A Search for Cyclin-Dependent Kinase 4/6 Inhibitors by Pharmacophore-Based Virtual Screening, Molecular Docking, and Molecular Dynamic Simulations
Abstract
:1. Introduction
2. Results and Discussion
2.1. Pharmacophore Modelling
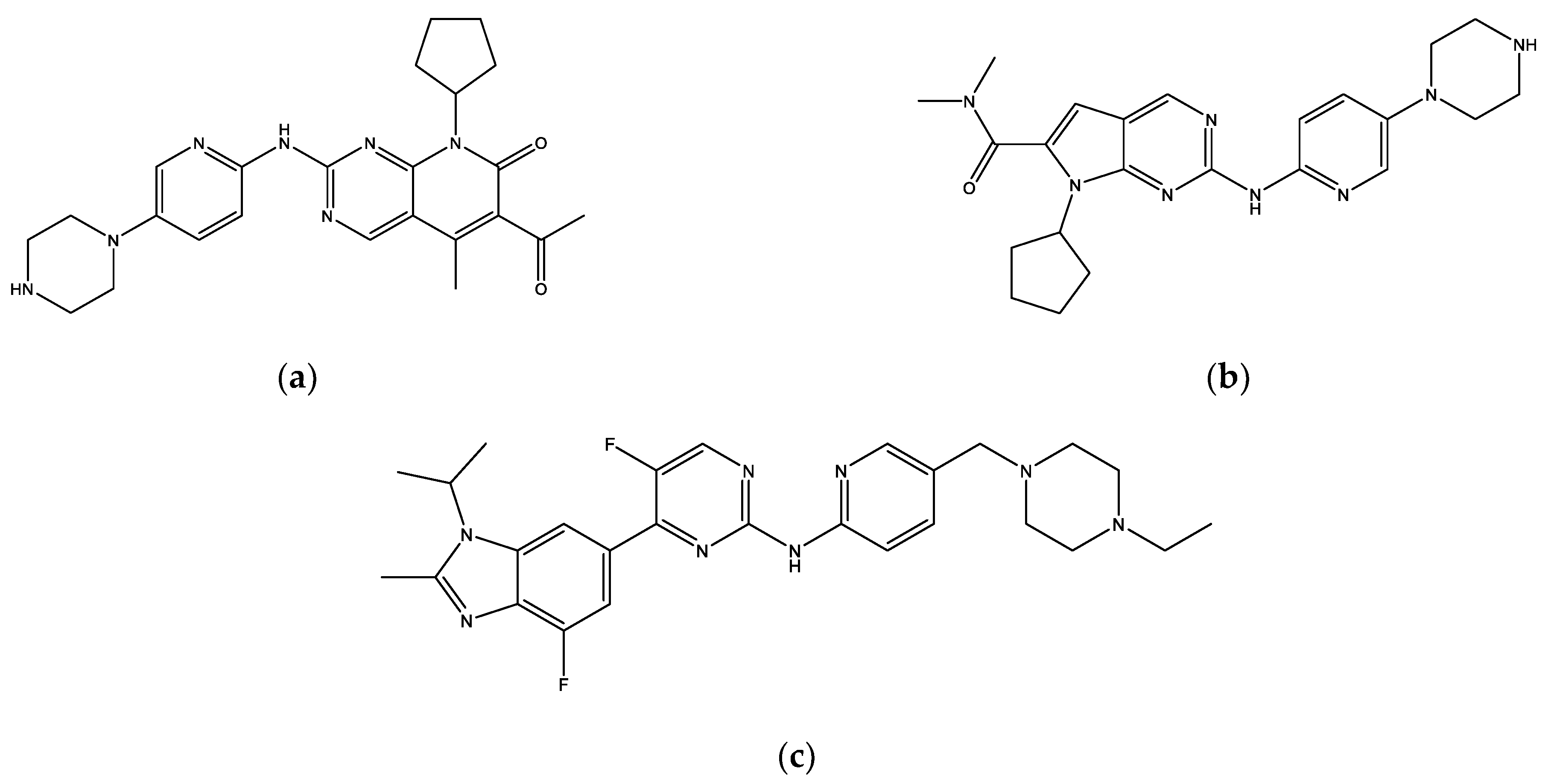
2.2. Molecular Docking Screening
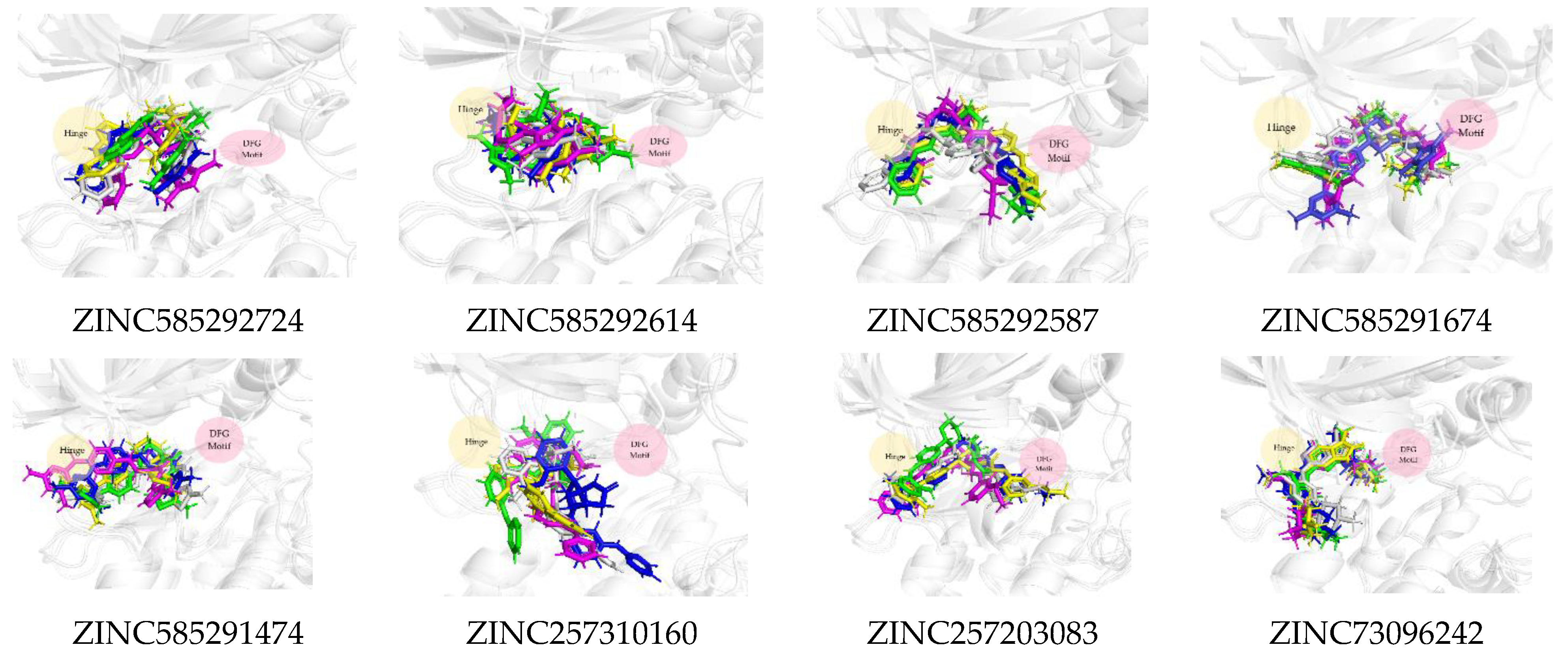
2.3. Molecular Docking Analysis
2.4. Molecular Dynamic Simulations
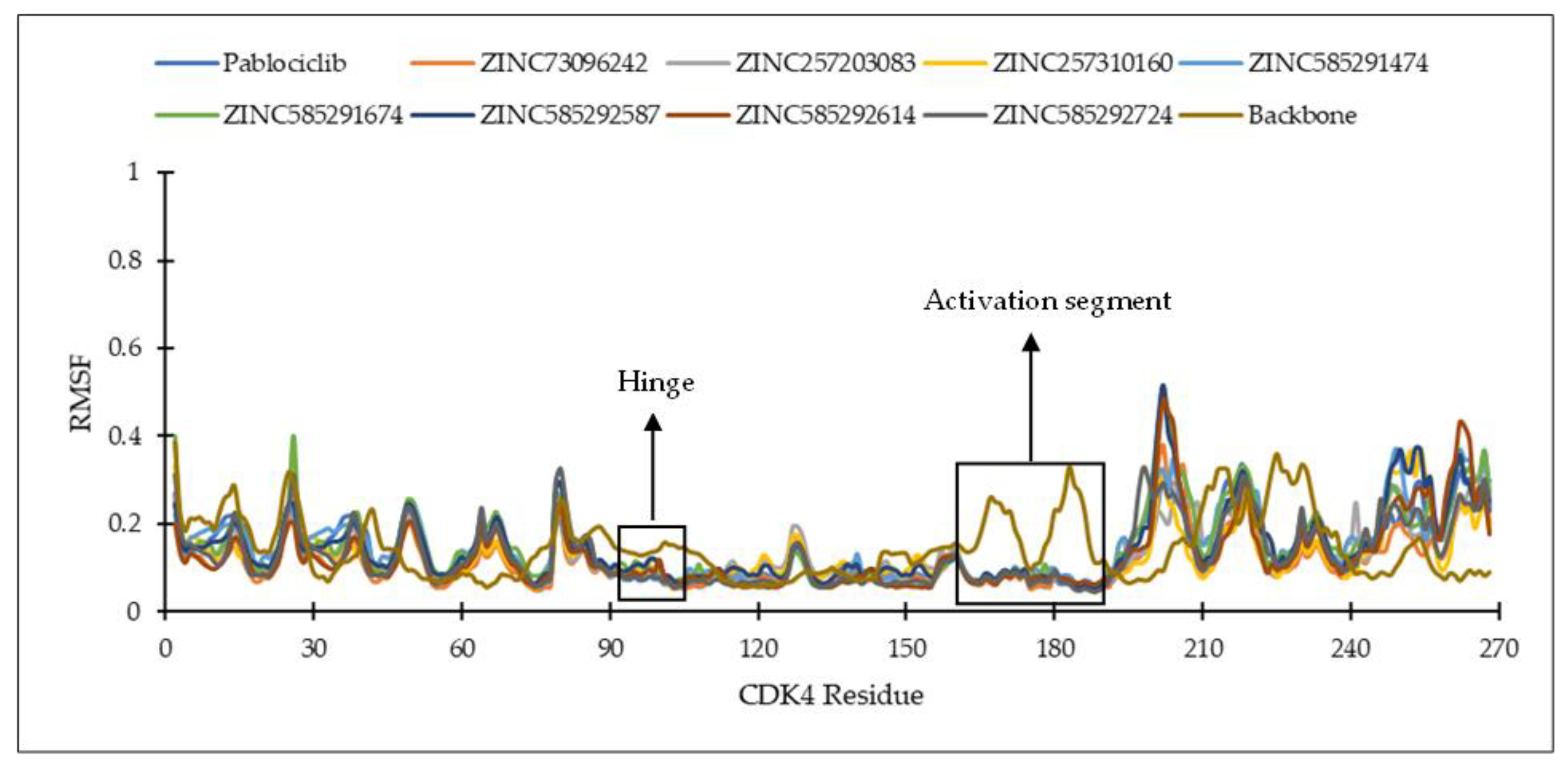
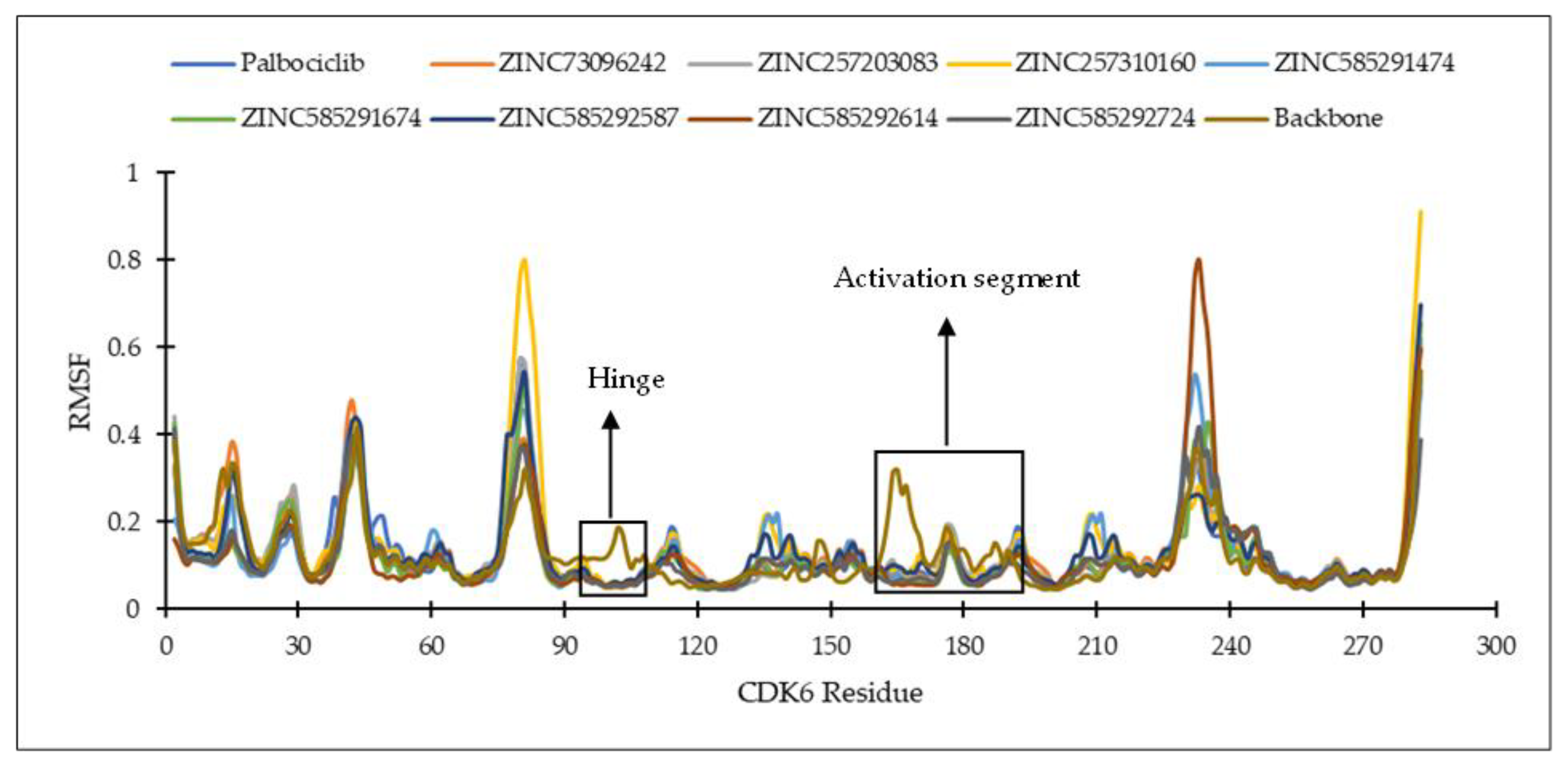
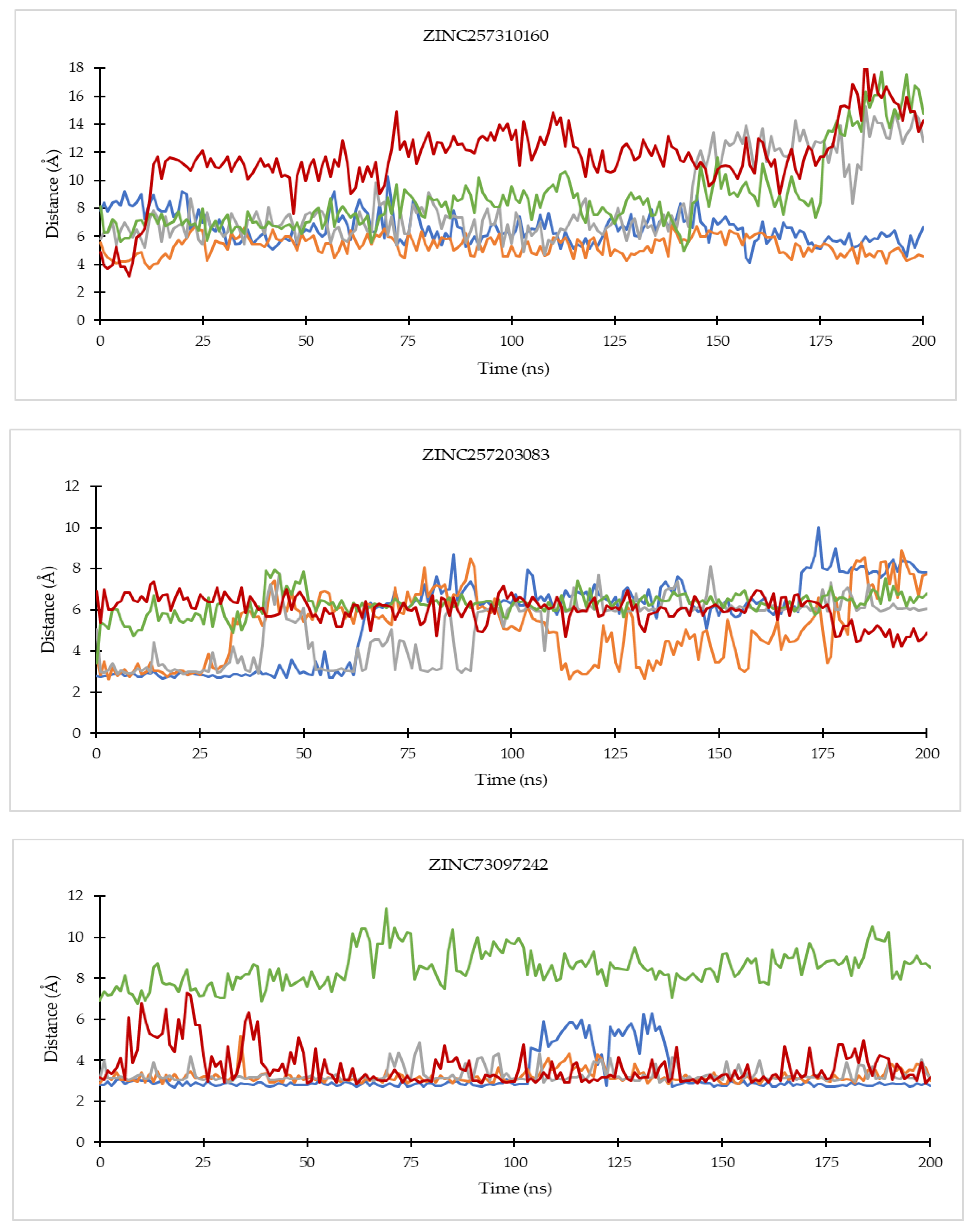
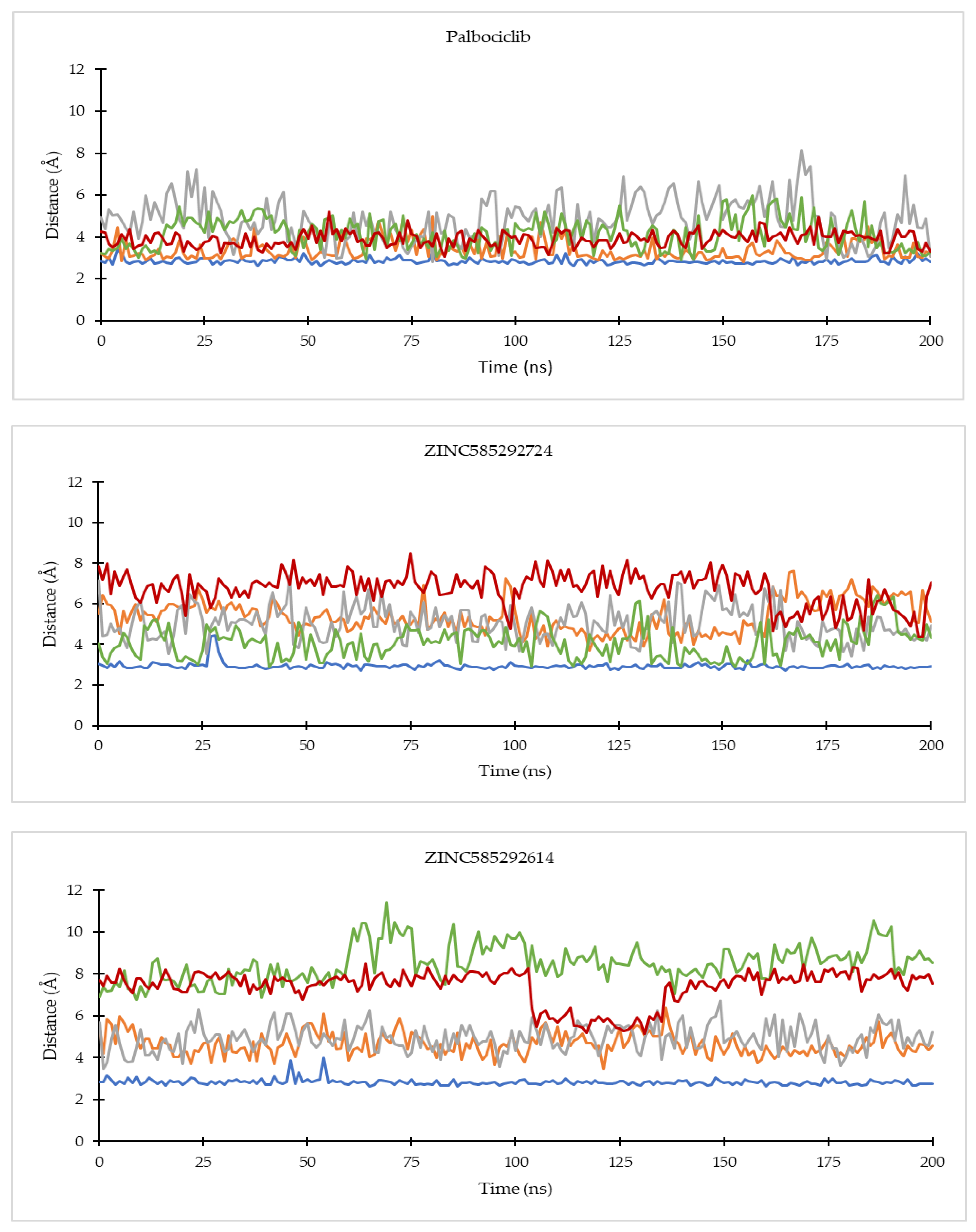
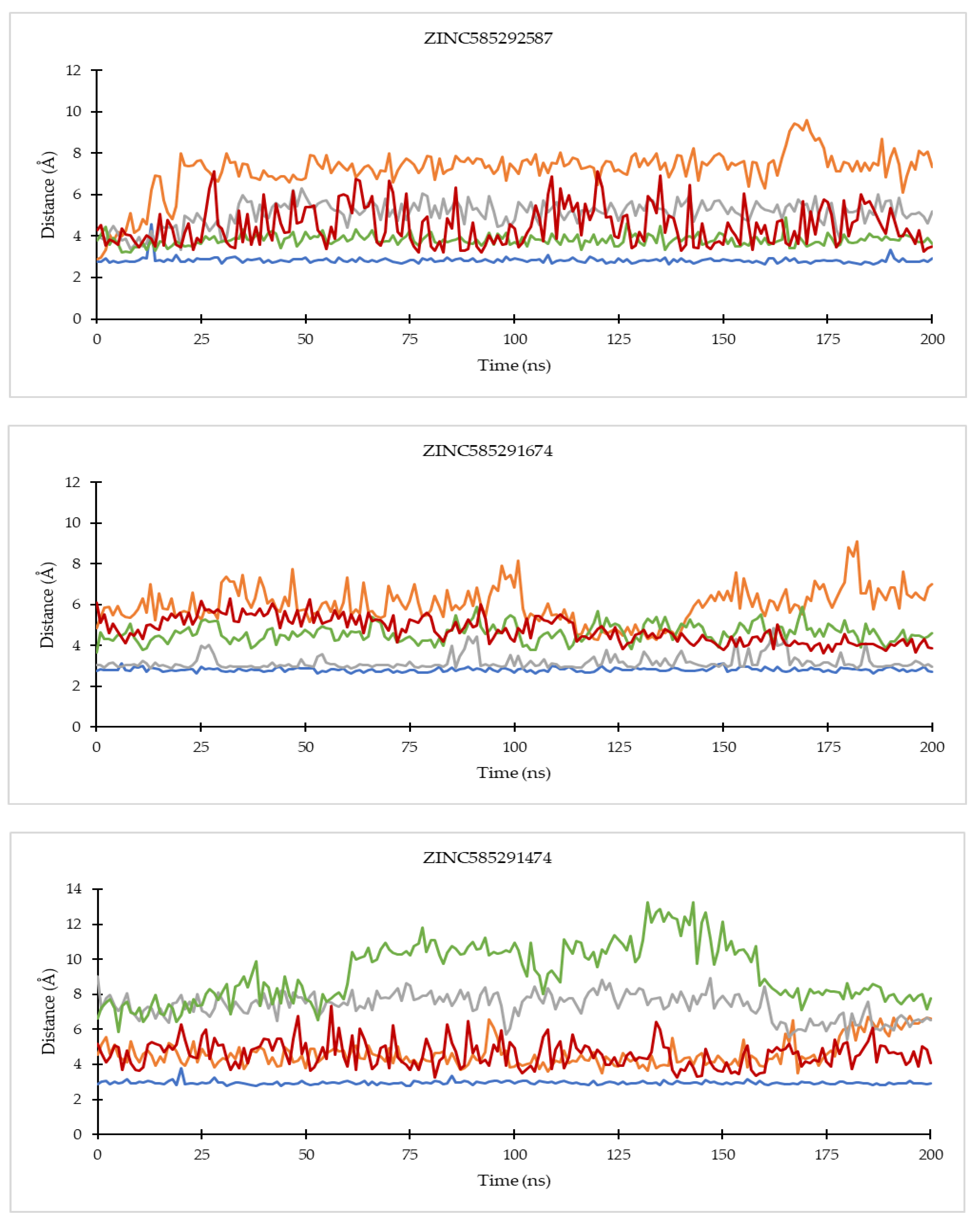
2.4.1. Ligand Binding Stability
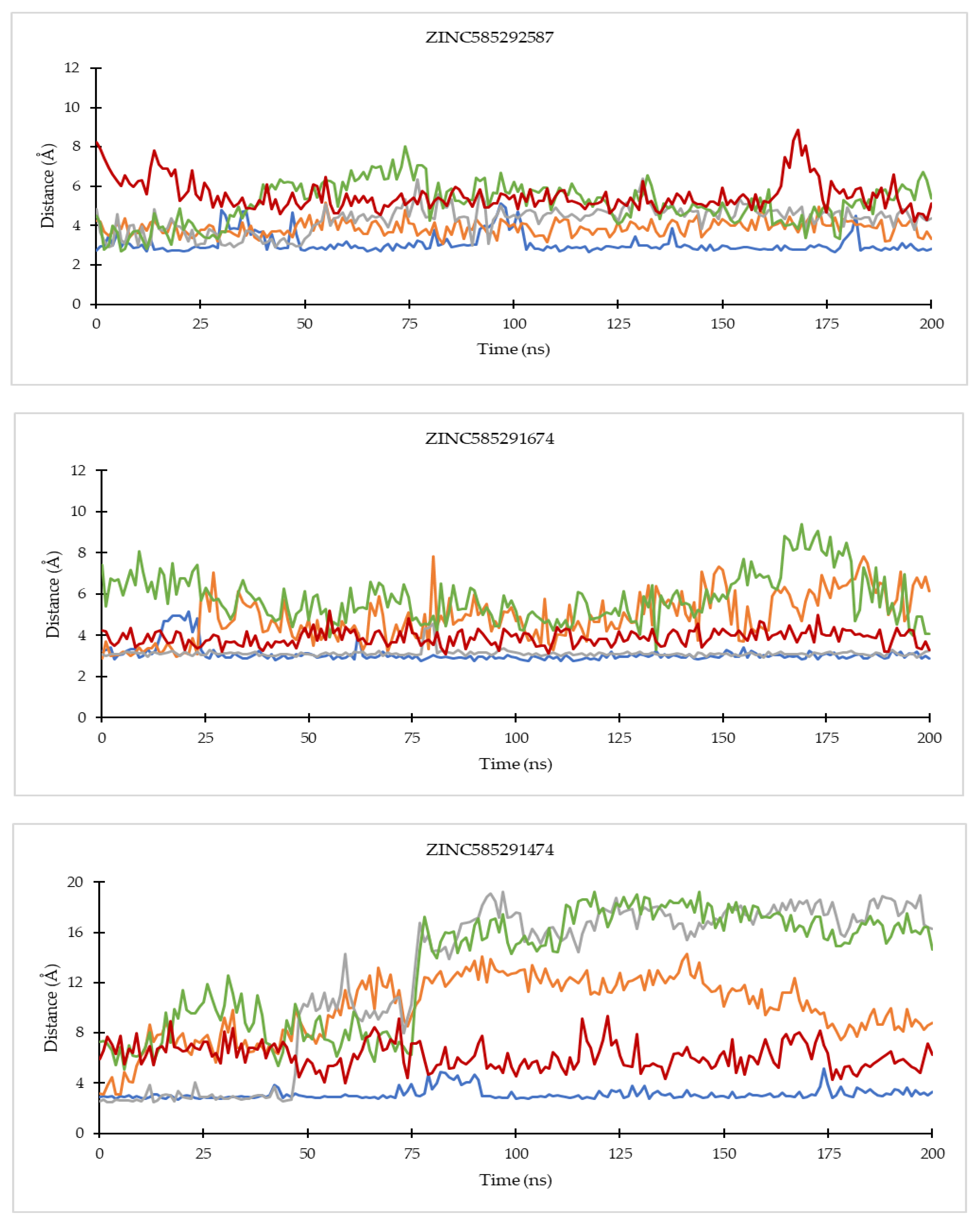
2.4.2. Hydrogen Bond Analysis
2.4.3. MMPBSA Analysis
3. Materials and Method
3.1. Ligand-Based Pharmacophore Modelling
3.2. Molecular Docking
3.3. Molecular Dynamic Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Barnum, K.J.; O’Connell, M.J. Cell cycle regulation by checkpoints. Methods Mol. Biol. 2014, 1170, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.; Kaldis, P. CDKs, cyclins and CKIs: Roles beyond cell cycle regulation. Development 2013, 140, 3079–3093. [Google Scholar] [CrossRef] [Green Version]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Cyclin-dependent protein kinase inhibitors including palbociclib as anticancer drugs. Pharmacol. Res. 2016, 111, 784–803. