Control of TRPM3 Ion Channels by Protein Kinase CK2-Mediated Phosphorylation in Pancreatic β-Cells of the Line INS-1
Abstract
1. Introduction
2. Results
2.1. CK2 Phosphorylates TRPM3
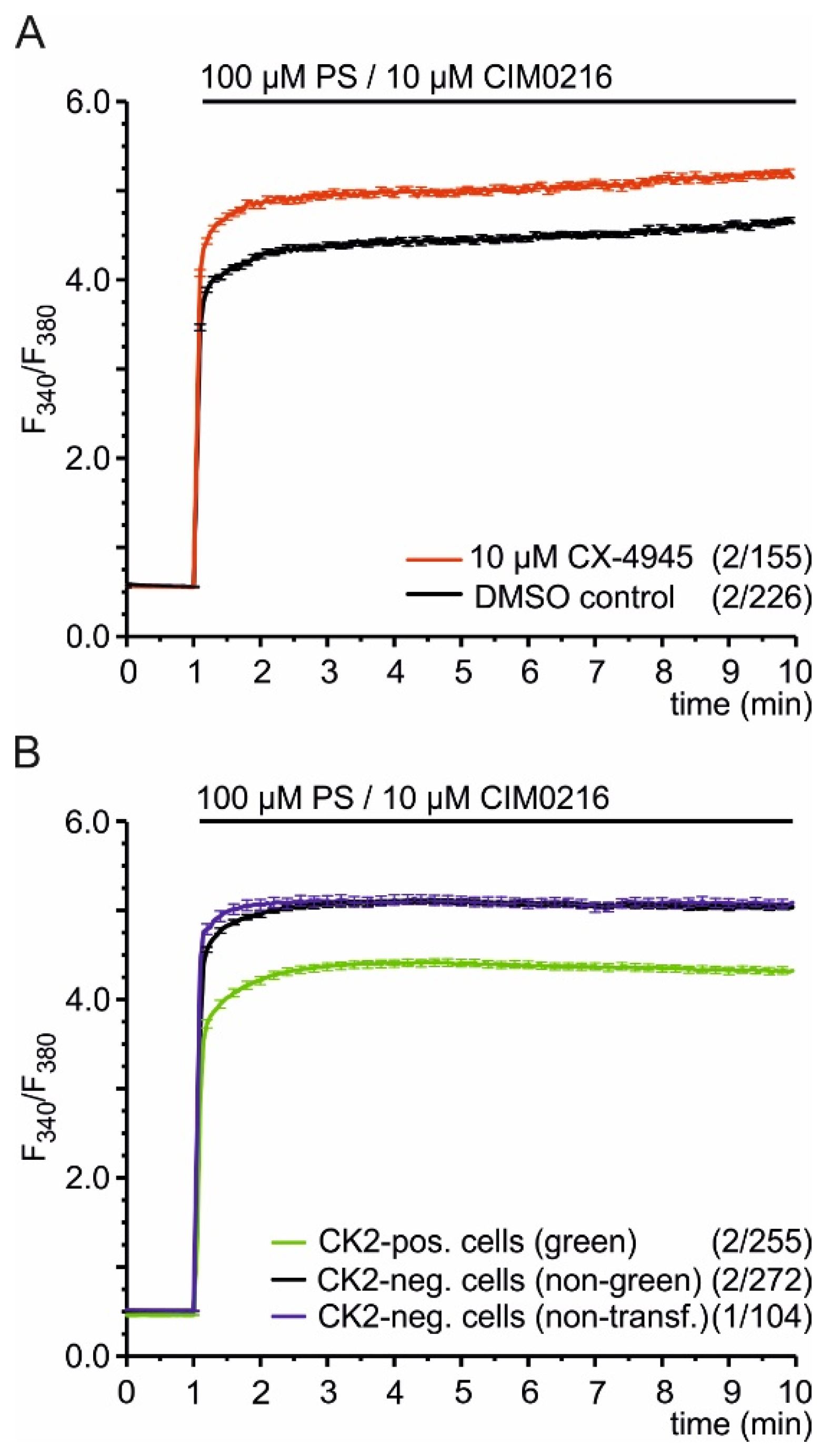
2.2. CK2 Phosphorylation Inhibits TRPM3-Mediated Ca2+ Entry
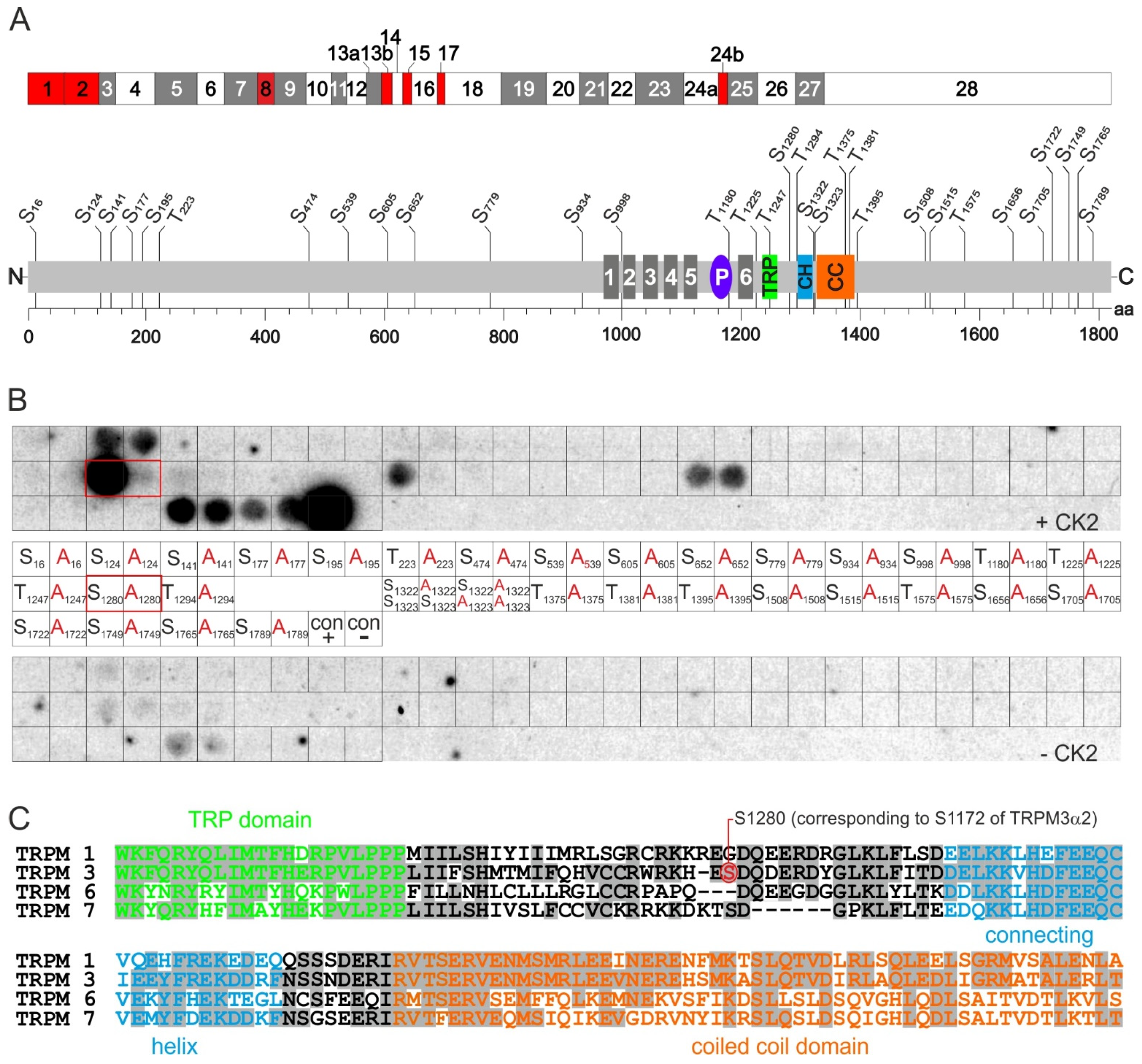
2.3. Identification of CK2 Phosphorylation Sites in TRPM3 Proteins
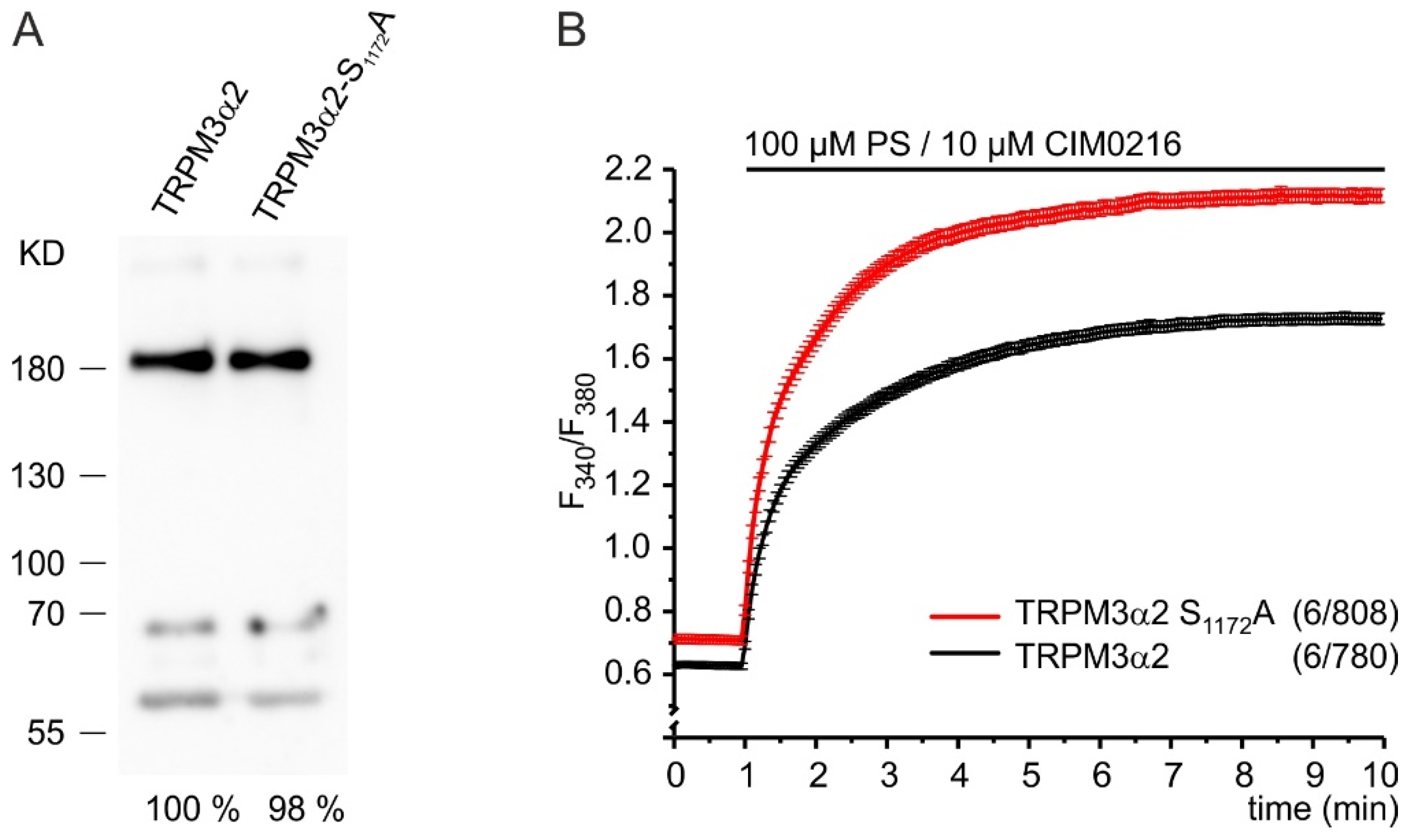
2.4. Phosphorylation of S1280 Reduces TRPM3 Channel Activity
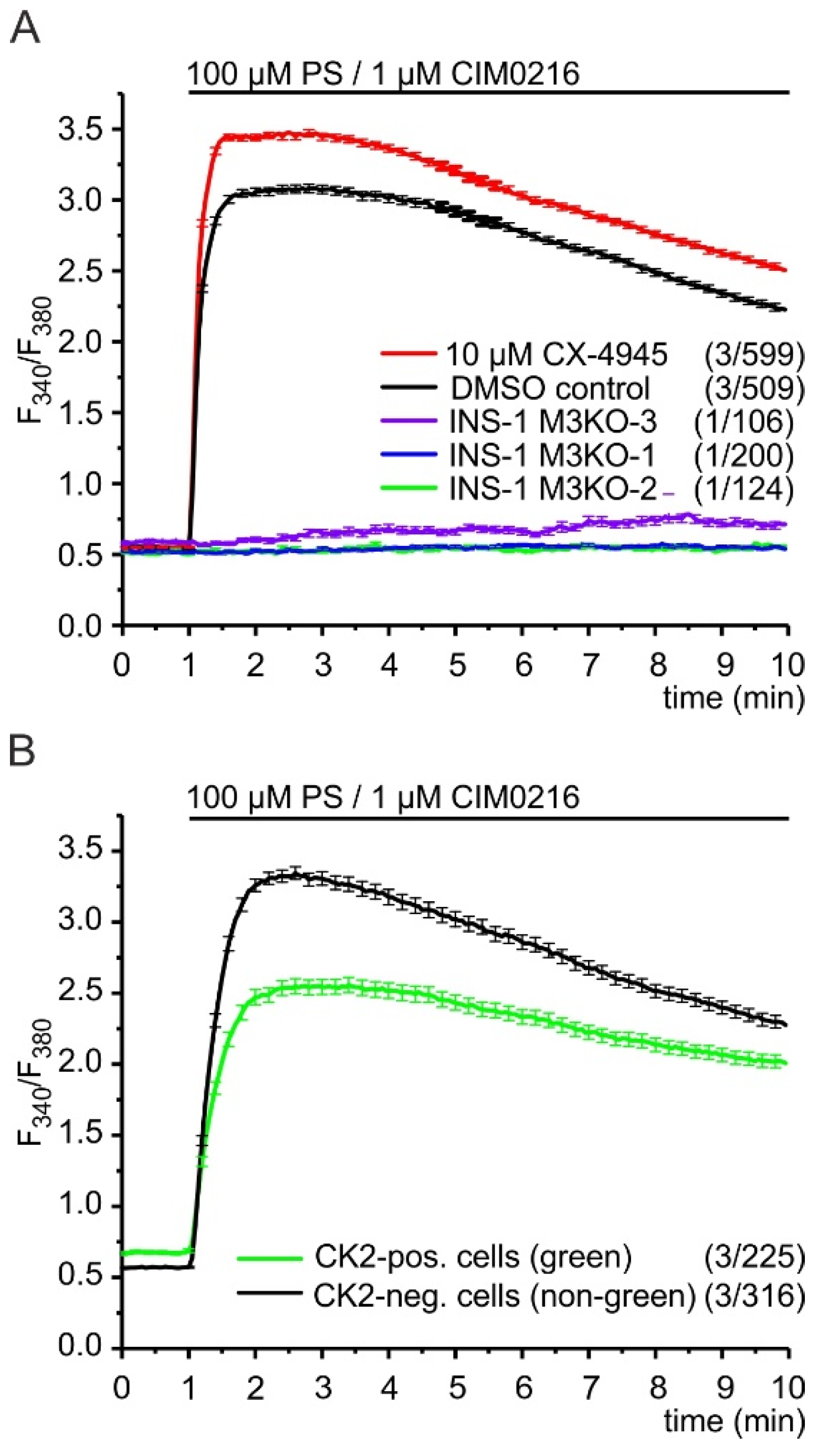
2.5. CK2 Controls TRPM3 Channels in INS-1 β-Cells
3. Discussion
3.1. TRPM3 Channels Are Controlled by Phosphorylation
3.2. Contribution of CK2-Mediated Phosphorylation of TRPM3 to the Control of Insulin Release
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. Mutagenesis of TRPM3, Plasmids, Transfection, and Fluorescence-Activated Cell Sorting (FACS)
4.3. Fura-2 Ca2+ Imaging
4.4. In Vitro Phosphorylation of Immunopurified Proteins
4.5. In Vitro Phosphorylation of a TRPM3 Peptide Library
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Klec, C.; Ziomek, G.; Pichler, M.; Malli, R.; Graier, W.F. Calcium Signaling in ß-cell Physiology and Pathology: A Revisit. Int. J. Mol. Sci. 2019, 20, 6110. [Google Scholar] [CrossRef]
- Yang, S.-N.; Shi, Y.; Yang, G.; Li, Y.; Yu, J.; Berggren, P.-O. Ionic mechanisms in pancreatic β cell signaling. Cell. Mol. Life Sci. 2014, 71, 4149–4177. [Google Scholar] [CrossRef]
- Catterall, W.A.; Wisedchaisri, G.; Zheng, N. The chemical basis for electrical signaling. Nat. Chem. Biol. 2017, 13, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Mannebach, S.; Mathar, I.; Weissgerber, P.; Freichel, M.; Loodin, A.P.; Fecher-Trost, C.; Belkacemi, A.; Beck, A.; Philipp, S.E. Control of Insulin Release by Transient Receptor Potential Melastatin 3 (TRPM3) Ion Channels. Cell. Physiol. Biochem. 2020, 54, 1115–1131. [Google Scholar] [CrossRef]
