The Role of Topoisomerase II in DNA Repair and Recombination in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
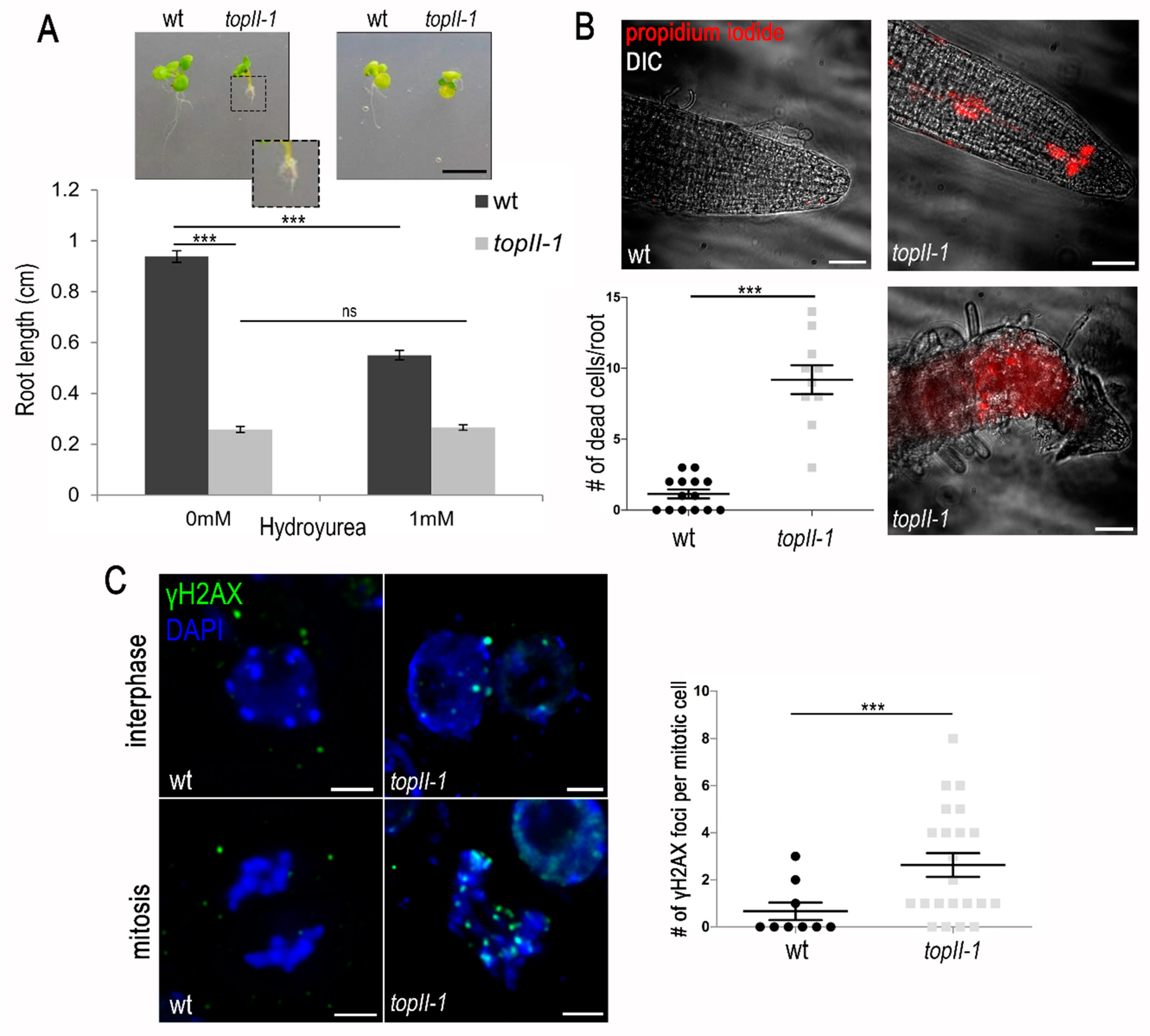
2.1. TOPII Plays an Important Role in Mitotic DNA Replication
2.2. HR-Mediated DNA Repair Problems Are Observed in TopII-1
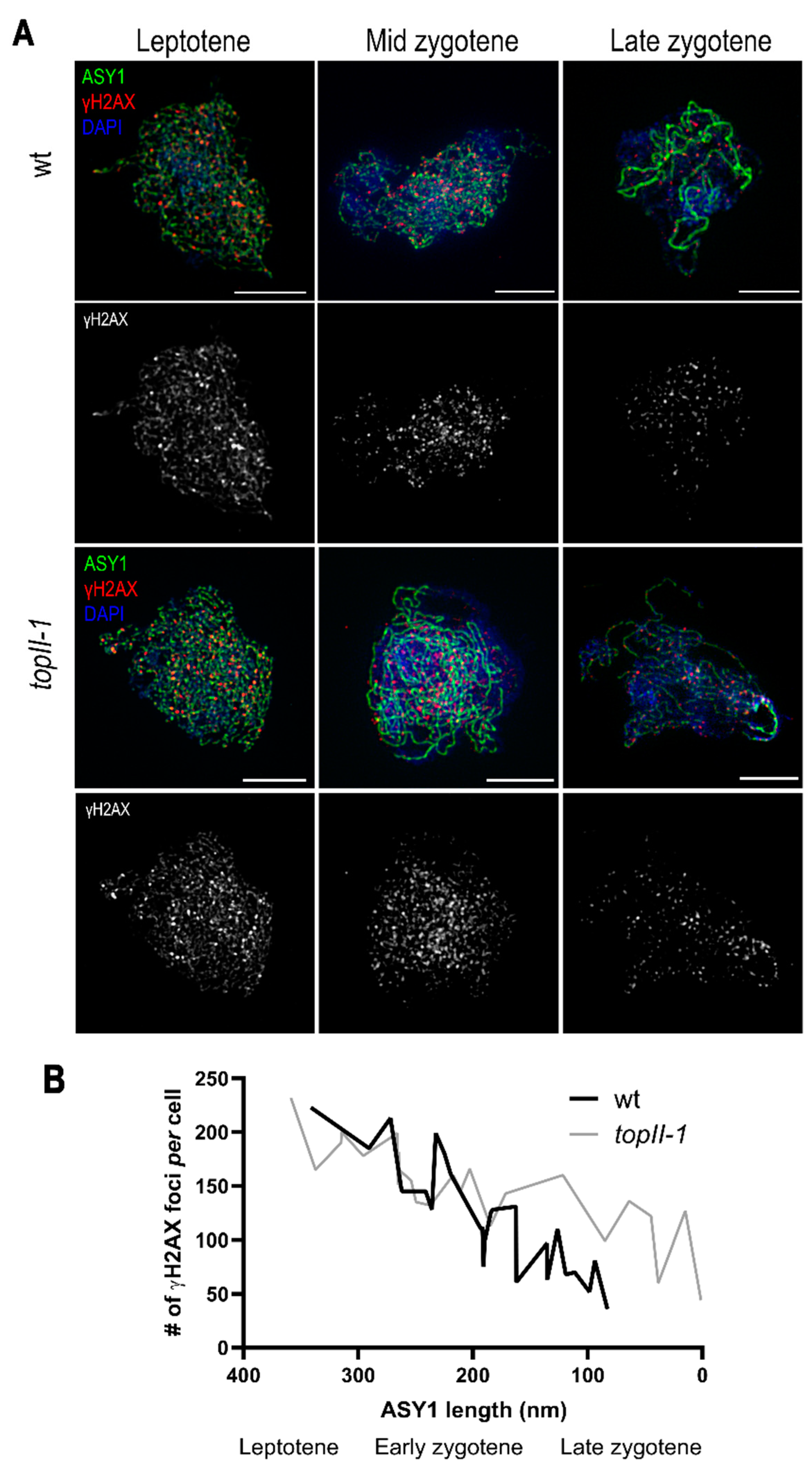
2.3. The Dynamics of Meiotic DSB Repair Are Affected in TopII-1
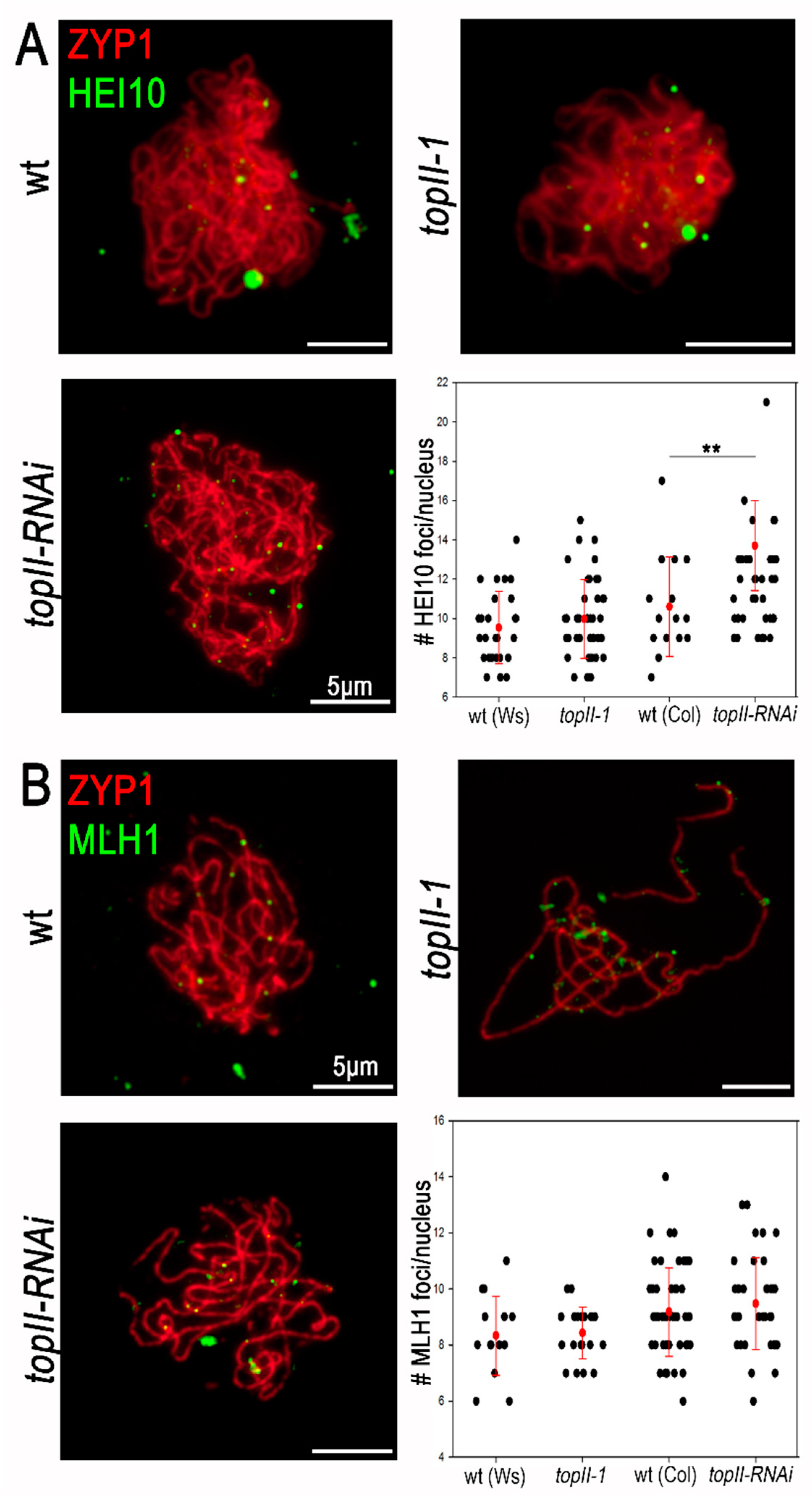
2.4. Participation of TOPII in Meiotic CO Formation
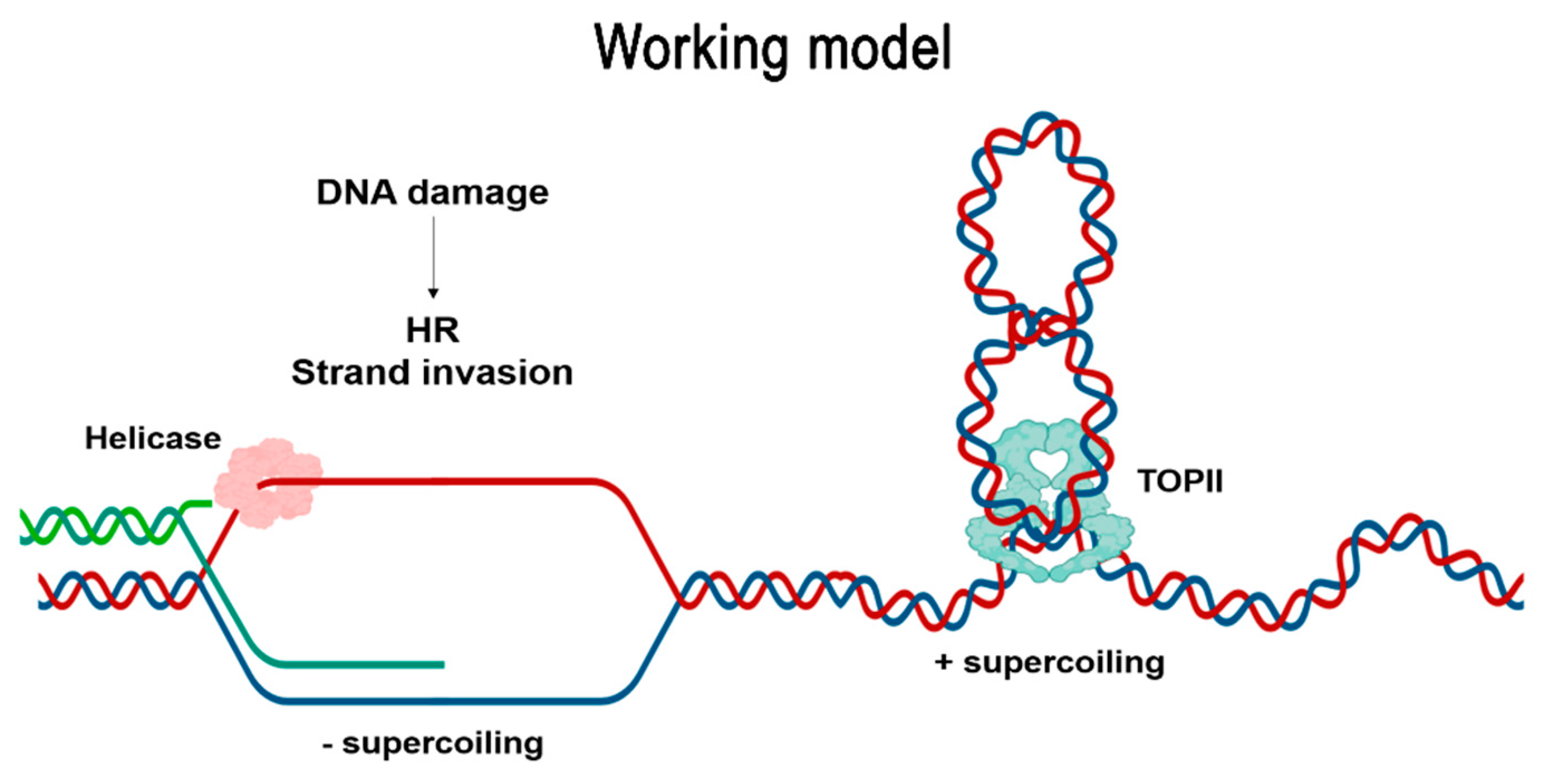
3. Discussion
3.1. Mitotic DNA Replication Is Affected More Than Meiotic DNA Replication in TopII-1
3.2. CO Formation Appears Normal in TopII-1 and TopII-RNAi Plants
3.3. Mitotic and Meiotic DSBs Can Remain Unrepaired in TopII-1
4. Materials and Methods
4.1. Plant Material
4.2. Plant Growth and Genotoxicity Experiments
4.3. Cytogenetic Techniques
4.4. Statistical and Graphic Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Germe, T.; Miller, K.; Cooper, J.P. A non-canonical function of topoisomerase II in disentangling dysfunctional telomeres. EMBO J. 2009, 28, 2803–2811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branzei, D.; Foiani, M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 2010, 11, 208–219. [Google Scholar] [CrossRef]
- Hughes, S.E.; Hawley, R.S. Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Drosophila melanogaster Female Meiosis. Copenhaver GP, editor. PLoS Genet. 2014, 10, e1004650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shintomi, K.; Takahashi, T.S.; Hirano, T. Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat. Cell Biol. 2015, 17, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Wu, J.; Modrich, P.; Hsieh, T. The C-terminal 20 Amino Acids of Drosophila Topoisomerase 2 Are Required for Binding to a BRCA1 C Terminus (BRCT) Domain-containing Protein, Mus101, and Fidelity of DNA