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J.Y.S.; Tse, G.M. Molecular classification of breast cancer. Adv. Anat. Pathol. 2020, 27, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Hammond, M.E.; Hayes, D.F.; Wolff, A.C.; Mangu, P.B.; Temin, S. American society of clinical oncology/college of American pathologist guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Oncol. Pract. 2010, 6, 195–197. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.S.; Dering, J.; Conklin, D.; Kalous, O.; Cohen, D.J.; Desai, A.J.; Ginther, C.; Atefi, M.; Chen, I.; Fowst, C.; et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009, 11, R77. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.S.; Crown, J.P.; Lang, I.; Boer, K.; Bondarenko, I.M.; Kulyk, S.O.; Ettl, J.; Patel, R.; Pinter, T.; Schmidt, M.; et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of estrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): A randomised phase 2 study. Lancet Oncol. 2015, 16, 25–35. [Google Scholar] [CrossRef]
- Finn, R.S.; Martin, M.; Rugo, H.S.; Jones, S.; Im, S.A.; Gelmon, K.; Harbeck, N.; Lipatov, O.N.; Walshe, J.M.; Moulder, S.; et al. Palbociclib and letrozole in advanced breast cancer. N. Engl. J. Med. 2016, 375, 1925–1936. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Turner, N.C.; Bondarenko, I.; Ro, J.; Im, S.A.; Masuda, N.; Colleoni, M.; DeMichele, A.; Loi, S.; Verma, S.; et al. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): Final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol. 2016, 17, 425–439. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.S.; Angove, H.; Brain, C.; Chen, C.H.; Cheng, H.; Cheng, R.; Chopra, R.; Chung, K.; Congreve, M.; Dagostin, C.; et al. Fragment-based discovery of 7-azabenzimidazoles as potent, highly selective, and orally active cdk4/6 inhibitors. ACS Med. Chem. Lett. 2012, 3, 445–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tadesse, S.; Yu, M.; Mekonnen, L.B.; Lam, F.; Islam, S.; Tomusange, K.; Rahaman, M.H.; Noll, B.; Basnet, S.K.C.; Teo, T.; et al. Highly potent, selective, and orally bioavailable 4-thiazol-N-(pyridin-2-yl)pyrimidin-2-amine Cyclin-Dependent Kinases 4 and 6 inhibitors as anticancer drug candidates: Design, synthesis, and evaluation. J. Med. Chem. 2017, 60, 1892–1995. [Google Scholar] [CrossRef] [Green Version]