- Oberwinkler, J.; Lis, A.; Giehl, K.M.; Flockerzi, V.; Philipp, S.E. Alternative Splicing Switches the Divalent Cation Selectivity of TRPM3 Channels. J. Biol. Chem. 2005, 280, 22540–22548. [Google Scholar] [CrossRef]
- Grimm, C.; Kraft, R.; Sauerbruch, S.; Schultz, G.; Harteneck, C. Molecular and Functional Characterization of the Melastatin-related Cation Channel TRPM3. J. Biol. Chem. 2003, 278, 21493–21501. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Chen, J.; Sun, L.; Wu, S.; Gray, K.R.; Rich, A.; Huang, M.; Lin, J.-H.; Feder, J.N.; Janovitz, E.B.; et al. Expression and Characterization of Human Transient Receptor Potential Melastatin 3 (hTRPM3). J. Biol. Chem. 2003, 278, 20890–20897. [Google Scholar] [CrossRef]
- Wagner, T.F.; Loch, S.; Lambert, S.; Straub, I.; Mannebach, S.; Mathar, I.; Düfer, M.; Lis, A.; Flockerzi, V.; Philipp, S.E.; et al. Transient receptor potential M3 channels are ionotropic steroid receptors in pancreatic β cells. Nat. Cell Biol. 2008, 10, 1421–1430. [Google Scholar] [CrossRef]
- Held, K.; Kichko, T.; De Clercq, K.; Klaassen, H.; Van Bree, R.; Vanherck, J.-C.; Marchand, A.; Reeh, P.; Chaltin, P.; Voets, T.; et al. Activation of TRPM3 by a potent synthetic ligand reveals a role in peptide release. Proc. Natl. Acad. Sci. USA 2015, 112, E1363–E1372. [Google Scholar] [CrossRef] [PubMed]
- Held, K.; Aloi, V.D.; Freitas, A.C.N.; Janssens, A.; Segal, A.; Przibilla, J.; Philipp, S.E.; Wang, Y.T.; Voets, T.; Vriens, J. Pharmacological properties of TRPM3 isoforms are determined by the length of the pore loop. Br. J. Pharmacol. in press. 2020. [Google Scholar] [CrossRef]