Segregation. J. Biol. Chem. 2016, 291, 13216–13228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Cussiol, J.R.; Dibitetto, D.; Sims, J.R.; Twayana, S.; Weiss, R.S.; Freire, R.; Marini, F.; Pellicioli, A.; Smolka, M.B. TOPBP1 Dpb11 plays a conserved role in homologous recombination DNA repair through the coordinated recruitment of 53BP1 Rad9. J. Cell Biol. 2017, 216, 623–639. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Garcia, M.; Schubert, V.; Osman, K.; Darbyshire, A.; Sanchez-Moran, E.; Franklin, F.C.H. TOPII and chromosome movement help remove interlocks between entangled chromosomes during meiosis. J. Cell Biol. 2018, 217, 4070–4079. [Google Scholar] [CrossRef]
- Heldrich, J.; Sun, X.; Vale-Silva, L.A.; Markowitz, T.E.; Hochwagen, A. Topoisomerases Modulate the Timing of Meiotic DNA Breakage and Chromosome Morphogenesis in Saccharomyces cerevisiae. Genetics 2020, 215, 59–73. [Google Scholar] [CrossRef] [Green Version]
- Bush, N.G.; Evans-Roberts, K.; Maxwell, A. DNA Topoisomerases. EcoSal Plus 2015, 6. [Google Scholar] [CrossRef]
- Hacker, L.; Dorn, A.; Enderle, J.; Puchta, H. The repair of topoisomerase 2 cleavage complexes in Arabidopsis. Plant Cell 2021, koab228. [Google Scholar] [CrossRef]
- Vologodskii, A. Disentangling DNA molecules. Phys. Life Rev. 2016, 18, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, A.I.; Cozzarelli, N.R.; Holmes, V.F.; Khodursky, A.B.; Peter, B.J.; Postow, L.; Rybenkov, V.; Vologodskii, A.V. Mechanisms of separation of the complementary strands of DNA during replication. Genetica 1999, 106, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Mundbjerg, K.; Jørgensen, S.W.; Fredsøe, J.; Nielsen, I.; Pedersen, J.M.; Bentsen, I.B.; Lisby, M.; Bjergbaek, L.; Andersen, A.H. Top2 and Sgs1-Top3 Act Redundantly to Ensure rDNA Replication Termination. Copenhaver GP, editor. PLoS Genet. 2015, 11, e1005697. [Google Scholar] [CrossRef] [Green Version]
