Elucidating the Molecular Mechanisms by which Seed-Borne Endophytic Fungi, Epichloë gansuensis, Increases the Tolerance of Achnatherum inebrians to NaCl Stress
Abstract
:1. Introduction
2. Results
2.1. SMRT-Based RNA Sequencing
2.2. Predictions of ORFs and SSRs
2.3. Functional Annotation of Transcripts and Sorting of Transcription Factors
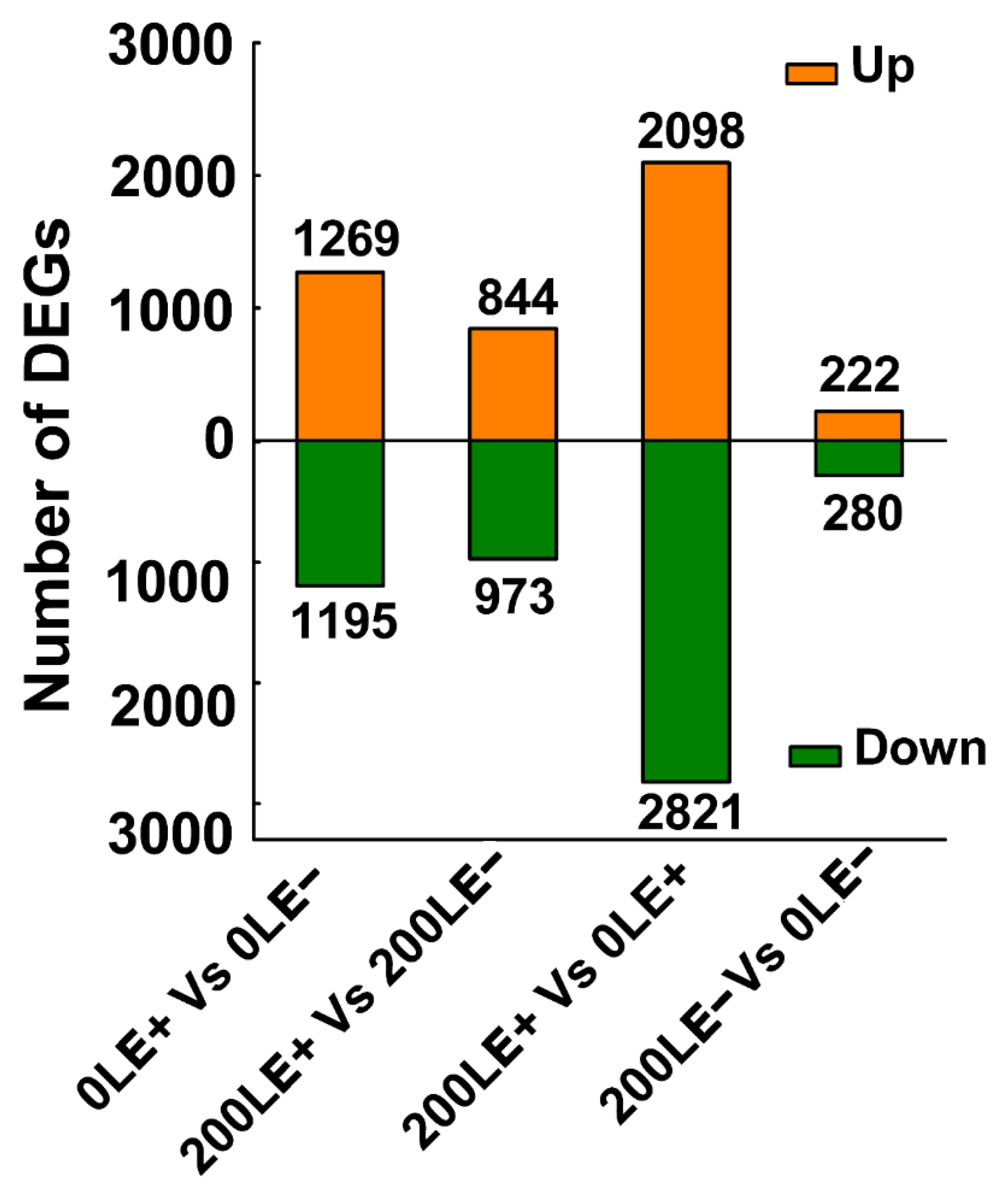
2.4. E. Gansuensis Altered Transcriptome Reprogramming of A. Inebrians at Various NaCl Concentrations
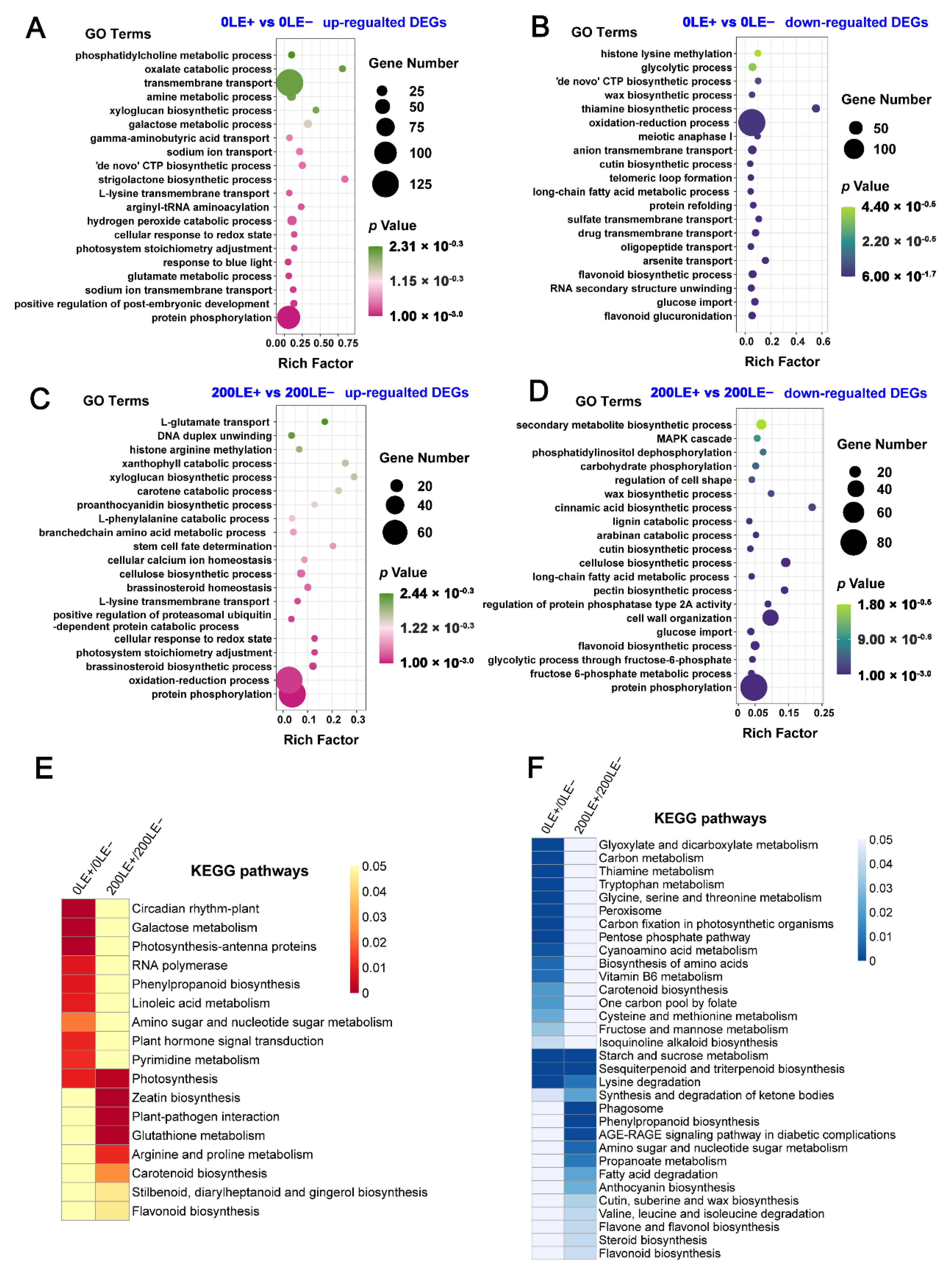
2.5. E. Gansuensis Regulated the Different Biological Processes of A. Inebrians to Maintain Balance between Growth and Resistance to NaCl Stress
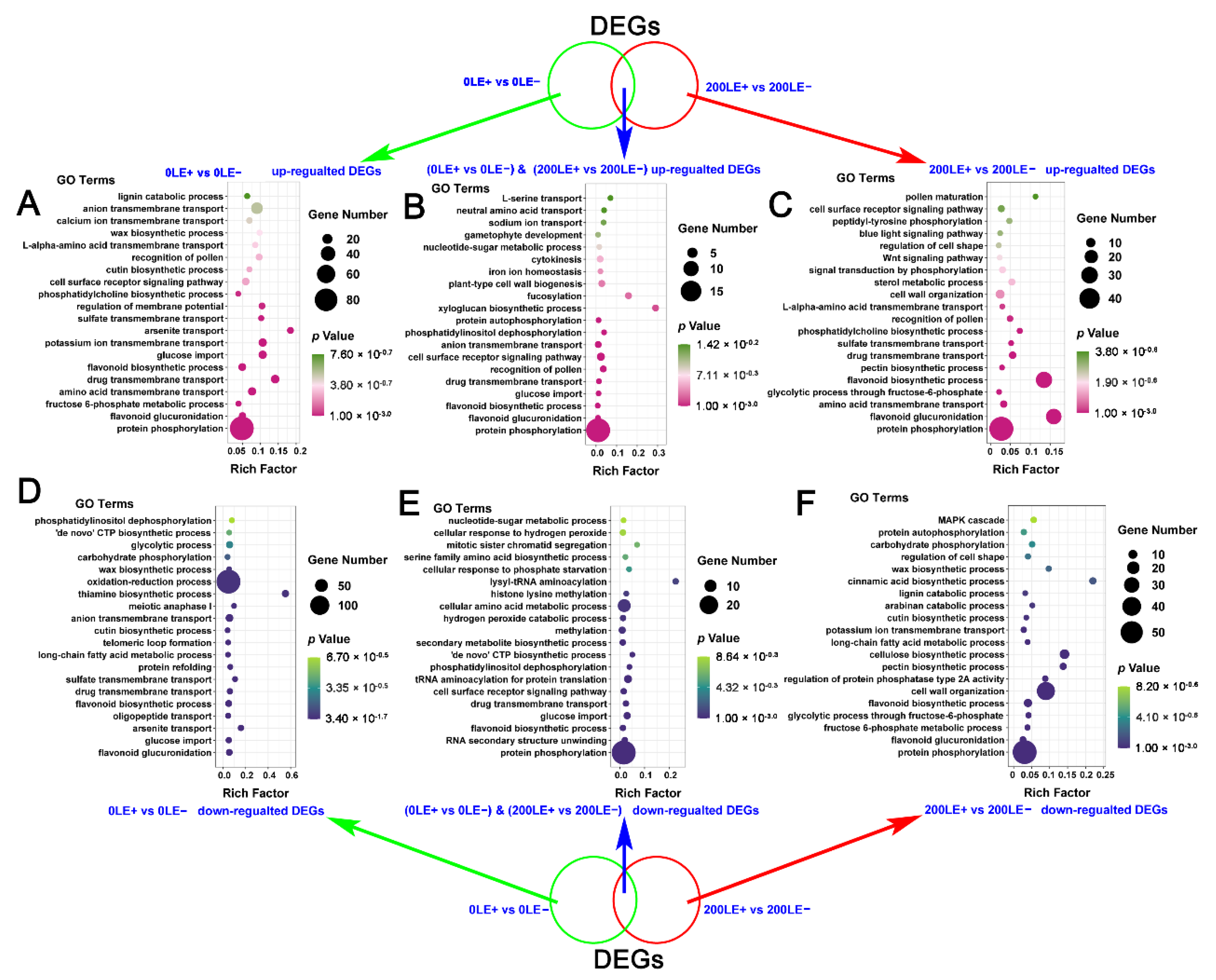
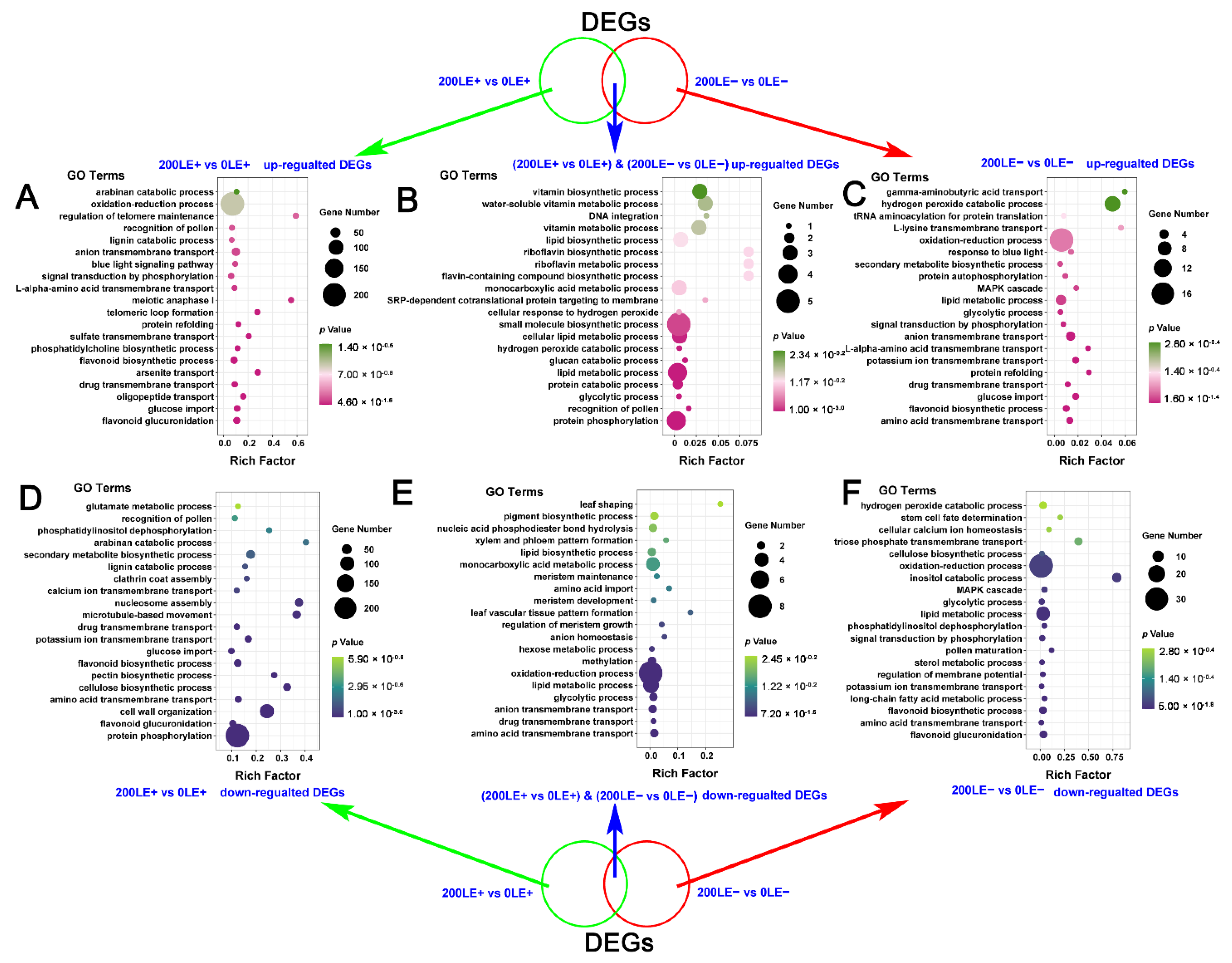
2.6. The Unique Role of E. Gansuensis on Transcriptome Reprogramming of A. Inebrians at Two NaCl Concentrations
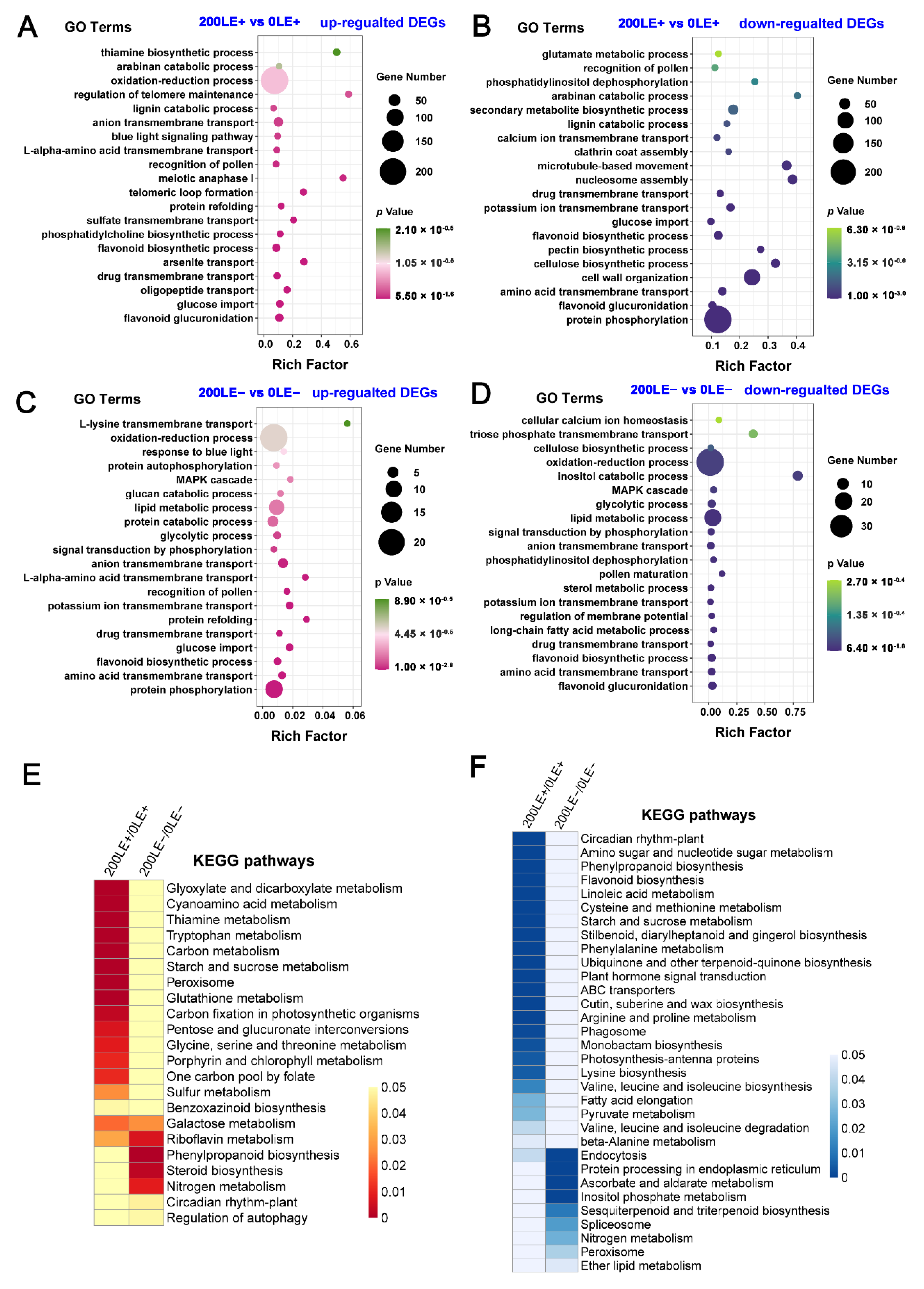
2.7. The Unique Effect of NaCl Stress on Transcriptome Reprogramming of A. Inebrians in E+ Plants and E− Plants
2.8. The Effect of E. Gansuensis on the Expression Pattern of Transcription Factors of A. Inebrians under NaCl Stress
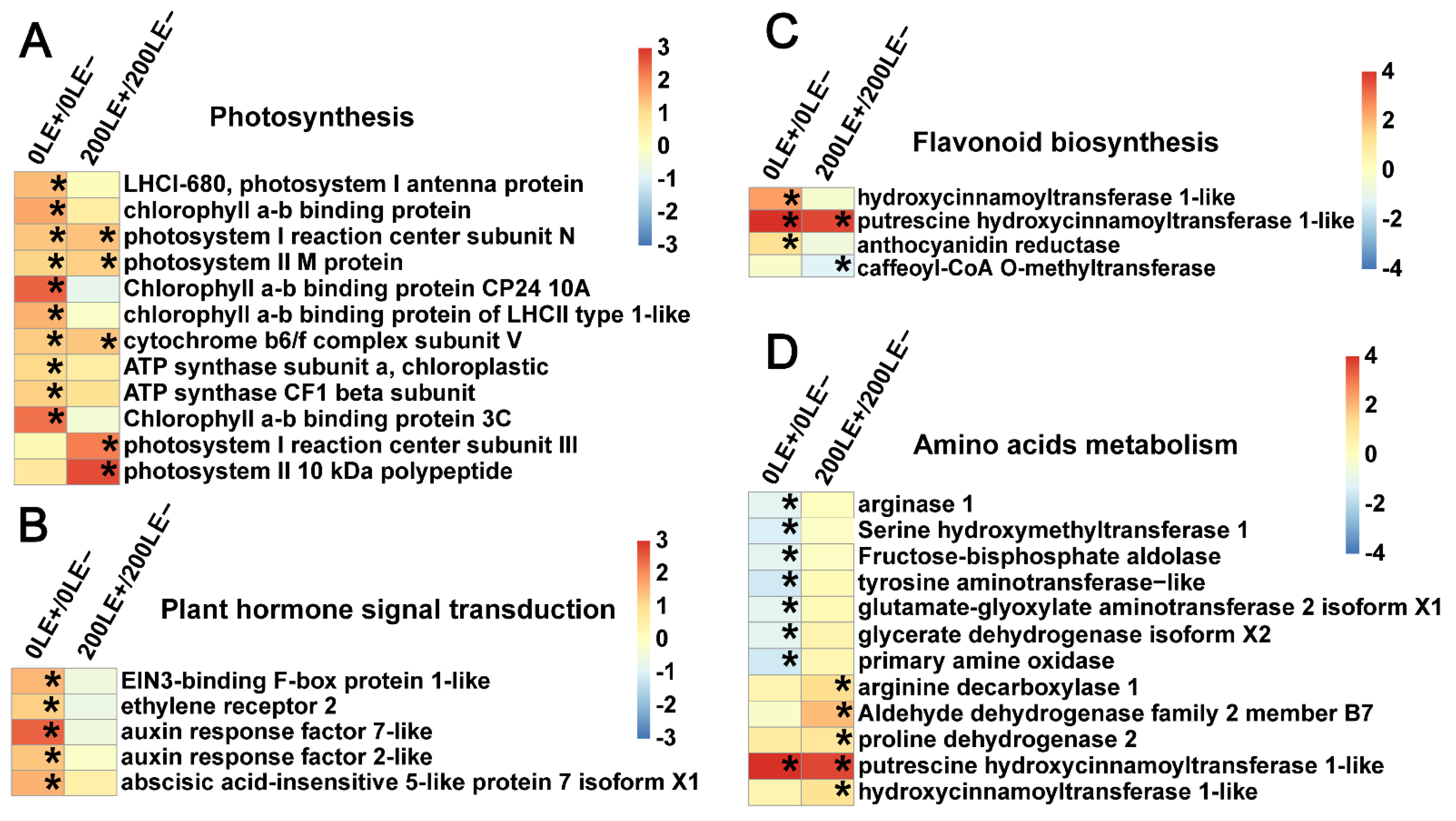
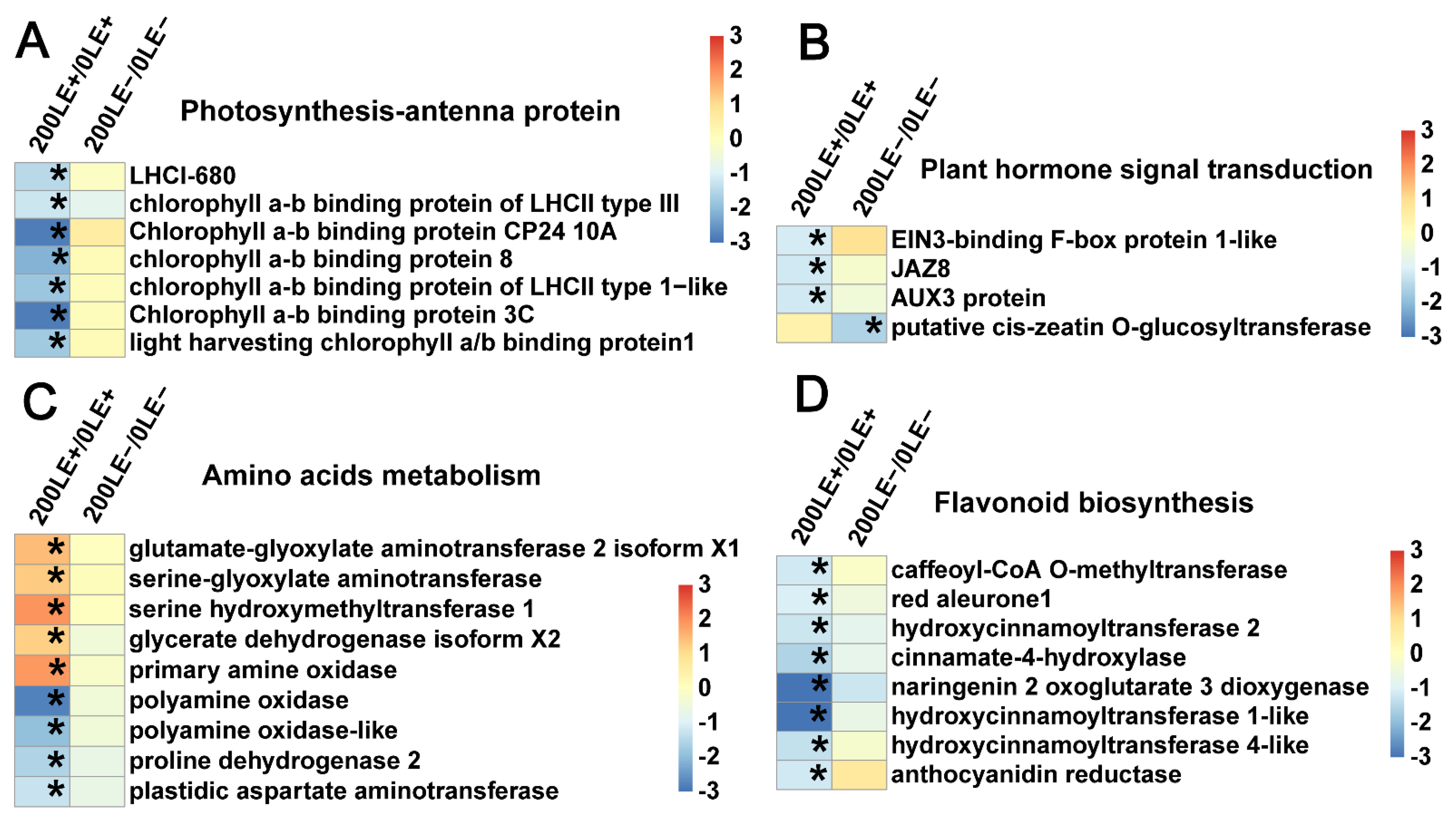
2.9. E. Gansuensis Alters Expression of Key Genes Associated with Flavonoid Biosynthesis, Photosynthesis, Plant Hormone Signal Transduction and Amino Acids Metabolism under NaCl Stress
3. Discussion
4. Materials and Methods
4.1. Plant Material and RNA Extraction
4.2. PacBio Library Preparation, SMRT Sequencing and Raw Data Analysis
4.3. Illumina Sequencing and Gene Annotation
4.4. Identification of Differentially Expressed Genes
4.5. The Analysis of GO Enrichment and KEGG Pathway
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Claeys, H.; Inzé, D. The agony of choice: How plants balance growth and survival under water-limiting conditions. Plant Physiol. 2013, 162, 1768–1779. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.-J.; Zhou, Y.-H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.-Q. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, J.; Wang, M.; Shi, Z.; Miao, X. OsEXPA10 mediates the balance between growth and resistance to biotic stress in rice. Plant Cell Rep. 2018, 37, 993–1002. [Google Scholar] [CrossRef]
- Li, C.; Nan, Z.; Gao, J.; Tian, P. Detection and distribution of Neotyphodium-Achnatherum inebrians association in China. In Proceedings of the 5th International Neotyphodium/Grass Interactions Symposium, Arkansas, AR, USA, 23–26 May 2004. [Google Scholar]
- Chen, L.; Li, X.; Li, C.; Swoboda, G.A.; Young, C.A.; Sugawara, K.; Leuchtmann, A.; Schardl, C.L. Two distinct Epichloë species symbiotic with Achnatherum inebrians, drunken horse grass. Mycologia 2015, 107, 863–873. [Google Scholar] [CrossRef] [Green Version]