- Tóth, B.I.; Konrad, M.; Ghosh, D.; Mohr, F.; Halaszovich, C.R.; Leitner, M.; Vriens, J.; Oberwinkler, J.; Voets, T. Regulation of the transient receptor potential channel TRPM3 by phosphoinositides. J. Gen. Physiol. 2015, 146, 51–63. [Google Scholar] [CrossRef]
- Badheka, D.; Borbiro, I.; Rohacs, T. Transient receptor potential melastatin 3 is a phosphoinositide-dependent ion channel. J. Gen. Physiol. 2015, 146, 65–77. [Google Scholar] [CrossRef]
- Przibilla, J.; Dembla, S.; Rizun, O.; Lis, A.; Jung, M.; Oberwinkler, J.; Beck, A.; Philipp, S.E. Ca2+-dependent regulation and binding of calmodulin to multiple sites of Transient Receptor Potential Melastatin 3 (TRPM3) ion channels. Cell Calcium 2018, 73, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Holakovska, B.; Grycova, L.; Jirku, M.; Sulc, M.; Bumba, L.; Teisinger, J. Calmodulin and S100A1 Protein Interact with N Terminus of TRPM3 Channel. J. Biol. Chem. 2012, 287, 16645–16655. [Google Scholar] [CrossRef]
- Koivisto, A.-P.; Belvisi, M.G.; Gaudet, R.; Szallasi, A. Advances in TRP channel drug discovery: From target validation to clinical studies. Nat. Rev. Drug Discov. 2021, 1–19. [Google Scholar] [CrossRef]
- Dembla, S.; Behrendt, M.; Mohr, F.; Goecke, C.; Sondermann, J.; Schneider, F.M.; Schmidt, M.; Stab, J.; Enzeroth, R.; Leitner, M.G.; et al. Anti-nociceptive action of peripheral mu-opioid receptors by G-beta-gamma protein-mediated inhibition of TRPM3 channels. eLife 2017, 6, 26280. [Google Scholar] [CrossRef]
