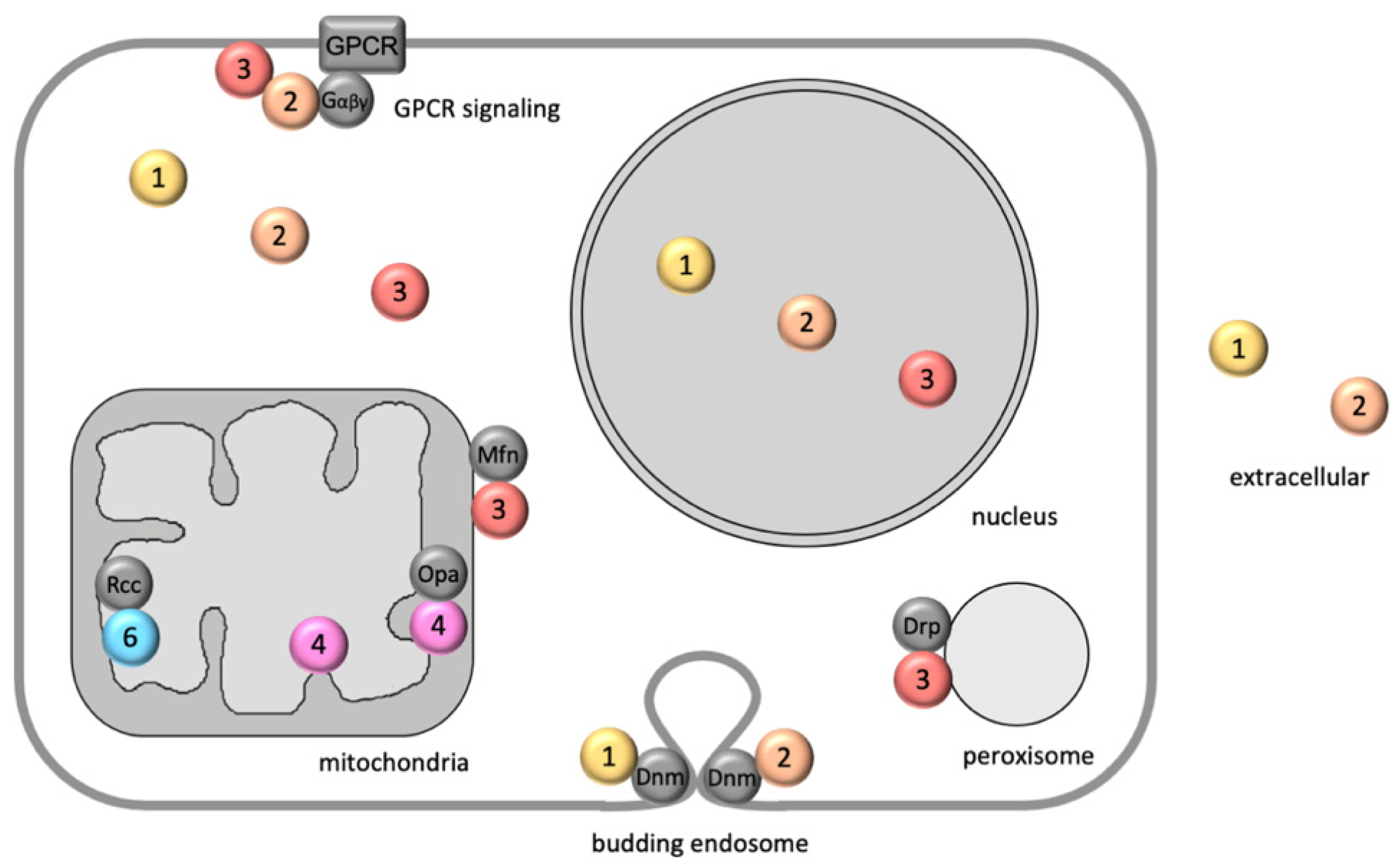
The Complex Functions of the NME Family—A Matter of Location and Molecular Activity
Funding
Acknowledgments
Conflicts of Interest
References
- Desvignes, T.; Pontarotti, P.; Fauvel, C.; Bobe, J. Nme protein family evolutionary history, a vertebrate perspective. BMC Evol. Biol. 2009, 9, 256. [Google Scholar] [CrossRef] [Green Version]
- Boissan, M.; Schlattner, U.; Lacombe, M.L. The NDPK/NME superfamily: State of the art. Lab. Investig. 2018, 98, 164–174. [Google Scholar] [CrossRef] [PubMed]
- The Nucleoside Diphosphate Kinase Superfamily: State of the Art. Available online: https://www.nature.com/collections/ttxdrtxsls (accessed on 3 October 2021).
