubMed]
- Dormitzer, P.R.; Nason, E.B.; Prasa, B.V.V.; Harrison, S.C. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 2004, 26, 1053–1058. [Google Scholar] [CrossRef]
- Chandran, K.; Farsetta, D.L.; Nibert, M.L. Strategy for Nonenveloped Virus Entry: A Hydrophobic Conformer of the Reovirus Membrane Penetration Protein μ1 Mediates Membrane Disruption. J. Virol. 2002, 76, 9920–9933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiethoff, C.M.; Wodrich, H.; Gerace, L.; Nemerow, G.R. Adenovirus Protein VI Mediates Membrane Disruption following Capsid Disassembly. J. Virol. 2005, 79, 1992–2000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniels, R.; Sadowicz, D.; Hebert, D.N. A very late viral protein triggers the lytic release of SV40. PLoS Pathog. 2007, 3, e98. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Orba, Y.; Okada, Y.; Sunden, Y.; Kimura, T.; Tanaka, S.; Nagashima, K.; Hall, W.W.; Sawa, H. The human polyoma JC virus agnoprotein acts as a viroporin. PLoS Pathog. 2010, 6, e1000801. [Google Scholar] [CrossRef] [Green Version]
- Panou, M.M.; Prescott, E.L.; Hurdiss, D.L.; Swinscoe, G.; Hollinshead, M.; Caller, L.G.; Morgan, E.L.; Carlisle, L.; Müller, M.; Antoni, M.; et al. Agnoprotein is an essential egress factor during BK Polyomavirus infection. Int. J. Mol. Sci. 2018, 19, 902. [Google Scholar] [CrossRef] [Green Version]
- Coric, P.; Saribas, A.S.; Abou-Gharbia, M.; Childers, W.; White, M.K.; Bouaziz, S.; Safak, M. Nuclear Magnetic Resonance Structure Revealed that the Human Polyomavirus JC Virus Agnoprotein Contains an α-Helix Encompassing the Leu/Ile/Phe-Rich Domain. J. Virol. 2014, 88, 6556–6575. [Google Scholar] [CrossRef] [Green Version]
- Strunze, S.; Engelke, M.F.; Wang, I.H.; Puntener, D.; Boucke, K.; Schleich, S.; Way, M.; Schoenenberger, P.; Burckhardt, C.J.; Greber, U.F. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe 2011, 10, 210–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mühlbauer, D.; Dzieciolowski, J.; Hardt, M.; Hocke, A.; Schierhorn, K.L.; Mostafa, A.; Müller, C.; Wisskirchen, C.; Herold, S.; Wolff, T.; et al. Influenza Virus-Induced Caspase-Dependent Enlargement of Nuclear Pores Promotes Nuclear Export of Viral Ribonucleoprotein Complexes. J. Virol. 2015, 89, 6009–6021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lammerding, J.; Stewart, C.L.; Lee, R.T.; Lammerding, J.; Schulze, P.C.; Takahashi, T.; Kozlov, S.; Sullivan, T.; Kamm, R.D.; Stewart, C.L.; et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Investig. 2004, 113, 370–378. [Google Scholar] [CrossRef] [Green Version]
- Gupta, P.; Bilinska, Z.T.; Sylvius, N.; Boudreau, E.; Veinot, J.P.; Labib, S.; Bolongo, P.M.; Hamza, A.; Jackson, T.; Ploski, R.; et al. Genetic and ultrastructural studies in dilated cardiomyopathy patients: A large deletion in the lamin A/C gene is associated with cardiomyocyte nuclear envelope disruption. Basic Res. Cardiol. 2010, 105, 365–377. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.Y.; Kim, P.; Weston, T.A.; Edillo, L.; Tu, Y.; Fong, L.G.; Young, S.G. Fibroblasts lacking nuclear lamins do not have nuclear blebs or protrusions but nevertheless have frequent nuclear membrane ruptures. Proc. Natl. Acad. Sci. USA 2018, 115, 10100–10105. [Google Scholar] [CrossRef] [Green Version]
- Villinger, C.; Neusser, G.; Kranz, C.; Walther, P.; Mertens, T. 3D Analysis of HCMV Induced-Nuclear Membrane Structures by FIB/SEM Tomography: Insight into an Unprecedented Membrane Morphology. Viruses 2015, 7, 5686–5704. [Google Scholar] [CrossRef]
- Ruchaud, S.; Korfali, N.; Villa, P.; Kottke, T.J.; Dingwall, C.; Kaufmann, S.H.; Earnshaw, W.C. Caspase-6 gene disruption reveals a requirement for lamin A cleavage in apoptotic chromatin condensation. EMBO J. 2002, 21, 1967–1977. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.H.; Ornelles, D.A.; Shenk, T. The adenovirus L3 23-kilodalton proteinase cleaves the amino-terminal head domain from cytokeratin 18 and disrupts the cytokeratin network of HeLa cells. J. Virol. 1993, 67, 3507–3514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kindsmuller, K.; Groitl, P.; Hartl, B.; Blanchette, P.; Hauber, J.; Dobner, T. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. USA 2007, 104, 6684–6689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reich, N.C.; Sarnow, P.; Duprey, E.; Levine, A.J. Monoclonal Antibodies Which Recognize Native and Denatured Forms of the Adenovirus DNA-Binding Protein. Virology 1983, 128, 480–484. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676. [Google Scholar] [CrossRef] [Green Version]