- Badheka, D.; Yudin, Y.; Borbiro, I.; Hartle, C.M.; Yazici, A.; Mirshahi, T.; Rohacs, T. Inhibition of Transient Receptor Potential Melastatin 3 ion channels by G-protein βγ subunits. eLife 2017, 6, e26147. [Google Scholar] [CrossRef] [PubMed]
- Quallo, T.; Alkhatib, O.; Gentry, C.; A Andersson, D.; Bevan, S. G protein βγ subunits inhibit TRPM3 ion channels in sensory neurons. eLife 2017, 6, e26138. [Google Scholar] [CrossRef] [PubMed]
- Alkhatib, O.; Da Costa, R.; Gentry, C.; Quallo, T.; Mannebach, S.; Weissgerber, P.; Freichel, M.; Philipp, S.E.; Bevan, S.; Andersson, D.A. Promiscuous G-Protein-Coupled Receptor Inhibition of Transient Receptor Potential Melastatin 3 Ion Channels by Gβγ Subunits. J. Neurosci. 2019, 39, 7840–7852. [Google Scholar] [CrossRef]
- Vriens, J.; Owsianik, G.; Hofmann, T.; Philipp, S.E.; Stab, J.; Chen, X.; Benoit, M.; Xue, F.; Janssens, A.; Kerselaers, S.; et al. TRPM3 Is a Nociceptor Channel Involved in the Detection of Noxious Heat. Neuron 2011, 70, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.F.J.; Drews, A.; Loch, S.; Mohr, F.; Philipp, S.E.; Lambert, S.; Oberwinkler, J. TRPM3 channels provide a regulated influx pathway for zinc in pancreatic beta cells. Pflüger’s Arch. Gesamte Physiol. Menschen Tiere 2010, 460, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Scheuer, R.; Philipp, S.E.; Becker, A.; Nalbach, L.; Ampofo, E.; Montenarh, M.; Götz, C. Protein Kinase CK2 Controls CaV2.1-Dependent Calcium Currents and Insulin Release in Pancreatic β-Cells. Int. J. Mol. Sci. 2020, 21, 4668. [Google Scholar] [CrossRef]