- Special Issue “Selected Papers from the 11th International Conference on the NME/NDPK/NM23/AWD Gene Family (NME 2019)”. Available online: https://www.mdpi.com/journal/ijms/special_issues/NME_2019 (accessed on 3 October 2021).
- Georgescauld, F.; Song, Y.; Dautant, A. Structure, folding and stability of nucleoside diphosphate kinases. Int. J. Mol. Sci. 2020, 21, 6779. [Google Scholar] [CrossRef]
- Radic, M.; Sostar, M.; Weber, I.; Cetkovic, H.; Slade, N.; Herak Bosnar, M. The subcellular localization and oligomerization preferences of NME1/NME2 upon radiation-induced DNA damage. Int. J. Mol. Sci. 2020, 21, 2363. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.C.; Su, Y.A.; Chiu, H.Y.; Chen, C.W.; Huang, X.R.; Tei, R.; Wang, H.C.; Chuang, M.C.; Lin, Y.C.; Hsu, J.C.; et al. NME3 Binds to Phosphatidic Acid and Tethers Mitochondria for Fusion. 2021. Available online: https://ssrn.com/abstract=3808299 (accessed on 1 December 2021). [CrossRef]
- Abu-Taha, I.H.; Heijman, J.; Hippe, H.J.; Wolf, N.M.; El-Armouche, A.; Nikolaev, V.O.; Schafer, M.; Wurtz, C.M.; Neef, S.; Voigt, N.; et al. Nucleoside diphosphate kinase-C suppresses cAMP formation in human heart failure. Circulation 2017, 135, 881–897. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.W.; Wang, H.L.; Huang, C.W.; Huang, C.Y.; Lim, W.K.; Tu, I.C.; Koorapati, A.; Hsieh, S.T.; Kan, H.W.; Tzeng, S.R.; et al. Two separate functions of NME3 critical for cell survival underlie a neurodegenerative disorder. Proc. Natl. Acad. Sci. USA 2019, 116, 566–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honsho, M.; Abe, Y.; Imoto, Y.; Chang, Z.F.; Mandel, H.; Falik-Zaccai, T.C.; Fujiki, Y. Mammalian homologue NME3 of DYNAMO1 regulates peroxisome division. Int. J. Mol. Sci. 2020, 21, 8040. [Google Scholar] [CrossRef]
- Tokarska-Schlattner, M.; Boissan, M.; Munier, A.; Borot, C.; Mailleau, C.; Speer, O.; Schlattner, U.; Lacombe, M.L. The nucleoside diphosphate kinase D (NM23-H4) binds the inner mitochondrial membrane with high affinity to cardiolipin and couples nucleotide transfer with respiration. J. Biol. Chem. 2008, 283, 26198–26207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proust, B.; Radic, M.; Vidacek, N.S.; Cottet, C.; Attia, S.; Lamarche, F.; Ackar, L.; Mikulcic, V.G.; Tokarska-Schlattner, M.; Cetkovic, H.; et al. NME6 is a phosphotransfer-inactive, monomeric NME/NDPK family member and functions in complexes at the interface of mitochondrial inner membrane and matrix. Cell Biosci. 2021, 11, 195. [Google Scholar] [CrossRef]
- Romani, P.; Ignesti, M.; Gargiulo, G.; Hsu, T.; Cavaliere, V. Extracellular NME proteins: A player or a bystander? Lab. Investig. 2018, 98, 248–257. [Google Scholar] [CrossRef] [Green Version]