- Seligman, A.M.; Wasserkrug, H.L.; Hanker, J.S. A new staining method (OTO) for enhancing contrast of lipid-containing membranes and droplets in osmium tetroxide-fixed tissue with osmiophilic thiocarbohydrazide (TCH). J. Cell Biol. 1966, 30, 424–432. [Google Scholar] [CrossRef] [Green Version]
- Carlemalm, E.; Garavito, R.M.; Villiger, W. Resin development for electron microscopy and an analysis of embedding at low temperature. J. Microsc. 1982, 126, 123–143. [Google Scholar] [CrossRef]
- Flomm, F.J.; Soh, T.K.; Schneider, C.; Britt, H.M.; Thalassinos, K.; Pfitzner, S.; Reimer, R.; Grünewald, K.; Bosse, J.B. Intermittent bulk release of human cytomegalovirus through multivesicular bodies. bioRxiv 2021. [Google Scholar] [CrossRef]
- Hayles, M.F.; Stokes, D.J.; Phifer, D.; Findlay, K.C. A technique for improved focused ion beam milling of cryo-prepared life science specimens. J. Microsc. 2007, 226, 263–269. [Google Scholar] [CrossRef]
- Wolff, G.; Limpens, R.W.A.L.; Zheng, S.; Snijder, E.J.; Agard, D.A.; Koster, A.J.; Bárcena, M. Mind the gap: Micro-expansion joints drastically decrease the bending of FIB-milled cryo-lamellae. J. Struct. Biol. 2019, 208, 107389. [Google Scholar] [CrossRef] [PubMed]
- Hagen, W.J.H.; Wan, W.; Briggs, J.A.G. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 2017, 197, 191–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005, 152, 36–51. [Google Scholar] [CrossRef]
- Scheres, S.H.W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012, 180, 519–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, S.Q.; Palovcak, E.; Armache, J.-P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2—Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef] [Green Version]


| Name | Use | Sequence |
|---|---|---|
| ADP ccdB amp fw | PCR | GCTTAGAAAACCCTTAGGGTATTAGGCCAAAGGCGCAGCTACTGTGGGGTTTGCCAGTATACACTCCGCTAG |
| ADP ccdB amp rv | PCR | GGACAGAAATTTGCTAACTGATTTTAAGTAAGTGATGCTTTATTATTTTTTTTTACAGCCCCATACGATATAAGTTG |
| ADP rescue fw | PCR | GCTTAGAAAACCCTTAGGGTATTAGGCCAAAGGCGCAGCTACTGTGGGGTTTATGACCAACACAACCAACG |
| ADP rescue rv | PCR | GGACAGAAATTTGCTAACTGATTTTAAGTAAGTGATGCTTTATTATTTTTTTTTATCATACTGTAAGAGAAAAGAACATGTG |
| ADP-HA ccdB amp fw | PCR | GAATCCATAGATTGGACGGACTGAAACACATGTTCTTTTCTCTTACAGTAGCCAGTATACACTCCGCTAG |
| ADP-HA ccdB amp rv | PCR | ATTTGCTAACTGATTTTAAGTAAGTGATGCTTTATTATTTTTTTTTATCACAGCCCCATACGATATAAGTTG |
| ADP-HA rescue | Direct Red Rec. | GAATCCATAGATTGGACGGACTGAAACACATGTTCTTTTCTCTTACAGTATACCCATACGATGTTCCAGATTACGCTTGATAAAAAAAAATAATAAAGCATCACTTACTTAAAATCAGTTAGCAAATTTCTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pfitzner, S.; Bosse, J.B.; Hofmann-Sieber, H.; Flomm, F.; Reimer, R.; Dobner, T.; Grünewald, K.; Franken, L.E. Human Adenovirus Type 5 Infection Leads to Nuclear Envelope Destabilization and Membrane Permeability Independently of Adenovirus Death Protein. Int. J. Mol. Sci. 2021, 22, 13034. https://doi.org/10.3390/ijms222313034
Pfitzner S, Bosse JB, Hofmann-Sieber H, Flomm F, Reimer R, Dobner T, Grünewald K, Franken LE. Human Adenovirus Type 5 Infection Leads to Nuclear Envelope Destabilization and Membrane Permeability Independently of Adenovirus Death Protein. International Journal of Molecular Sciences. 2021; 22(23):13034. https://doi.org/10.3390/ijms222313034
Chicago/Turabian StylePfitzner, Søren, Jens B. Bosse, Helga Hofmann-Sieber, Felix Flomm, Rudolph Reimer, Thomas Dobner, Kay Grünewald, and Linda E. Franken. 2021. "Human Adenovirus Type 5 Infection Leads to Nuclear Envelope Destabilization and Membrane Permeability Independently of Adenovirus Death Protein" International Journal of Molecular Sciences 22, no. 23: 13034. https://doi.org/10.3390/ijms222313034
APA StylePfitzner, S., Bosse, J. B., Hofmann-Sieber, H., Flomm, F., Reimer, R., Dobner, T., Grünewald, K., & Franken, L. E. (2021). Human Adenovirus Type 5 Infection Leads to Nuclear Envelope Destabilization and Membrane Permeability Independently of Adenovirus Death Protein. International Journal of Molecular Sciences, 22(23), 13034. https://doi.org/10.3390/ijms222313034