- Baxter, J. “Breaking up is hard to do”: The formation and resolution of sister chromatid intertwines. J. Mol. Biol. 2015, 427, 590–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeney, S.; Giroux, C.N.; Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 1997, 88, 375–384. [Google Scholar] [CrossRef] [Green Version]
- Manova, V.; Gruszka, D. DNA damage and repair in plants—From models to crops. Front. Plant Sci. 2015, 6, 885. [Google Scholar] [CrossRef] [Green Version]
- Jasin, M.; Rothstein, R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013, 5, a012740. [Google Scholar] [CrossRef]
- Wang, Y.; Copenhaver, G.P. Meiotic Recombination: Mixing It Up in Plants. Annu. Rev. Plant Biol. 2018, 69, 577–609. [Google Scholar] [CrossRef]
- Brown, M.S.; Bishop, D.K. DNA Strand Exchange and RecA Homologs in Meiosis. Cold Spring Harb. Perspect. Biol. 2015, 7, a016659. [Google Scholar] [CrossRef] [Green Version]
- Pradillo, M.; Varas, J.; Oliver, C.; Santos, J.L. On the role of AtDMC1, AtRAD51 and its paralogs during Arabidopsis meiosis. Front. Plant Sci. 2014, 5, 23. [Google Scholar] [CrossRef] [Green Version]
- Holliday, R. A mechanism for gene conversion in fungi. Genet. Res. Camb. 1964, 5, 282–304. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Mao, C.; Iwasaki, H.; Kemper, B.; Seeman, N.C. No braiding of Holliday junctions in positively supercoiled DNA molecules. J. Mol. Biol. 1999, 294, 683–699. [Google Scholar] [CrossRef]
- Lu, C.-H.; Li, H.-W. DNA with Different Local Torsional States Affects RecA-Mediated Recombination Progression. ChemPhysChem 2017, 18, 584–590. [Google Scholar] [CrossRef]
- Morotomi-Yano, K.; Saito, S.; Adachi, N.; Yano, K. Dynamic behavior of DNA topoisomerase IIβ in response to DNA double-strand breaks. Sci. Rep. 2018, 8, 10344. [Google Scholar] [CrossRef] [Green Version]