- Tadesse, S.; Zhu, G.; Mekonnen, L.B.; Lenjisa, J.L.; Yu, M.; Brown, M.P.; Wang, S. A novel series of N-(pyridin-2-yl)-4-(thiazol-5-yl) pyrimidin-2-amines as highly potent CDK4/6 inhibitors. Future Med. Chem. 2017, 13, 1495–1506. [Google Scholar] [CrossRef]
- Freeman-Cook, K.D.; Hoffman, R.L.; Behenna, D.C.; Boras, B.; Carelli, J.; Diehl, W.; Ferre, R.A.; He, Y.; Hui, A.; Huang, B.; et al. Discovery of PF-06873600, a CDK2/4/6 inhibitor for the treatment of cancer. J. Med. Chem. 2021, 64, 9056–9077. [Google Scholar] [CrossRef] [PubMed]
- Lavecchia, A.; Giovanni, C.D. Virtual screening strategies in drug discovery: A critical review. Curr. Med. Chem. 2013, 20, 2839–2860. [Google Scholar] [CrossRef]
- Lionta, E.; Spyrou, G.; Vassilatis, D.K.; Cournia, Z. Structure-based virtual screening for drug discovery: Principles, applications and recent advances. Curr. Top. Med. Chem. 2014, 14, 1923–1938. [Google Scholar] [CrossRef] [PubMed]
- Svensson, F.; Karlén, A.; Sköld, C. Virtual screening data fusion using both structure- and ligand-based methods. J. Chem. Inf. Model. 2012, 52, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Molecular Operating Environment (MOE), 2014.01; Chemical Computing Group ULC: Montreal, QC, Canada, 2015.