- Allende, J.E.; Allende, C.C. Protein kinase CK2: An enzyme with multiple substrates and a puzzling regulation. FASEB J. 1995, 9, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Frühwald, J.; Londoño, J.C.; Dembla, S.; Mannebach, S.; Lis, A.; Drews, A.; Wissenbach, U.; Oberwinkler, J.; Philipp, S.E. Alternative Splicing of a Protein Domain Indispensable for Function of Transient Receptor Potential Melastatin 3 (TRPM3) Ion Channels. J. Biol. Chem. 2012, 287, 36663–36672. [Google Scholar] [CrossRef]
- Ampofo, E.; Kietzmann, T.; Zimmer, A.; Jakupovic, M.; Montenarh, M.; Götz, C. Phosphorylation of the von Hippel–Lindau protein (VHL) by protein kinase CK2 reduces its protein stability and affects p53 and HIF-1α mediated transcription. Int. J. Biochem. Cell Biol. 2010, 42, 1729–1735. [Google Scholar] [CrossRef]
- Oberwinkler, J.; Philipp, S.E. TRPM3. In Mediators and Drugs in Gastrointestinal Motility I; Springer: Berlin/Heidelberg, Germany, 2014; Volume 222, pp. 427–459. [Google Scholar] [CrossRef]
- Duan, J.; Li, Z.; Li, J.; Hulse, R.; Santa-Cruz, A.; Valinsky, W.C.; Abiria, S.A.; Krapivinsky, G.; Zhang, J.; Clapham, D.E. Structure of the mammalian TRPM7, a magnesium channel required during embryonic development. Proc. Natl. Acad. Sci. USA 2018, 115, E8201–E8210. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Minor, D.L. X-ray Crystal Structure of a TRPM Assembly Domain Reveals an Antiparallel Four-stranded Coiled-coil. J. Mol. Biol. 2008, 383, 854–870. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.-S.; Zhong, L.; Hsieh, T.-H.; Abooj, M.; Bishnoi, M.; Hughes, L.; Premkumar, L.S. Expression of Transient Receptor Potential Ankyrin 1 (TRPA1) and Its Role in Insulin Release from Rat Pancreatic Beta Cells. PLOS ONE 2012, 7, e38005. [Google Scholar] [CrossRef]
- Yao, X.; Kwan, H.-Y.; Huang, Y. Regulation of TRP Channels by Phosphorylation. Neurosignals 2005, 14, 273–280. [Google Scholar] [CrossRef]