- Lambing, C.; Franklin, F.C.H.; Wang, C.-J.R. Understanding and Manipulating Meiotic Recombination in Plants. Plant Physiol. 2017, 173, 1530–1542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, L.B.; Hunsicker, P.R.; Kerley, M.; Pyle, A.; Saxton, A.M. Etoposide exposure during male mouse pachytene has complex effects on crossing-over and causes nondisjunction. Mutat. Res./Genet. Toxicol. Environ. Mutagenesis 2004, 565, 61–77. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, S.; Yin, S.; Hong, S.; Kim, K.P.; Kleckner, N. Topoisomerase II mediates meiotic crossover interference. Nature 2014, 511, 551–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Z.; Zickler, D.; Prentiss, M.; Chang, F.S.; Witz, G.; Maeshima, K.; Kleckner, N. Chromosomes Progress to Metaphase in Multiple Discrete Steps via Global Compaction/Expansion Cycles. Cell 2015, 161, 1124–1137. [Google Scholar] [CrossRef] [Green Version]
- Klein, F.; Laroche, T.; E Cardenas, M.; Hofmann, J.F.; Schweizer, D.; Gasser, S. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J. Cell Biol. 1992, 117, 935–948. [Google Scholar] [CrossRef] [Green Version]
- Fukata, H.; Ohgami, K.; Fukasawa, H. Isolation and characterization of DNA topoisomerase II from cauliflower inflorescences. Plant Mol. Biol. 1986, 6, 137–144. [Google Scholar] [CrossRef]
- Carballo, M.; Giné, R.; Santos, M.; Puigdomènech, P. Characterization of topoisomerase I and II activities in nuclear extracts during callogenesis in immature embryos of Zea mays. Plant Mol. Biol. 1991, 16, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.N.; Mudgil, Y.; Sopory, S.K.; Reddy, M.K. Molecular characterization of a nuclear topoisomerase II from Nicotiana tabacum that functionally complements a temperature-sensitive topoisomerase II yeast mutant. Plant Mol. Biol. 2003, 52, 1063–1076. [Google Scholar] [CrossRef]