- Xia, C.; Li, N.; Zhang, Y.; Li, C.; Zhang, X.; Nan, Z. Role of Epichloë endophytes in defense responses of cool-season grasses to pathogens: A Review. Plant Dis. 2018, 102, 2061–2073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Hou, W.; Christensen, M.J.; Li, X.; Xia, C.; Li, C.; Nan, Z. Role of Epichloë endophytes in improving host grass resistance ability and soil properties. J. Agric. Food Chem. 2020, 68, 6944–6955. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.P.; Xia, C.; Christensen, M.J.; Wang, J.F.; Li, X.Z.; Chen, T.; Nan, Z.B. Effect of Epichloë gansuensis endophyte on rhizosphere bacterial communities and nutrient concentrations and ratios in the perennial grass species Achnatherum inebrians during three growth seasons. Crop Pasture Sci. 2020, 71, 1050–1066. [Google Scholar] [CrossRef]
- Xia, C.; Christensen, M.J.; Zhang, X.; Nan, Z. Effect of Epichloë gansuensis endophyte and transgenerational effects on the water use efficiency, nutrient and biomass accumulation of Achnatherum inebrians under soil water deficit. Plant Soil 2018, 424, 555–571. [Google Scholar] [CrossRef]
- Wang, J.; Tian, P.; Christensen, M.J.; Zhang, X.; Li, C.; Nan, Z. Effect of Epichloë gansuensis endophyte on the activity of enzymes of nitrogen metabolism, nitrogen use efficiency and photosynthetic ability of Achnatherum inebrians under various NaCl concentrations. Plant Soil 2019, 435, 57–68. [Google Scholar] [CrossRef]
- Wang, J.; Nan, Z.; Christensen, M.J.; Li, C. Glucose-6-phosphate dehydrogenase plays a vital role in Achnatherum inebrians plants host to Epichloë gansuensis by improving growth under nitrogen deficiency. Plant Soil 2018, 430, 37–48. [Google Scholar] [CrossRef]
- Wang, J.; Nan, Z.; Christensen, M.J.; Zhang, X.; Tian, P.; Zhang, Z.; Niu, X.; Gao, P.; Chen, T.; Ma, L. Effect of Epichloë gansuensis endophyte on the nitrogen metabolism, nitrogen use efficiency, and stoichiometry of Achnatherum inebrians under nitrogen limitation. J. Agric. Food Chem. 2018, 66, 4022–4031. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, C.; Nan, Z. Effects of cadmium stress on growth and anti-oxidative systems in Achnatherum inebrians symbiotic with Neotyphodium gansuense. J. Hazard. Mater. 2010, 175, 703–709. [Google Scholar] [CrossRef]
- Li, C.; Zhang, X.; Li, F.; Nan, Z.; Schardl, C. Disease and pests resistance of endophyte infected and non-infected drunken horse grass. In Proceedings of the 6th International Symposium on Fungal Endophytes of Grasses, Dunedin, New Zealand, 25–28 March 2007. [Google Scholar]
- Gibert, A.; Tozer, W.; Westoby, M. Plant performance response to eight different types of symbiosis. New Phytol. 2019, 222, 526–542. [Google Scholar] [CrossRef]
- Bastías, D.A.; Gianoli, E.; Gundel, P.E. Fungal endophytes can eliminate the plant growth-defence trade-off. New Phytol. 2021, 230, 2105–2113. [Google Scholar] [CrossRef]
- Malinowski, D.P.; Brauer, D.K.; Belesky, D.P. The endophyte Neotyphodium coenophialum affects root morphology of Tall Fescue grown under phosphorus deficiency. J. Agron. Crop Sci. 1999, 183, 53–60. [Google Scholar] [CrossRef]
- Chen, T.; White, J.F.; Li, C. Fungal endophyte Epichloë bromicola infection regulates anatomical changes to account for salt stress tolerance in wild barley (Hordeum brevisubulatum). Plant Soil 2021, 461, 533–546. [Google Scholar] [CrossRef]