- Bunce, C.M.; Khanim, F.L. The ‘known-knowns’, and ‘known-unknowns’ of extracellular Nm23-H1/NDPK proteins. Lab. Investig. 2018, 98, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Zala, D.; Schlattner, U.; Desvignes, T.; Bobe, J.; Roux, A.; Chavrier, P.; Boissan, M. The advantage of channeling nucleotides for very processive functions. F1000Research 2017, 6, 724. [Google Scholar] [CrossRef]
- Boissan, M.; Montagnac, G.; Shen, Q.; Griparic, L.; Guitton, J.; Romao, M.; Sauvonnet, N.; Lagache, T.; Lascu, I.; Raposo, G.; et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science 2014, 344, 1510–1515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imoto, Y.; Abe, Y.; Honsho, M.; Okumoto, K.; Ohnuma, M.; Kuroiwa, H.; Kuroiwa, T.; Fujiki, Y. Onsite GTP fuelling via DYNAMO1 drives division of mitochondria and peroxisomes. Nat. Commun. 2018, 9, 4634. [Google Scholar] [CrossRef]
- Chen, C.W.; Tsao, N.; Zhang, W.; Chang, Z.F. NME3 regulates mitochondria to reduce ROS-mediated genome instability. Int. J. Mol. Sci. 2020, 21, 5048. [Google Scholar] [CrossRef]
- Imoto, Y.; Itoh, K.; Fujiki, Y. Molecular basis of mitochondrial and peroxisomal division machineries. Int. J. Mol. Sci. 2020, 21, 5452. [Google Scholar] [CrossRef]
- Kalagiri, R.; Hunter, T. The many ways that nature has exploited the unusual structural and chemical properties of phosphohistidine for use in proteins. Biochem. J. 2021, 478, 3575–3596. [Google Scholar] [CrossRef] [PubMed]
- Adam, K.; Ning, J.; Reina, J.; Hunter, T. NME/NM23/NDPK and histidine phosphorylation. Int. J. Mol. Sci. 2020, 21, 5848. [Google Scholar] [CrossRef] [PubMed]
- Adam, K.; Lesperance, J.; Hunter, T.; Zage, P.E. The potential functional roles of NME1 histidine kinase activity in neuroblastoma pathogenesis. Int. J. Mol. Sci. 2020, 21, 3319. [Google Scholar] [CrossRef] [PubMed]
- Puts, G.S.; Leonard, M.K.; Pamidimukkala, N.V.; Snyder, D.E.; Kaetzel, D.M. Nuclear functions of NME proteins. Lab. Investig. 2018, 98, 211–218. [Google Scholar] [CrossRef]
- Puts, G.; Jarrett, S.; Leonard, M.; Matsangos, N.; Snyder, D.; Wang, Y.; Vincent, R.; Portney, B.; Abbotts, R.; McLaughlin, L.; et al. Metastasis suppressor NME1 modulates choice of double-strand break repair pathways in melanoma cells by enhancing alternative NHEJ while inhibiting NHEJ and HR. Int. J. Mol. Sci. 2020, 21, 5896. [Google Scholar] [CrossRef]
- Sharma, S.; Sengupta, A.; Chowdhury, S. Emerging molecular connections between NM23 proteins, telomeres and telomere-associated factors: Implications in cancer metastasis and ageing. Int. J. Mol. Sci. 2021, 22, 3457. [Google Scholar] [CrossRef]
- Yu, B.Y.K.; Tossounian, M.A.; Hristov, S.D.; Lawrence, R.; Arora, P.; Tsuchiya, Y.; Peak-Chew, S.Y.; Filonenko, V.; Oxenford, S.; Angell, R.; et al. Regulation of metastasis suppressor NME1 by a key metabolic cofactor coenzyme A. Redox Biol. 2021, 44, 101978. [Google Scholar] [CrossRef] [PubMed]
- Schlattner, U.; Tokarska-Schlattner, M.; Ramirez, S.; Tyurina, Y.Y.; Amoscato, A.A.; Mohammadyani, D.; Huang, Z.; Jiang, J.; Yanamala, N.; Seffouh, A.; et al. Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: A cardiolipin-dependent switch. J. Biol. Chem. 2013, 288, 111–121. [Google Scholar] [CrossRef] [Green Version]