- Singh, B.N.; Achary, V.M.M.; Panditi, V.; Sopory, S.K.; Reddy, M.K. Dynamics of tobacco DNA topoisomerases II in cell cycle regulation: To manage topological constrains during replication, transcription and mitotic chromosome condensation and segregation. Plant Mol. Biol. 2017, 94, 595–607. [Google Scholar] [CrossRef]
- Xie, S.; Lam, E. Characterization of a DNA Topoisomerase II cDNA from Arabidopsis thaliana. Plant Physiol. 1994, 106, 1701–1702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, S.; Lam, E. Abundance of nuclear DNA topoisomerase II is correlated with proliferation in Arabidopsis thaliana. Nucleic Acids Res. 1994, 22, 5729–5736. [Google Scholar] [CrossRef] [Green Version]
- Nyberg, K.A.; Michelson, R.J.; Putnam, C.W.; Weinert, T.A. Toward Maintaining the Genome: DNA Damage and Replication Checkpoints. Annu. Rev. Genet. 2002, 36, 617–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paull, T.T.; Rogakou, E.P.; Yamazaki, V.; Kirchgessner, C.U.; Gellert, M.; Bonner, W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000, 10, 886–895. [Google Scholar] [CrossRef] [Green Version]
- Benítez-Bribiesca, L.; Sánchez-Suárez, P. Oxidative Damage, Bleomycin, and Gamma Radiation Induce Different Types of DNA Strand Breaks in Normal Lymphocytes and Thymocytes: A Comet Assay Study. Ann. N. Y. Acad. Sci. 1999, 887, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Charbonnel, C.; Allain, E.; Gallego, M.E.; White, C.I. Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. DNA Repair 2011, 10, 611–619. [Google Scholar] [CrossRef]
- Deans, A.J.; West, S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 2011, 11, 467–480. [Google Scholar] [CrossRef] [Green Version]
- Tomasz, M.; Lipman, R.; Chowdary, D.; Pawlak, J.; Verdine, G.; Nakanishi, K. Isolation and structure of a covalent cross-link adduct between mitomycin C and DNA. Science 1987, 235, 1204–1208. [Google Scholar] [CrossRef] [Green Version]