- Richardson, M.D.; Hoveland, C.S.; Bacon, C.W. Photosynthesis and stomatal conductance of symbiotic and nonsymbiotic Tall Fescue. Crop Sci. 1993, 33, 145–149. [Google Scholar] [CrossRef]
- Chen, T.; Li, C.; White, J.F.; Nan, Z. Effect of the fungal endophyte Epichloë bromicola on polyamines in wild barley (Hordeum brevisubulatum) under salt stress. Plant Soil 2019, 436, 29–48. [Google Scholar] [CrossRef]
- Raven, J.A. Plant resource allocation. Trends Ecol. Evol. 1998, 13, 84–85. [Google Scholar] [CrossRef]
- Leuchtmann, A.; Schardl, C.L.; Siegel, M.R. Sexual compatibility and taxonomy of a new species of Epichloë symbiotic with fine fescue grasses. Mycologia 1994, 86, 802–812. [Google Scholar]
- Schardl, C.; Scott, B.; Florea, S.; Zhang, D. Epichloë endophytes: Clavicipitaceous symbionts of grasses. Plant Relationships 2009, 5, 275–306. [Google Scholar]
- Dinkins, R.D.; Nagabhyru, P.; Graham, M.A.; Boykin, D.; Schardl, C.L. Transcriptome response of Lolium arundinaceum to its fungal endophyte Epichloë coenophiala. New Phytol. 2017, 213, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.Y.; Spiering, M.J.; Scott, V.; Lane, G.A.; Christensen, M.J.; Schmid, J. In planta regulation of extension of an endophytic fungus and maintenance of high metabolic rates in its mycelium in the absence of apical extension. Appl. Environ. Microbiol. 2001, 67, 5377–5383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christensen, M.J.; Bennett, R.J.; Ansari, H.A.; Koga, H.; Johnson, R.D.; Bryan, G.T.; Simpson, W.R.; Koolaard, J.P.; Nickless, E.M.; Voisey, C.R. Epichloë endophytes grow by intercalary hyphal extension in elongating grass leaves. Fungal Genet. Biol. 2008, 45, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, D.; Tanaka, A.; Scott, B. A p67Phox-like regulator is recruited to control hyphal branching in a fungal-grass mutualistic symbiosis. Plant Cell 2006, 18, 2807–2821. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Christensen, M.J.; Takemoto, D.; Park, P.; Scott, B. Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction. Plant Cell 2006, 18, 1052–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, A.; Takemoto, D.; Hyon, G.S.; Park, P.; Scott, B. NoxA activation by the small GTPase RacA is required to maintain a mutualistic symbiotic association between Epichloë festucae and perennial ryegrass. Mol. Microbiol. 2008, 68, 1165–1178. [Google Scholar] [CrossRef] [PubMed]
- Becker, Y.; Eaton, C.; Brasell, E.; May, K.; Becker, M.; Hassing, B.; Cartwright, G.; Reinhold, L.; Scott, B. The fungal cell-wall integrity MAPK cascade is crucial for hyphal network formation and maintenance of restrictive growth of Epichloë festucae in symbiosis with Lolium perenne. Mol. Plant-Microbe Interact. 2015, 28, 69–85. [Google Scholar]
- Berens, M.L.; Berry, H.M.; Mine, A.; Argueso, C.T.; Tsuda, K. Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 2017, 55, 401–425. [Google Scholar] [CrossRef] [PubMed]