- Broertjes, J.; Klarenbeek, J.; Habani, Y.; Langeslag, M.; Jalink, K. TRPM7 residue S1269 mediates cAMP dependence of Ca2+ influx. PLOS ONE 2019, 14, e0209563. [Google Scholar] [CrossRef]
- Nilius, B.; Prenen, J.; Tang, J.; Wang, C.; Owsianik, G.; Janssens, A.; Voets, T.; Zhu, M.X. Regulation of the Ca2+ Sensitivity of the Nonselective Cation Channel TRPM4. J. Biol. Chem. 2005, 280, 6423–6433. [Google Scholar] [CrossRef] [PubMed]
- Cerda, O.; Cáceres, M.; Park, K.-S.; Leiva-Salcedo, E.; Romero, A.; Varela, D.; Trimmer, J.S.; Stutzin, A. Casein kinase-mediated phosphorylation of serine 839 is necessary for basolateral localization of the Ca2+-activated non-selective cation channel TRPM4. Pflüger’s Arch. Gesamte Physiol. Menschen Tiere 2015, 467, 1723–1732. [Google Scholar] [CrossRef][Green Version]
- Rivera, B.; Moreno, C.; Lavanderos, B.; Hwang, J.Y.; Fernández-Trillo, J.; Park, K.-S.; Orio, P.; Viana, F.; Madrid, R.; Pertusa, M. Constitutive phosphorylation as a key regulator of TRPM8 channel function. J. Neurosci. 2021, 41, 8475–84963. [Google Scholar] [CrossRef]
- Sacco, F.; Humphrey, S.J.; Cox, J.; Mischnik, M.; Schulte, A.; Klabunde, T.; Schäfer, M.; Mann, M. Glucose-regulated and drug-perturbed phosphoproteome reveals molecular mechanisms controlling insulin secretion. Nat. Commun. 2016, 7, 13250. [Google Scholar] [CrossRef]
- Han, H.; Moon, S.; Kim, Y.; Ho, W.-K.; Kim, K.; Kang, Y.; Jun, H.; Kim, Y. Comprehensive Phosphoproteome Analysis of INS-1 Pancreatic Beta-Cells using Various Digestion Strategies Coupled with Liquid Chromatography–Tandem Mass Spectrometry. J. Proteome Res. 2012, 11, 2206–2223. [Google Scholar] [CrossRef]