- Higgins, J.D.; Buckling, E.F.; Franklin, F.C.H.; Jones, G.H. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over: AtMUS81 expression and function. Plant J. 2008, 54, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Mannuss, A.; Dukowic-Schulze, S.; Suer, S.; Hartung, F.; Pacher, M.; Puchta, H. RAD5A, RECQ4A, and MUS81 Have Specific Functions in Homologous Recombination and Define Different Pathways of DNA Repair in Arabidopsis thaliana. Plant Cell 2010, 22, 3318–3330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eastman, A. Interstrand cross-links and sequence specificity in the reaction of cis-dichloro(ethylenediamine)platinum(II) with DNA. Biochemistry 1985, 24, 5027–5032. [Google Scholar] [CrossRef] [PubMed]
- De Silva, I.U. Defects in interstrand cross-link uncoupling do not account for the extreme sensitivity of ERCC1 and XPF cells to cisplatin. Nucleic Acids Res. 2002, 30, 3848–3856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olivier, M.; Da Ines, O.; Amiard, S.; Serra, H.; Goubely, C.; White, C.I.; Gallego, M.E. The Structure-Specific Endonucleases MUS81 and SEND1 Are Essential for Telomere Stability in Arabidopsis. Plant Cell 2016, 28, 74–86. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Moran, E.; Santos, J.-L.; Jones, G.H.; Franklin, F.C.H. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007, 21, 2220–2233. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, S.J.; Caryl, A.P.; Jones, G.H.; Franklin, F.C. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 2002, 115, 3645–3655. [Google Scholar] [CrossRef] [Green Version]
- Hetherington, D. The Exponential Function and Its Applications in Science. Pwiki. 2011. Available online: http://gauss.vaniercollege.qc.ca/pwiki/index.php?title=The_Exponential_Function_and_Its_Applications_in_Science&oldid=1269 (accessed on 15 September 2020).