- Van Butselaar, T.; Van den Ackerveken, G. Salicylic acid steers the growth-immunity tradeoff. Trends Plant Sci. 2020, 25, 566–576. [Google Scholar] [CrossRef]
- Johnson, L.J.; Johnson, R.D.; Schardl, C.L.; Panaccione, D.G. Identification of differentially expressed genes in the mutualistic association of tall fescue with Neotyphodium coenophialum. Physiol. Mol. Plant Pathol. 2003, 63, 305–317. [Google Scholar] [CrossRef]
- Felitti, S.; Shields, K.; Ramsperger, M.; Tian, P.; Sawbridge, T.; Webster, T.; Logan, E.; Erwin, T.; Forster, J.; Edwards, D.; et al. Transcriptome analysis of Neotyphodium and Epichloë grass endophytes. Fungal Genet. Biol. 2006, 43, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Dinkins, R.D.; Barnes, A.; Waters, W. Microarray analysis of endophyte-infected and endophyte-free tall fescue. J. Plant Physiol. 2010, 167, 1197–1203. [Google Scholar] [CrossRef]
- Ambrose, K.V.; Belanger, F.C. SOLiD-SAGE of endophyte-infected red fescue reveals numerous effects on host transcriptome and an abundance of highly expressed fungal secreted proteins. PLoS ONE 2012, 7, e53214. [Google Scholar] [CrossRef] [Green Version]
- Dupont, P.-Y.; Eaton, C.J.; Wargent, J.J.; Fechtner, S.; Solomon, P.; Schmid, J.; Day, R.C.; Scott, B.; Cox, M.P. Fungal endophyte infection of ryegrass reprograms host metabolism and alters development. New Phytol. 2015, 208, 1227–1240. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.D.; Byrne, S.; Paina, C.; Asp, T. De Novo assembly of the perennial ryegrass transcriptome using an RNA-Seq strategy. PLoS ONE 2014, 9, e103567. [Google Scholar] [CrossRef]
- Talukder, S.K.; Azhaguvel, P.; Mukherjee, S.; Young, C.A.; Tang, Y.; Krom, N.; Saha, M.C. De Novo assembly and characterization of tall fescue transcriptome under water stress. Plant Genome 2015, 8. [Google Scholar] [CrossRef]
- Hou, W.; Wang, J.; Christensen, M.J.; Liu, J.; Zhang, Y.; Liu, Y.; Cheng, C. Metabolomics insights into the mechanism by which Epichloë gansuensis endophyte increased Achnatherum inebrians tolerance to low nitrogen stress. Plant Soil 2021, 463, 487–508. [Google Scholar] [CrossRef]
- Liu, Y.; Hou, W.; Jin, J.; Christensen, M.J.; Gu, L.; Cheng, C.; Wang, J. Epichloë gansuensis increases the tolerance of Achnatherum inebrians to low-P stress by modulating amino acids metabolism and phosphorus utilization efficiency. J. Fungi 2021, 7, 390. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Han, C.; Gao, H.; Cao, Y. Comparative transcriptome analysis of the garden asparagus (Asparagus officinalis L.) reveals the molecular mechanism for growth with arbuscular mycorrhizal fungi under salinity stress. Plant Physiol. Biochem. 2019, 141, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Alikhani, M.; Khatabi, B.; Sepehri, M.; Nekouei, M.K.; Mardi, M.; Salekdeh, G.H. A proteomics approach to study the molecular basis of enhanced salt tolerance in barley (Hordeum vulgare L.) conferred by the root mutualistic fungus Piriformospora indica. Mol. BioSyst. 2013, 9, 1498–1510. [Google Scholar] [CrossRef]