- Meggio, F.; Marin, O.; A Pinna, L. Substrate specificity of protein kinase CK2. Cell. Mol. Boil. Res. 1994, 40, 401–409. [Google Scholar]
- Rekha, N.; Srinivasan, N. Structural basis of regulation and substrate specificity of protein kinase CK2 deduced from the modeling of protein-protein interactions. BMC Struct. Biol. 2003, 3, 4. [Google Scholar] [CrossRef][Green Version]
- Köttgen, M.; Benzing, T.; Simmen, T.; Tauber, R.; Buchholz, B.; Feliciangeli, S.; Huber, T.B.; Schermer, B.; Kramer-Zucker, A.; Höpker, K.; et al. Trafficking of TRPP2 by PACS proteins represents a novel mechanism of ion channel regulation. EMBO J. 2005, 24, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Hellmich, U.A.; Gaudet, R. Structural Biology of TRP Channels. In Mediators and Drugs in Gastrointestinal Motility I; Springer: Berlin/Heidelberg, Germany, 2014; Volume 223, pp. 963–990. [Google Scholar] [CrossRef]
- Gregorio-Teruel, L.; Valente, P.; Liu, B.; Fernández-Ballester, G.; Qin, F.; Ferrer-Montiel, A. The Integrity of the TRP Domain Is Pivotal for Correct TRPV1 Channel Gating. Biophys. J. 2015, 109, 529–541. [Google Scholar] [CrossRef]
- Rohács, T.; Lopes, C.M.B.; Michailidis, I.; E Logothetis, D. PI(4,5)P2 regulates the activation and desensitization of TRPM8 channels through the TRP domain. Nat. Neurosci. 2005, 8, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Klose, C.; Straub, I.; Riehle, M.; Ranta, F.; Krautwurst, D.; Ullrich, S.; Meyerhof, W.; Harteneck, C. Fenamates as TRP channel blockers: Mefenamic acid selectively blocks TRPM3. Br. J. Pharmacol. 2011, 162, 1757–1769. [Google Scholar] [CrossRef] [PubMed]