- Schlattner, U.; Tokarska-Schlattner, M.; Epand, R.M.; Boissan, M.; Lacombe, M.L.; Kagan, V.E. NME4/nucleoside diphosphate kinase D in cardiolipin signaling and mitophagy. Lab. Investig. 2018, 98, 228–232. [Google Scholar] [CrossRef]
- Kagan, V.E.; Jiang, J.; Huang, Z.; Tyurina, Y.Y.; Desbourdes, C.; Cottet-Rousselle, C.; Dar, H.H.; Verma, M.; Tyurin, V.A.; Kapralov, A.A.; et al. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death Differ. 2016, 23, 1140–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, S.S.; He, Q.; Janczy, J.R.; Elliott, E.I.; Zhong, Z.; Olivier, A.K.; Sadler, J.J.; Knepper-Adrian, V.; Han, R.; Qiao, L.; et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 2013, 39, 311–323. [Google Scholar] [CrossRef] [Green Version]
- Ernst, O.; Sun, J.; Lin, B.; Banoth, B.; Dorrington, M.G.; Liang, J.; Schwarz, B.; Stromberg, K.A.; Katz, S.; Vayttaden, S.J.; et al. A genome-wide screen uncovers multiple roles for mitochondrial nucleoside diphosphate kinase D in inflammasome activation. Sci. Signal. 2021, 14. [Google Scholar] [CrossRef] [PubMed]
- Huna, A.; Nawrocki-Raby, B.; Padilla-Benavides, T.; Gavard, J.; Coscoy, S.; Bernard, D.; Boissan, M. Loss of the metastasis suppressor NME1, but not of its highly related isoform NME2, induces a hybrid epithelial-mesenchymal state in cancer cells. Int. J. Mol. Sci. 2021, 22, 3718. [Google Scholar] [CrossRef]
- Lacombe, M.L.; Lamarche, F.; De Wever, O.; Padilla-Benavides, T.; Carlson, A.; Khan, I.; Huna, A.; Vacher, S.; Calmel, C.; Desbourdes, C.; et al. The mitochondrially-localized nucleoside diphosphate kinase D (NME4) is a novel metastasis suppressor. BMC Biol. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Gril, B.; Steeg, P.S. Metastasis suppressors NME1 and NME2 promote dynamin 2 oligomerization and regulate tumor cell endocytosis, motility, and metastasis. Cancer Res. 2019, 79, 4689–4702. [Google Scholar] [CrossRef] [PubMed]
- Lodillinsky, C.; Fuhrmann, L.; Irondelle, M.; Pylypenko, O.; Li, X.Y.; Bonsang-Kitzis, H.; Reyal, F.; Vacher, S.; Calmel, C.; De Wever, O.; et al. Metastasis-suppressor NME1 controls the invasive switch of breast cancer by regulating MT1-MMP surface clearance. Oncogene 2021, 40, 4019–4032. [Google Scholar] [CrossRef] [PubMed]
- Serafini, G.; Giordani, G.; Grillini, L.; Andrenacci, D.; Gargiulo, G.; Cavaliere, V. The impact of Drosophila Awd/NME1/2 levels on notch and Wg signaling pathways. Int. J. Mol. Sci. 2020, 21, 7257. [Google Scholar] [CrossRef]
- Pennino, F.P.; Murakami, M.; Zollo, M.; Robertson, E.S. The metastasis suppressor protein NM23-H1 modulates the PI3K-AKT axis through interaction with the p110alpha catalytic subunit. Oncogenesis 2021, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Felix, I.; Lomada, S.K.; Barth, H.; Wieland, T. Bacillus anthracis’ PA63 delivers the tumor metastasis suppressor protein NDPK-A/NME1 into breast cancer cells. Int. J. Mol. Sci. 2020, 21, 3295. [Google Scholar] [CrossRef]