- Morán-Diez, E.; Rubio, B.; Domínguez, S.; Hermosa, R.; Monte, E.; Nicolás, C. Transcriptomic response of Arabidopsis thaliana after 24h incubation with the biocontrol fungus Trichoderma harzianum. J. Plant Physiol. 2012, 169, 614–620. [Google Scholar] [CrossRef]
- Zouari, I.; Salvioli, A.; Chialva, M.; Novero, M.; Miozzi, L.; Tenore, G.C.; Bagnaresi, P.; Bonfante, P. From root to fruit: RNA-Seq analysis shows that arbuscular mycorrhizal symbiosis may affect tomato fruit metabolism. BMC Genom. 2014, 15, 221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gazara, R.K.; Khan, S.; Iqrar, S.; Ashrafi, K.; Abdin, M.Z. Comparative transcriptome profiling of rice colonized with beneficial endophyte, Piriformospora indica, under high salinity environment. Mol. Biol. Rep. 2020, 47, 7655–7673. [Google Scholar]
- Koga, H.; Christensen, M.J.; Bennett, R.J. Incompatibility of some grass-Acremonium endophyte associations. Mycol. Res. 1993, 97, 1237–1244. [Google Scholar] [CrossRef]
- Kim, K.; Jang, Y.J.; Lee, S.M.; Oh, B.T.; Chae, J.C.; Lee, K.J. Alleviation of salt stress by enterobacter sp. EJ01 in tomato and Arabidopsis is accompanied by up-regulation of conserved salinity responsive factors in plants. Mol. Cells 2014, 37, 109–117. [Google Scholar] [CrossRef]
- Liu, S.; Hao, H.; Lu, X.; Zhao, X.; Wang, Y.; Zhang, Y.; Xie, Z.; Wang, R. Transcriptome profiling of genes involved in induced systemic salt tolerance conferred by Bacillus amyloliquefaciens FZB42 in Arabidopsis thaliana. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef]
- Baskar, V.; Venkatesh, R.; Ramalingam, S. Flavonoids (antioxidants systems) in higher plants. In Antioxidants and Antioxidant Enzymes in Higher Plants; Springer: Berlin, Germany, 2018; pp. 253–268. [Google Scholar]
- Sirrenberg, A.; Göbel, C.; Grond, S.; Czempinski, N.; Ratzinger, A.; Karlovsky, P.; Santos, P.; Feussner, I.; Pawlowski, K. Piriformospora indica affects plant growth by auxin production. Physiol. Plant 2007, 131, 581–589. [Google Scholar] [CrossRef]
- Ghaffari, M.R.; Ghabooli, M.; Khatabi, B.; Hajirezaei, M.R.; Schweizer, P.; Salekdeh, G.H. Metabolic and transcriptional response of central metabolism affected by root endophytic fungus Piriformospora indica under salinity in barley. Plant Mol. Biol. 2016, 90, 699–717. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gao, H.; Liang, Y.; Cao, Y. Full-length transcriptome analysis of asparagus roots reveals the molecular mechanism of salt tolerance induced by arbuscular mycorrhizal fungi. Environ. Exp. Bot. 2021, 185, 104402. [Google Scholar] [CrossRef]
- Leschevin, M.; Ismael, M.; Quero, A.; San Clemente, H.; Roulard, R.; Bassard, S.; Marcelo, P.; Pageau, K.; Jamet, E.; Rayon, C. Physiological and biochemical traits of two major Arabidopsis accessions, Col-0 and Ws, under salinity. Front. Plant Sci. 2021, 12, 639154. [Google Scholar] [CrossRef]
- Gall, H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef]
- Wahid, A.; Ghazanfar, A. Possible involvement of some secondary metabolites in salt tolerance of sugarcane. J. Plant Physiol. 2006, 163, 723–730. [Google Scholar] [CrossRef]