- Ampofo, E.; Nalbach, L.; Menger, M.D.; Montenarh, M.; Götz, C. Protein Kinase CK2—A Putative Target for the Therapy of Diabetes Mellitus? Int. J. Mol. Sci. 2019, 20, 4398. [Google Scholar] [CrossRef] [PubMed]
- Asfari, M.; Janjic, D.; Meda, P.; Li, G.; A Halban, P.; Wollheim, C.B. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinol. 1992, 130, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Grankowski, N.; Boldyreff, B.; Issinger, O.-G. Isolation and characterization of recombinant human casein kinase II subunits alpha and beta from bacteria. JBIC J. Biol. Inorg. Chem. 1991, 198, 25–30. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Becker, A.; Götz, C.; Montenarh, M.; Philipp, S.E. Control of TRPM3 Ion Channels by Protein Kinase CK2-Mediated Phosphorylation in Pancreatic β-Cells of the Line INS-1. Int. J. Mol. Sci. 2021, 22, 13133. https://doi.org/10.3390/ijms222313133
Becker A, Götz C, Montenarh M, Philipp SE. Control of TRPM3 Ion Channels by Protein Kinase CK2-Mediated Phosphorylation in Pancreatic β-Cells of the Line INS-1. International Journal of Molecular Sciences. 2021; 22(23):13133. https://doi.org/10.3390/ijms222313133
Chicago/Turabian StyleBecker, Alexander, Claudia Götz, Mathias Montenarh, and Stephan E. Philipp. 2021. "Control of TRPM3 Ion Channels by Protein Kinase CK2-Mediated Phosphorylation in Pancreatic β-Cells of the Line INS-1" International Journal of Molecular Sciences 22, no. 23: 13133. https://doi.org/10.3390/ijms222313133
APA StyleBecker, A., Götz, C., Montenarh, M., & Philipp, S. E. (2021). Control of TRPM3 Ion Channels by Protein Kinase CK2-Mediated Phosphorylation in Pancreatic β-Cells of the Line INS-1. International Journal of Molecular Sciences, 22(23), 13133. https://doi.org/10.3390/ijms222313133