- Sunseri, J.; Koes, D.R. Pharmit: Interactive exploration of chemical space. Nucleic Acid Res. 2016, 44, W442–W448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guner, O.F. History and evolution of the pharmacophore concept in computer-aided drug design. Curr. Top. Med. Chem. 2002, 2, 1321–1332. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC15-ligand discovery for everyone. J. Chem. Inf. Model 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Lagorce, D.; Bouslama, L.; Becot, K.; Miteva, M.A.; Villoutrei, B.O. FAF-Drugs4: Free ADME-Tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics 2017, 33, 3658–3660. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexiblity. J. Comput. Chem. 2009, 16, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hevener, K.E.; Zhao, W.; Ball, D.M.; Babaoglu, K.; Qi, J.; White, S.W.; Lee, R.E. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J. Chem. Inf. Model. 2009, 49, 444–460. [Google Scholar] [CrossRef]
- Hosmer, D.W.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression, 3rd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; p. 177. [Google Scholar]
- Young, S.C.; Borland, M.; Brain, C.; Chen, C.H.T.; Cheng, H.; Chopra, R.; Chung, K.; Groarke, J.; He, G.; Hou, Y.; et al. 4-(pyrazol-4-yl)-pyrimidines as selective inhibitors of cyclin-dependent kinase 4/6. J. Med. Chem. 2010, 53, 7938–7957. [Google Scholar] [CrossRef]
- Susanti, N.M.P.; Tjahjono, D.H.T. Cyclin-dependent kinase 4 and 6 inhibitors in cell cycle dysregulation for breast cancer treatment. Molecules 2021, 26, 4462. [Google Scholar] [CrossRef] [PubMed]