- Vialaret, J.; Di Pietro, M.; Hem, S.; Maurel, C.; Rossignol, M.; Santoni, V. Phosphorylation dynamics of membrane proteins from Arabidopsis roots submitted to salt stress. Proteomics 2014, 14, 1058–1070. [Google Scholar] [CrossRef]
- Bal, H.B.; Nayak, L.; Das, S.; Adhya, T.K. Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant Soil 2013, 366, 93–105. [Google Scholar] [CrossRef]
- Shen, M.; Kang, Y.J.; Wang, H.L.; Zhang, X.S.; Zhao, Q.X. Effect of plant growth-promoting rhizobacteria (PGPRs) on plant growth, yield, and quality of tomato (Lycopersicon esculentum Mill.) under simulated seawater irrigation. J. Gen. Appl. Microbiol. 2012, 58, 253–262. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Dodd, I.C.; Theobald, J.C.; Belimov, A.A.; Davies, W.J. The rhizobacterium Variovorax paradoxus 5C-2, containing ACC deaminase, promotes growth and development of Arabidopsis thaliana via an ethylene-dependent pathway. J. Exp. Bot. 2013, 64, 1565–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadopoulos, A.P.; Saha, U.; Hao, X.; Khosla, S. Response of rockwool-grown greenhouse cucumber, tomato, and pepper to kinetin foliar sprays. Horttechnology 2006, 16, 493–501. [Google Scholar] [CrossRef] [Green Version]
- Bozcuk, S. Effects of kinetin and salinity on germination of tomato, barley and cotton Seeds. Ann. Bot. 1981, 48, 81–84. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Agarwal, R.M. Salinity stress induced alterations in antioxidant metabolism and nitrogen assimilation in wheat (Triticum aestivum L) as influenced by potassium supplementation. Plant Physiol. Biochem. 2017, 115, 449–460. [Google Scholar] [CrossRef]
- Sherameti, I.; Shahollari, B.; Venus, Y.; Altschmied, L.; Varma, A.; Oelmüller, R. The endophytic fungus Piriformospora indica stimulates the expression of nitrate reductase and the starch-degrading enzyme glucan-water dikinase in tobacco and Arabidopsis roots through a homeodomain transcription factor that binds to a conserved motif in their promoters. J. Biol. Chem. 2005, 280, 26241–26247. [Google Scholar]
- Hoshida, H.; Tanaka, Y.; Hibino, T.; Hayashi, Y.; Tanaka, A.; Takabe, T.; Takabe, T. Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol. Biol. 2000, 43, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Nagabhyru, P.; Dinkins, R.D.; Wood, C.L.; Bacon, C.W.; Schardl, C.L. Tall fescue endophyte effects on tolerance to water-deficit stress. BMC Plant Biol. 2013, 13, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shohan, M.; Sinha, S.; Nabila, F.H.; Sg, D.; Seraj, Z.I. Hkt1;5 transporter gene expression and association of amino acid substitutions with salt tolerance across rice genotypes. Front. Plant Sci. 2019, 10, 1420. [Google Scholar] [CrossRef] [Green Version]