- Chelysheva, L.; Vezon, D.; Chambon, A.; Gendrot, G.; Pereira, L.; Lemhemdi, A.; Vrielynck, N.; Le Guin, S.; Novatchkova, M.; Grelon, M. The Arabidopsis HEI10 Is a New ZMM Protein Related to Zip3. Franklin FCH, editor. PLoS Genet. 2012, 8, e1002799. [Google Scholar] [CrossRef] [Green Version]
- De Muyt, A.; Zhang, L.; Piolot, T.; Kleckner, N.; Espagne, E.; Zickler, D. E3 ligase Hei10: A multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev. 2014, 28, 1111–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Culligan, K.; Tissier, A.; Britt, A. ATR Regulates a G2-Phase Cell-Cycle Checkpoint in Arabidopsis thaliana. Plant Cell 2004, 16, 1091–1104. [Google Scholar] [CrossRef] [Green Version]
- Edgar, B.A.; Zielke, N.; Gutierrez, C. Endocycles: A recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 2014, 15, 197–210. [Google Scholar] [CrossRef]
- Kalve, S.; De Vos, D.; Beemster, G.T.S. Leaf development: A cellular perspective. Front. Plant Sci. 2014, 5, 362. [Google Scholar] [CrossRef] [Green Version]
- Hakovirta, H.; Parvinen, M.; Lähdetie, J. Effects of etoposide on stage-specific DNA synthesis during rat spermatogenesis. Mutat. Res. Lett. 1993, 301, 189–193. [Google Scholar] [CrossRef]
- Varas, J.; Sánchez-Morán, E.; Copenhaver, G.P.; Santos, J.L.; Pradillo, M. Analysis of the Relationships between DNA Double-Strand Breaks, Synaptonemal Complex and Crossovers Using the Atfas1-4 Mutant. PLoS Genet. 2015, 11, e1005301. [Google Scholar] [CrossRef] [Green Version]
- Varas, J.; Santos, J.L.; Pradillo, M. The Absence of the Arabidopsis Chaperone Complex CAF-1 Produces Mitotic Chromosome Abnormalities and Changes in the Expression Profiles of Genes Involved in DNA Repair. Front. Plant Sci. 2017, 8, 525. [Google Scholar] [CrossRef] [Green Version]
- Holm, P.B. The premeiotic DNA replication of euchromatin and heterochromatin in Lilium longiflorum (Thunb.). Carlsberg Res. Commun. 1977, 42, 249–281. [Google Scholar] [CrossRef] [Green Version]
- Cha, R.S.; Weiner, B.M.; Keeney, S.; Dekker, J.; Kleckner, N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 2000, 14, 493–503. [Google Scholar] [CrossRef]
- Stronghill, P.E.; Azimi, W.; Hasenkampf, C.A. A novel method to follow meiotic progression in Arabidopsis using confocal microscopy and 5-ethynyl-2′-deoxyuridine labeling. Plant Methods 2014, 10, 33. [Google Scholar] [CrossRef] [Green Version]
- Mickelson-Young, L.; Wear, E.; Mulvaney, P.; Lee, T.-J.; Szymanski, E.S.; Allen, G.; Hanley-Bowdoin, L.; Thompson, W. A flow cytometric method for estimating S-phase duration in plants. J. Exp. Bot. 2016, 67, 6077–6087. [Google Scholar] [CrossRef] [Green Version]
- Lambing, C.; Osman, K.; Nuntasoontorn, K.; West, A.; Higgins, J.D.; Copenhaver, G.P.; Yang, J.; Armstrong, S.; Mechtler, K.; Roitinger, E.; et al. Arabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers. Puchta, H., editor. PLoS Genet. 2015, 11, e1005372. [Google Scholar] [CrossRef] [Green Version]
- Makarevitch, I.; Somers, D.A. Purification and characterization of topoisomerase IIA from Arabidopsis thaliana. Plant Sci. 2005, 168, 1023–1033. [Google Scholar] [CrossRef]
- Povirk, L.F. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: Bleomycin, neocarzinostatin and other enediynes. Mutat. Res. 1996, 355, 71–89. [Google Scholar] [CrossRef]
- Parra-Nunez, P.; Cooper, C.; Sanchez-Moran, E. The Role of DNA Topoisomerase Binding Protein 1 (TopBP1) in Genome Stability in Arabidopsis. Plants 2021, 10, 2568. [Google Scholar] [CrossRef]
- Sasaki, M.S.; Takata, M.; Sonoda, E.; Tachibana, A.; Takeda, S. Recombination repair pathway in the maintenance of chromosomal integrity against DNA interstrand crosslinks. Cytogenet. Genome Res. 2004, 104, 28–34. [Google Scholar] [CrossRef]
- West, K.L.; Austin, C.A. Human DNA topoisomerase IIβ binds and cleaves four-way junction DNA in vitro. Nucleic Acids Res. 1999, 27, 984–992. [Google Scholar] [CrossRef]
- René, B.; Fermandjian, S.; Mauffret, O. Does topoisomerase II specifically recognize and cleave hairpins, cruciforms and crossovers of DNA? Biochimie 2007, 89, 508–515. [Google Scholar] [CrossRef]
- van der Heijden, T.; Modesti, M.; Hage, S.; Kanaar, R.; Wyman, C.; Dekker, C. Homologous Recombination in Real Time: DNA Strand Exchange by RecA. Mol. Cell 2008, 30, 530–538. [Google Scholar] [CrossRef]