- Linden, O.P.; Kooistra, A.J.; Leurs, R.; de Esch, I.J.; de Graaf, C. KLIFS: A knowledge-based structural database to navigate kinase-ligand interaction space. J. Med. Chem. 2014, 57, 249–277. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol. Res. 2015, 100, 1–23. [Google Scholar] [CrossRef]
- Desheng, L.; Jian, G.; Yuanhua, C.; Wei, C.; Huai, Z.; Mingjuan, J. Molecular dynamics simulations and MM/GBSA methods to investigate binding mechanisms of aminomethylpyrimidine inhibitors with DPP-IV. Bioorg. Med. Chem. Lett. 2011, 21, 6630–6635. [Google Scholar] [CrossRef] [PubMed]
- Labute, P.; Williams, C.; Feher, M.; Sourial, E.; Schmidt, J.M. Flexible alignment of small molecule. J. Med. Chem. 2001, 44, 1483–1490. [Google Scholar] [CrossRef] [PubMed]
- Cereto-Massagué, A.; Guasch, L.; Valls, C.; Mulero, M.; Pujadar, G.; Garcia-Vallveé, S. DecoyFinder: An easy-to-use phyton GUI application for building target-specific decoy sets. Bioinformatics 2012, 28, 1661–1662. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J. Community benchmarks for virtual screening. J. Comput. Aided Mol. Des. 2008, 22, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Kollman, P.A. AMBER 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]

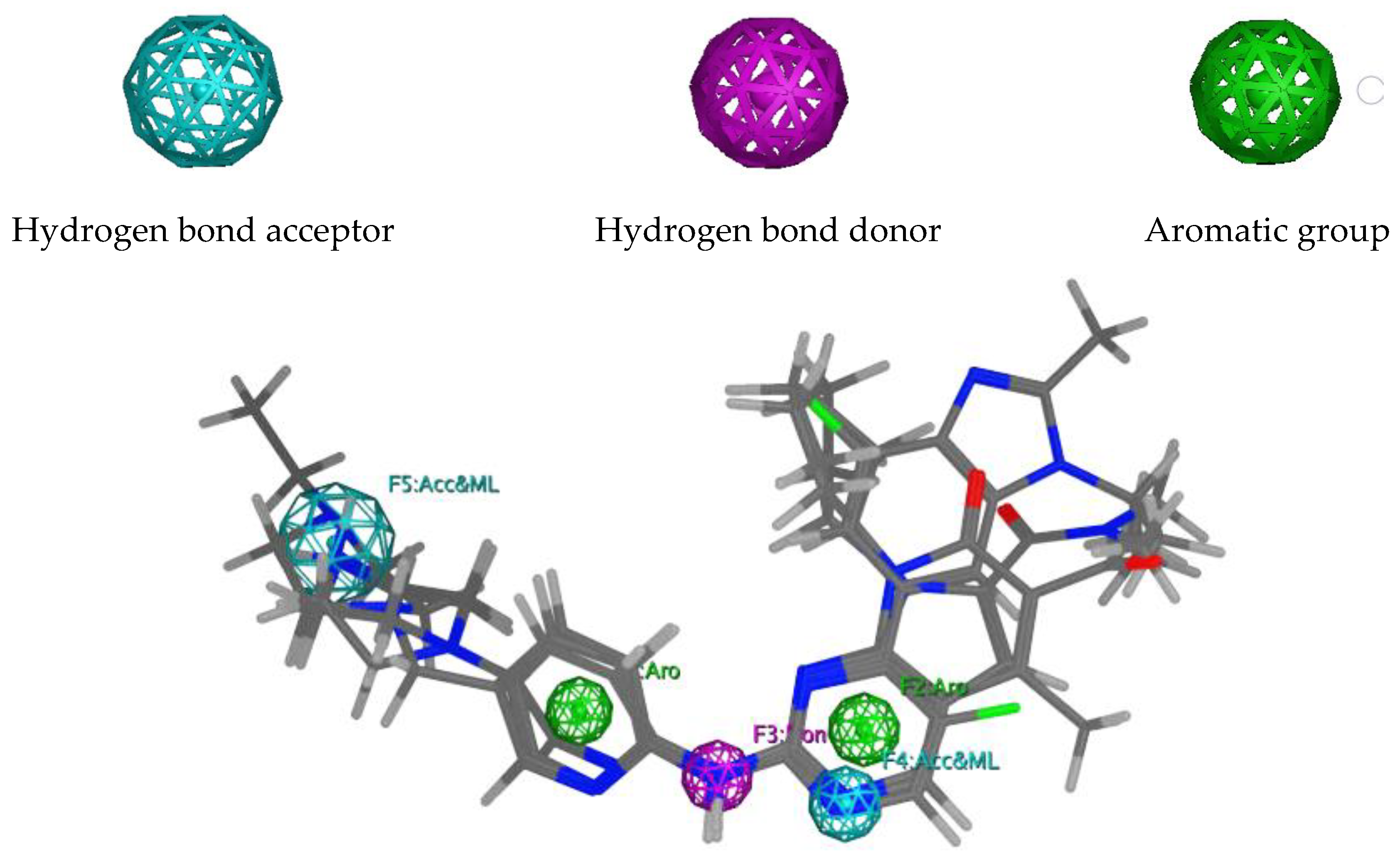
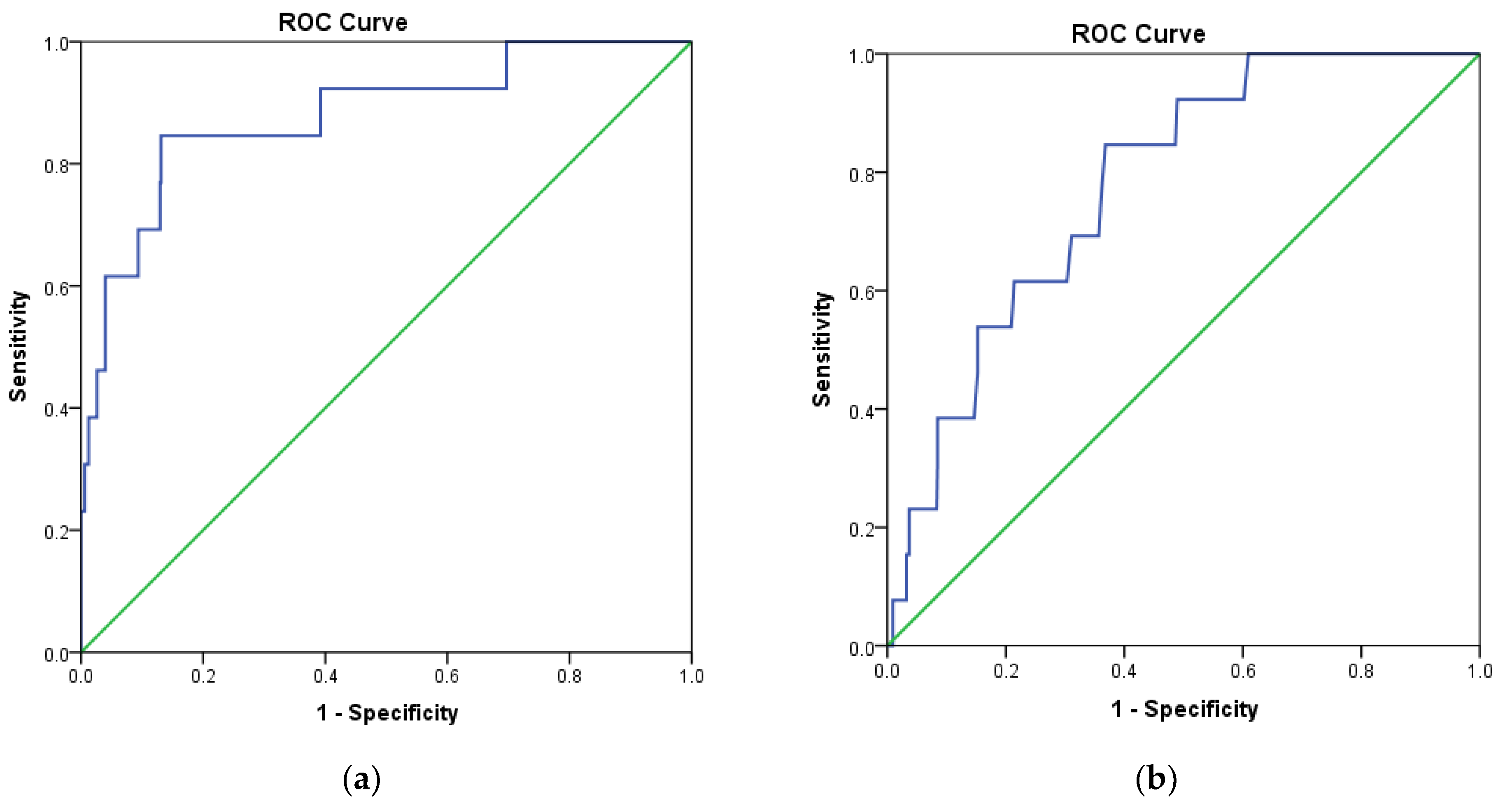
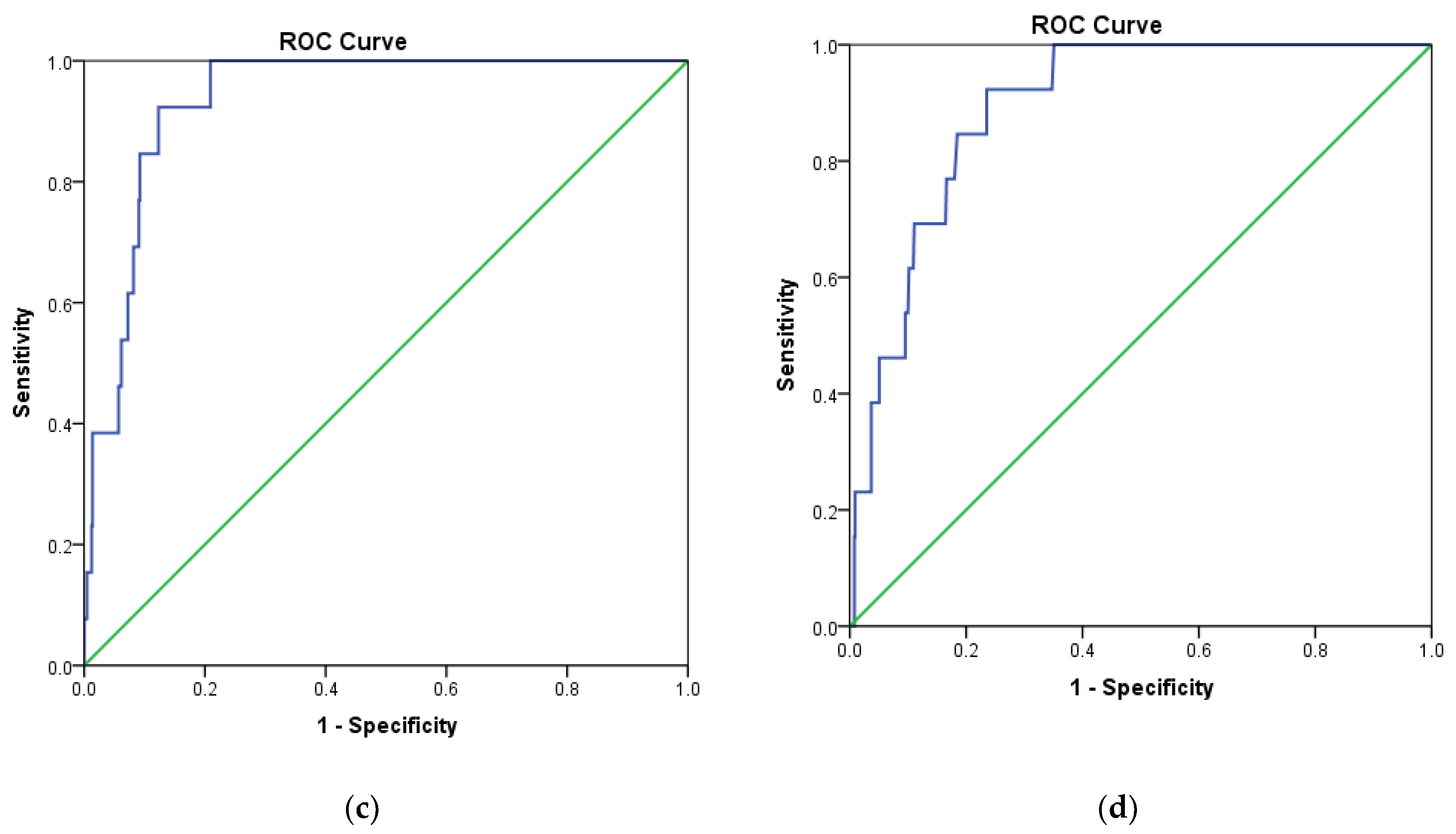




| Internal Validation | Independent Validation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GH | Se | Sp | Acc | Ya | GH | Se | Sp | Acc | Ya | |
| MOE2014 | 0.981 | 0.923 | 1.000 | 0.998 | 1.000 | 0.975 | 0.900 | 1.000 | 0.998 | 1.000 |
| Pharmitt | 0.981 | 0.923 | 1.000 | 0.998 | 1.000 | 0.975 | 0.900 | 1.000 | 0.998 | 1.000 |