- Ren, Z.H.; Gao, J.-P.; Li, L.-G.; Cai, X.-L.; Huang, W.; Chao, D.-Y.; Zhu, M.-Z.; Wang, Z.-Y.; Luan, S.; Lin, H.-X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Su, M.; Chi, X.; Zhang, Z.; Pan, L.; Chen, M.; Wang, T.; Wang, M.; Yang, Z.; Yu, S. Transcriptome analysis reveals salt-stress-regulated biological processes and key pathways in roots of peanut (Arachis hypogaea L.). Genes Genom. 2016, 38, 493–507. [Google Scholar] [CrossRef]
- Bastias, D.A.; Martínez-Ghersa, M.A.; Ballaré, C.L.; Gundel, P.E. Epichloë Fungal endophytes and plant defenses: Not just Alkaloids. Trends Plant Sci. 2017, 22, 939–948. [Google Scholar] [CrossRef]
- Huang, M.; Hu, Y.; Xu, L.; Li, Y.; Hou, X. Arabidopsis LEAFY COTYLEDON1 controls cell fate determination during post-embryonic development. Front. Plant Sci. 2015, 6, 955. [Google Scholar] [CrossRef] [Green Version]
- Solomos, G.T. Galactose metabolism and ethylene production during development and ripening of tomato fruit. Postharvest Biol. Technol. 1991, 1, 67–80. [Google Scholar]
- Libert, B.; Franceschi, V.R. Oxalate in crop plants. J. Agric. Food Chem. 1987, 35, 926–938. [Google Scholar] [CrossRef]
- Haslam, T.M.; Kunst, L. Extending the story of very-long-chain fatty acid elongation. Plant Sci. 2013, 210, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wu, Q.; Yang, L.; Hu, W.; Liu, Y. Ectopic expression of CsKCS6 from navel orange promotes the production of very-long-chain fatty acids (VLCFAs) and increases the abiotic stress tolerance of Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 564656. [Google Scholar] [CrossRef] [PubMed]
- Wieloch, Thomas. A cytosolic oxidation-reduction cycle in plant leaves. J. Exp. Bot. 2021, 72, 4186–4189. [Google Scholar] [CrossRef]
- Pang, C.H.; Wang, B.S. Oxidative Stress and Salt Tolerance in Plants; Springer: Berlin, Germany, 2008. [Google Scholar]
- Zhao, B.; Li, J. Regulation of brassinosteroid biosynthesis and inactivation. J. Integr. Plant Biol. 2012, 54, 746–759. [Google Scholar] [CrossRef]
- Ryu, H.; Cho, Y.-G. Plant hormones in salt stress tolerance. J. Plant Biol. 2015, 58, 147–155. [Google Scholar] [CrossRef]
- Mohsin, T.; Babar, S.; Ankit, S.; Sajitha, B.; Renu, B. 24-Epibrassinolide; an active brassinolide and its role in salt stress tolerance in plants: A review. Plant Physiol. Biochem. 2018, 130, 69–79. [Google Scholar]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef] [Green Version]
- Stone, S.L. The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci. 2014, 5, 135. [Google Scholar] [CrossRef] [PubMed]
- Kurepa, J.; Toh-e, A.; Smalle, J.A. 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 2008, 53, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Smalle, J. The pleiotropic role of the 26S proteasome dubunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 2003, 15, 965–980. [Google Scholar] [CrossRef] [Green Version]
- Dandan, L.; Xiaoqing, H.; Chenghao, L. Overexpression of the LoMYB29 gene of Larix olgensis contributes to the regulation of proanthocyanidin biosynthesis in Arabidopsis thaliana. J. For. Res. 2019, 30, 255–266. [Google Scholar]