- Wong, B.C.; Chiu, S.-K.; Chow, S.A. The Role of Negative Superhelicity and Length of Homology in the Formation of Paranemic Joints Promoted by RecA Protein. J. Biol. Chem. 1998, 273, 12120–12127. [Google Scholar] [CrossRef] [Green Version]
- Banda, S.; Tiwari, P.B.; Darici, Y.; Tse-Dinh, Y.-C. Investigating direct interaction between Escherichia coli topoisomerase I and RecA. Gene 2016, 585, 65–70. [Google Scholar] [CrossRef] [Green Version]
- Higgins, J.D.; Armstrong, S.J.; Franklin, F.C.H.; Jones, G.H. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes Dev. 2004, 18, 2557–2570. [Google Scholar] [CrossRef] [Green Version]
- Stacey, N.J.; Kuromori, T.; Azumi, Y.; Roberts, G.; Breuer, C.; Wada, T.; Maxwell, A.; Roberts, K.; Sugimoto-Shirasu, K. Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 2006, 48, 206–216. [Google Scholar] [CrossRef]
- Jackson, N.; Sanchez-Moran, E.; Buckling, E.; Armstrong, S.J.; Jones, G.H.; Franklin, F.C.H. Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J. 2006, 25, 1315–1323. [Google Scholar] [CrossRef] [Green Version]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Martinez-Garcia, M.; Pradillo, M. Functional Analysis of Arabidopsis ARGONAUTEs in Meiosis and DNA Repair. In Plant Argonaute Proteins; Carbonell, A., Ed.; Springer: New York, NY, USA, 2017; pp. 145–158. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Curtis, M.J.; Hays, J.B. Tolerance of dividing cells to replication stress in UVB-irradiated Arabidopsis roots: Requirements for DNA translesion polymerases eta and zeta. DNA Repair 2007, 6, 1341–1358. [Google Scholar] [CrossRef]
- Amiard, S.; Depeiges, A.; Allain, E.; White, C.I.; Gallego, M.E. Arabidopsis ATM and ATR Kinases Prevent Propagation of Genome Damage Caused by Telomere Dysfunction. Plant Cell 2011, 23, 4254–4265. [Google Scholar] [CrossRef]
- Charbonnel, C.; Gallego, M.E.; White, C.I. Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants: Double-strand break repair kinetics in Arabidopsis. Plant J. 2010, 64, 280–290. [Google Scholar] [CrossRef]
- Armstrong, S.J.; Sanchez-Moran, E.; Chris, F.; Franklin, H. Cytological Analysis of Arabidopsis thaliana Meiotic Chromosomes. In Meiosis; Keeney, S., Ed.; Humana Press: Totowa, NJ, USA, 2009; pp. 131–145. [Google Scholar] [CrossRef]
- Higgins, J.D.; Sanchez-Moran, E.; Armstrong, S.J.; Jones, G.H.; Franklin, F.C.H. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 2005, 19, 2488–2500. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Morán, E.; Jones, G.H.; Franklin, F.C.H.; Santos, J.L. A Puromycin-Sensitive Aminopeptidase Is Essential for Meiosis in Arabidopsis thaliana. Plant Cell 2004, 16, 2895–2909. [Google Scholar] [CrossRef] [Green Version]
- Moran, E.S.; Armstrong, S.J.; Santos, J.L.; Franklin, F.C.; Jones, G.H. Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res. 2001, 9, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Fransz, P.; Armstrong, S.; Alonso-Blanco, C.; Fischer, T.C.; Torres-Ruiz, R.A.; Jones, G. Cytogenetics for the model system Arabidopsis thaliana. Plant J. 1998, 13, 867–876. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez-Garcia, M.; White, C.I.; Franklin, F.C.H.; Sanchez-Moran, E. The Role of Topoisomerase II in DNA Repair and Recombination in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 13115. https://doi.org/10.3390/ijms222313115
Martinez-Garcia M, White CI, Franklin FCH, Sanchez-Moran E. The Role of Topoisomerase II in DNA Repair and Recombination in Arabidopsis thaliana. International Journal of Molecular Sciences. 2021; 22(23):13115. https://doi.org/10.3390/ijms222313115
Chicago/Turabian StyleMartinez-Garcia, Marina, Charles I. White, F. Chris. H. Franklin, and Eugenio Sanchez-Moran. 2021. "The Role of Topoisomerase II in DNA Repair and Recombination in Arabidopsis thaliana" International Journal of Molecular Sciences 22, no. 23: 13115. https://doi.org/10.3390/ijms222313115
APA StyleMartinez-Garcia, M., White, C. I., Franklin, F. C. H., & Sanchez-Moran, E. (2021). The Role of Topoisomerase II in DNA Repair and Recombination in Arabidopsis thaliana. International Journal of Molecular Sciences, 22(23), 13115. https://doi.org/10.3390/ijms222313115