- Kim, B.; Lee, K.J. Activation of Nm23-H1 to suppress breast cancer metastasis via redox regulation. Exp. Mol. Med. 2021, 53, 346–357. [Google Scholar] [CrossRef]
- Duan, S.; Nordmeier, S.; Byrnes, A.E.; Buxton, I.L.O. Extracellular vesicle-mediated purinergic signaling contributes to host microenvironment plasticity and metastasis in triple negative breast cancer. Int. J. Mol. Sci. 2021, 22, 597. [Google Scholar] [CrossRef]
- Gupta, A.; Sinha, K.M.; Abdin, M.Z.; Puri, N.; Selvapandiyan, A. NDK/NME proteins: A host-pathogen interface perspective towards therapeutics. Curr. Genet. 2021. [Google Scholar] [CrossRef] [PubMed]
- Abu-Taha, I.H.; Vettel, C.; Wieland, T. Targeting altered Nme heterooligomerization in disease? Oncotarget 2018, 9, 1492–1493. [Google Scholar] [CrossRef]
- Abu-Taha, I.H.; Heijman, J.; Feng, Y.; Vettel, C.; Dobrev, D.; Wieland, T. Regulation of heterotrimeric G-protein signaling by NDPK/NME proteins and caveolins: An update. Lab. Investig. 2018, 98, 190–197. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, A.; Eshwaran, R.; Poschet, G.; Lomada, S.; Halawa, M.; Wilhelm, K.; Schmidt, M.; Hammes, H.P.; Wieland, T.; Feng, Y. Involvement of NDPK-B in glucose metabolism-mediated endothelial damage via activation of the hexosamine biosynthesis pathway and suppression of O-GlcNAcase activity. Cells 2020, 9, 2324. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhao, D.; Butenschon, V.M.; Bauer, A.T.; Schneider, S.W.; Skolnik, E.Y.; Hammes, H.P.; Wieland, T.; Feng, Y. Nucleoside diphosphate kinase B deficiency causes a diabetes-like vascular pathology via up-regulation of endothelial angiopoietin-2 in the retina. Acta Diabetol. 2016, 53, 81–89. [Google Scholar] [CrossRef]
- Chatterjee, A.; Eshwaran, R.; Huang, H.; Zhao, D.; Schmidt, M.; Wieland, T.; Feng, Y. Role of the Ang2-Tie2 axis in vascular damage driven by high glucose or nucleoside diphosphate kinase B deficiency. Int. J. Mol. Sci. 2020, 21, 3713. [Google Scholar] [CrossRef]
- Anderegg, L.; Im Hof Gut, M.; Hetzel, U.; Howerth, E.W.; Leuthard, F.; Kyostila, K.; Lohi, H.; Pettitt, L.; Mellersh, C.; Minor, K.M.; et al. NME5 frameshift variant in Alaskan Malamutes with primary ciliary dyskinesia. PLoS Genet. 2019, 15, e1008378. [Google Scholar] [CrossRef]
- Cho, E.H.; Huh, H.J.; Jeong, I.; Lee, N.Y.; Koh, W.J.; Park, H.C.; Ki, C.S. A nonsense variant in NME5 causes human primary ciliary dyskinesia with radial spoke defects. Clin. Genet. 2020, 98, 64–68. [Google Scholar] [CrossRef]
- Sahabian, A.; von Schlehdorn, L.; Drick, N.; Pink, I.; Dahlmann, J.; Haase, A.; Gohring, G.; Welte, T.; Martin, U.; Ringshausen, F.C.; et al. Generation of two hiPSC clones (MHHi019-A, MHHi019-B) from a primary ciliary dyskinesia patient carrying a homozygous deletion in the NME5 gene (c.415delA (p.Ile139Tyrfs*8)). Stem Cell Res. 2020, 48, 101988. [Google Scholar] [CrossRef]