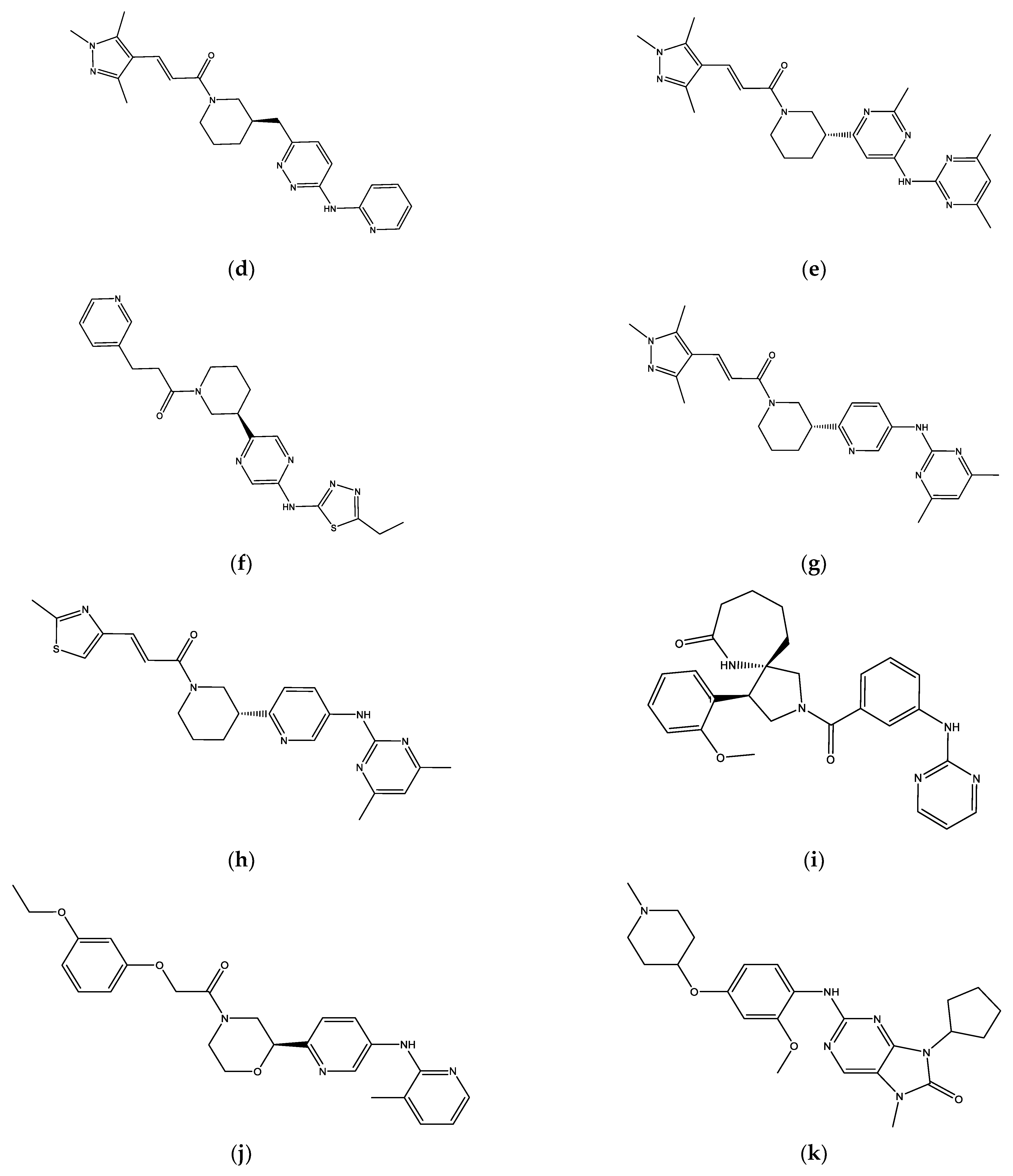
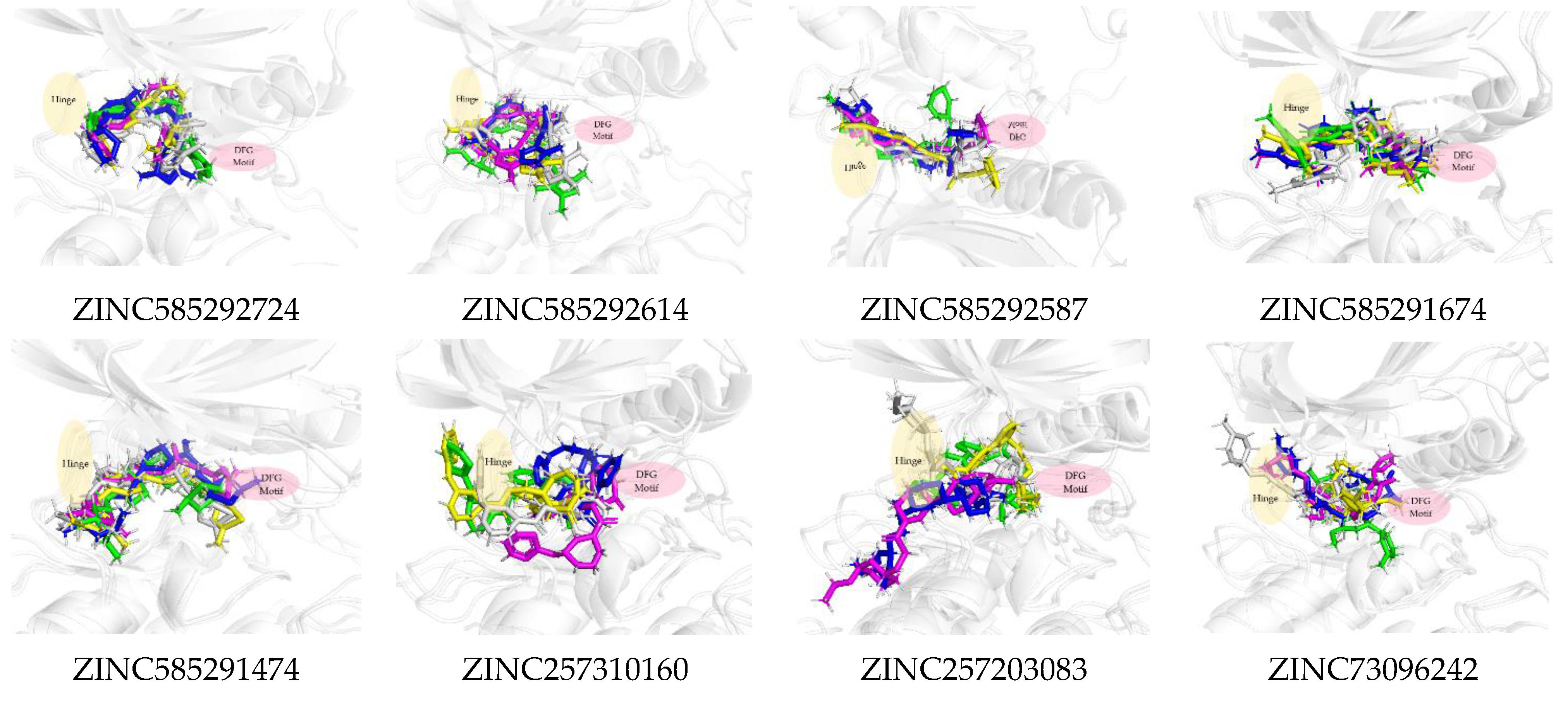
| Ligands | Docking Score MOE2014 | Docking Score Autoock4.2 | H-Bond | Hydrophobic Interaction |
|---|---|---|---|---|
| Palbociclib | −51.94 | −10.77 | Val96 Asp99 | Ile12, Val20, Ala33, Val96, Val72, Leu147, Ala157 |
| ZINC585292724 | −54.01 | −11.11 | Val96 | Ile12, Ala33, Leu147, Ala157 |
| ZINC585292614 | −52.13 | −10.84 | Asp158 | Ile12. Tyr17, Val20, Ala33, Val72, Phe93, His95, Leu147 |
| ZINC585292587 | −53.07 | −11.03 | Val96 Asn145 | Tyr17, Val20, Ala33, Val72, Leu147, Ala157 |
| ZINC585291674 | −54.88 | −11.29 | Val96 | Ile12, Val20, Ala157, Leu147 |
| ZINC585291474 | −53.03 | −11.15 | Val96 | Ile12, Lys22, Ala33, His95, Val96, Ala157 |
| ZINC257310160 | −52.86 | −10.82 | Asp99 | Ile12, Val20, Val96, Leu147 |
| ZINC257203083 | −52.62 | −11.02 | Val96 His95 | Ile12, Ala16, Val20, His95, Asp97, Leu147 |
| ZINC73096242 | −53.23 | −10.86 | Val96 His95 | Ile12, Val20, Ala33, Val72, Val96, Asp97, His95, Leu147, Ala157 |
| Ligands | Docking Score MOE2014 | Docking Score Autoock4.2 | H-Bond | Hydrophobic Interaction |
|---|---|---|---|---|
| Palbociclib | −57.16 | −11.50 | Val101 Asp163 | Val27, Ala41, Val77, Phe98, Ile19, Leu152, Ala162 |
| ZINC585292724 | −59.03 | −11.62 | Val101 | Ile19, Val27, Ala41, Lys43, Phe98, Gln103, Leu152, Ala162 |
| ZINC585292614 | −60.28 | −11.74 | Val101 Lys147 | Ile19, Val27, Asp102, Gln103 |
| ZINC585292587 | −59.23 | −11.62 | Val101 Lys43 | Ile19, Val27, Ala41, Val77, Phe98, Val101, Leu152, Ala162 |
| ZINC585291674 | −62.01 | −12.28 | Val101 Lys43 | Ile19, Val27, Ala41, Phe98, Val101, Gln103, Leu152, Ala162 |