- Udomchalothorn, T.; Maneeprasobsuk, S.; Bangyeekhun, E.; Boon-Long, P.; Chadchawan, S. The role of the bifunctional enzyme, fructose-6-phosphate-2-kinase/fructose-2,6-bisphosphatase, in carbon partitioning during salt stress and salt tolerance in Rice (Oryza sativa L.). Plant Sci. 2009, 176, 334–341. [Google Scholar] [CrossRef]
- Chow, W.S.; Anderson, M. Adjustment of photosystem stoichiometry in chloroplast improves the quantum efficiency of photosynthesis. Proc. Natl. Acad. Sci. USA 1990, 87, 7502–7506. [Google Scholar] [CrossRef] [Green Version]
- Yupsanis, T.; Moustakas, M.; Eleftheriou, P.; Damianidou, K. Protein phosphorylation-dephosphorylation in alfalfa seeds germinating under salt stress. J. Plant Physiol. 1994, 143, 234–240. [Google Scholar] [CrossRef]
- Trewavas, A. Post-Translational Modification of Proteins by Phosphorylation. Annu. Rev. Plant Physiol. 1976, 27, 349–374. [Google Scholar] [CrossRef]
- Ranjeva, R.; Graziana, A.; Ranty, B.; Cavalie, G.; Boudet, A. Phosphorylation of proteins in plants: A step in the integration of extra and intracellular stimuli? Physiol. Veg. 1984, 22, 365–376. [Google Scholar]
- Ranjeva, R.; Boudet, A.M. Phosphorylation of proteins in plants: Regulatory effects and potential involvement in stimulus/response coupling. Annu. Rev. Plant Biol. 2003, 38, 73–94. [Google Scholar] [CrossRef]
- Hardie, D.G.; Guy, P.S. The Regulation of enzyme activity by reversible phosphorylation. Prog. Brain Res. 1982, 56, 145–161. [Google Scholar]
- Fang, Z.; Hou, Z.; Wang, S.; Liu, Z.; Wei, S.; Zhang, Y.; Song, J.; Yin, J. Transcriptome analysis reveals the accumulation mechanism of anthocyanins in Buckwheat (Fagopyrum esculentum Moench) cotyledons and flowers. Int. J. Mol. Sci. 2019, 20, 1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Yin, J.-L.; Fang, Z.-W.; Sun, C.; Zhang, P.; Zhang, X.; Lu, C.; Wang, S.-P.; Ma, D.-F.; Zhu, Y.-X. Rapid identification of a stripe rust resistant gene in a space-induced wheat mutant using specific locus amplified fragment (SLAF) sequencing. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, C.; Wang, J.; Hou, W.; Malik, K.; Zhao, C.; Niu, X.; Liu, Y.; Huang, R.; Li, C.; Nan, Z. Elucidating the Molecular Mechanisms by which Seed-Borne Endophytic Fungi, Epichloë gansuensis, Increases the Tolerance of Achnatherum inebrians to NaCl Stress. Int. J. Mol. Sci. 2021, 22, 13191. https://doi.org/10.3390/ijms222413191
Cheng C, Wang J, Hou W, Malik K, Zhao C, Niu X, Liu Y, Huang R, Li C, Nan Z. Elucidating the Molecular Mechanisms by which Seed-Borne Endophytic Fungi, Epichloë gansuensis, Increases the Tolerance of Achnatherum inebrians to NaCl Stress. International Journal of Molecular Sciences. 2021; 22(24):13191. https://doi.org/10.3390/ijms222413191
Chicago/Turabian StyleCheng, Chen, Jianfeng Wang, Wenpeng Hou, Kamran Malik, Chengzhou Zhao, Xueli Niu, Yinglong Liu, Rong Huang, Chunjie Li, and Zhibiao Nan. 2021. "Elucidating the Molecular Mechanisms by which Seed-Borne Endophytic Fungi, Epichloë gansuensis, Increases the Tolerance of Achnatherum inebrians to NaCl Stress" International Journal of Molecular Sciences 22, no. 24: 13191. https://doi.org/10.3390/ijms222413191