- Sedova, L.; Bukova, I.; Bazantova, P.; Petrezselyova, S.; Prochazka, J.; Skolnikova, E.; Zudova, D.; Vcelak, J.; Makovicky, P.; Bendlova, B.; et al. Semi-lethal primary ciliary dyskinesia in rats lacking the Nme7 gene. Int. J. Mol. Sci. 2021, 22, 3810. [Google Scholar] [CrossRef] [PubMed]
- Sedova, L.; Skolnikova, E.; Hodulova, M.; Vcelak, J.; Seda, O.; Bendlova, B. Expression profiling of Nme7 interactome in experimental models of metabolic syndrome. Physiol. Res. 2018, 67, S543–S550. [Google Scholar] [CrossRef] [PubMed]
- Sedova, L.; Prochazka, J.; Zudova, D.; Bendlova, B.; Vcelak, J.; Sedlacek, R.; Seda, O. Heterozygous Nme7 mutation affects glucose tolerance in male rats. Genes 2021, 12, 1087. [Google Scholar] [CrossRef] [PubMed]
- Perina, D.; Korolija, M.; Mikoc, A.; Halasz, M.; Herak Bosnar, M.; Cetkovic, H. Characterization of Nme5-like gene/protein from the red alga Chondrus crispus. Mar. Drugs 2019, 18, 13. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.; Rong, Z.; Liu, X.; Gao, J.; Xu, X.; Zi, Y.; Mu, Y.; Guan, Y.; Cao, Z.; Zhang, Y.; et al. The protein kinase activity of NME7 activates Wnt/beta-Catenin signaling to promote one-carbon metabolism in hepatocellular carcinoma. Cancer Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.; Steeg, P.S.; Zollo, M.; Wieland, T.; The Steering Committee on Nme-related Research; The Organizers of the International Congresses of the NDP Kinase/Nm23/awd Gene Family. Progress on Nme (NDP kinase/Nm23/Awd) gene family-related functions derived from animal model systems: Studies on development, cardiovascular disease, and cancer metastasis exemplified. Naunyn-Schmiedeberg’s Arch Pharm. 2015, 388, 109–117. [Google Scholar] [CrossRef] [Green Version]
- Potel, C.M.; Fasci, D.; Heck, A.J.R. Mix and match of the tumor metastasis suppressor Nm23 protein isoforms in vitro and in vivo. FEBS J. 2018, 285, 2856–2868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schlattner, U. The Complex Functions of the NME Family—A Matter of Location and Molecular Activity. Int. J. Mol. Sci. 2021, 22, 13083. https://doi.org/10.3390/ijms222313083
Schlattner U. The Complex Functions of the NME Family—A Matter of Location and Molecular Activity. International Journal of Molecular Sciences. 2021; 22(23):13083. https://doi.org/10.3390/ijms222313083
Chicago/Turabian StyleSchlattner, Uwe. 2021. "The Complex Functions of the NME Family—A Matter of Location and Molecular Activity" International Journal of Molecular Sciences 22, no. 23: 13083. https://doi.org/10.3390/ijms222313083
APA StyleSchlattner, U. (2021). The Complex Functions of the NME Family—A Matter of Location and Molecular Activity. International Journal of Molecular Sciences, 22(23), 13083. https://doi.org/10.3390/ijms222313083