| ZINC585291474 | −61.65 | −12.18 | Val101 Lys43 | Val27, Ala41, Lys43, Val77, Phe98, His100, Leu152, Ala162 |
| ZINC257310160 | −59.03 | −11.52 | Asp163 | Ile19, Val27, Ala41, Phe98, Val101, Leu152, Ala162 |
| ZINC257203083 | −59.26 | −11.50 | Val101 Lys43 | Ile19, Val27, Tyr108, Lys111, Leu152, Val153 |
| ZINC73096242 | −61.85 | −11.81 | Val101 | Ile19, Tyr24, Val27, Lys43, Ala41, His100, Leu152 |
| Ligands | Target | |||
|---|---|---|---|---|
| CDK4 | CDK6 | |||
| Donor-Acceptor | Occupancy (%) | Donor-Acceptor | Occupancy (%) | |
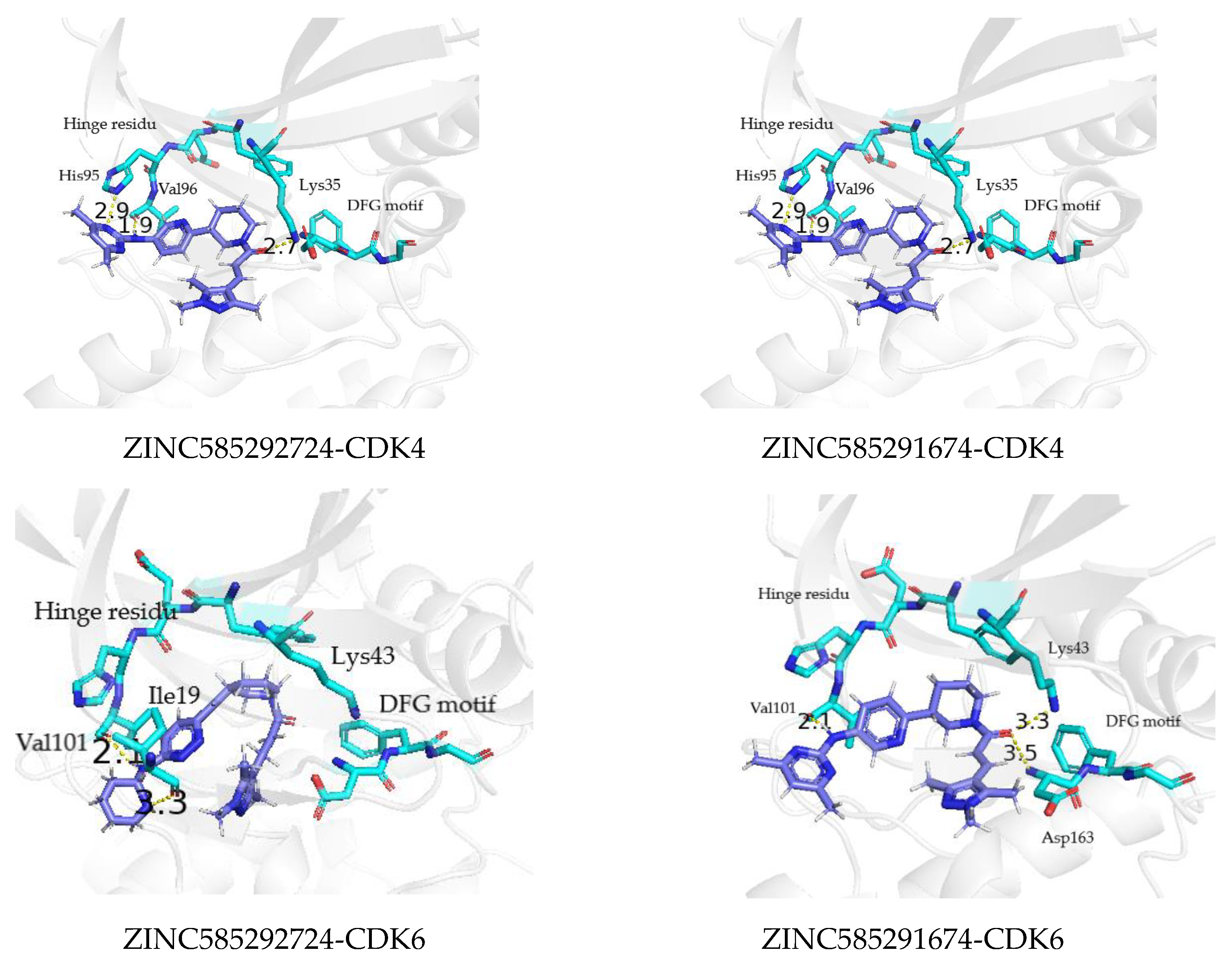
| Palbociclib | (N21)-VAL96(O) (N31)-Thr102(O) Val96(N)-(N9) | 71.65 26.04 13.88 | (N21)-Val101(O) His100(H2)-(N23) (N31)-Thr107(O) | 82.15 28.36 9.74 |
| ZINC585292724 | (N56)-Val96(O) Lys35(N)-(O61) | 52.26 20.26 | (N56)-Val101(O) | 67.38 |
| ZINC585292614 | Asp158(N)-(N64) (N60)-Asp158(O) | 2.54 2.47 | (N60)-Val101(O) | 73.29 |
| ZINC585292587 | Val96(N)-(O52) Lys35(N)-N51 | 51.14 10.69 | (N46)-Val101(O) | 80.43 |
| ZINC585291674 | (N59)-Val96(O) Lys35(N)-(O64) | 55.78 29.39 | (N59)-Val101(O) Lys43(N)-(O64) | 83.61 26.59 |
| ZINC585291474 | (N51)-Val96(O) Lys35(N)-(O56) | 57.26 17.46 | (N51)-Val101(O) | 68.33 |
| ZINC257310160 | (N61)-Glu144(O) Tyr17(OH)-(O56) | 10.42 8.94 | (N59)-Asp104(O) (N61)-Asp102(O) | 34.39 13.66 |
| ZINC257203083 | (N58)-Val96(O) Lys35(N)-(O55) His95(N)-(N60) | 33.22 24.5 22.02 | (N58)-Val101(O) (N58)-Asp144(O) | 35.25 17.31 |
| ZINC73096242 | (N60)-Val96(O) His95(N)-(o56) Lys35(N)-(o58) Val96(N)-(N62) | 53.45 35.0 24.02 9.83 | (N60)-Asp104(O) (N60)-Val101(O) (N60)-Asp163(O) | 23.05 17.21 9.82 |
| Ligands | Van der Waals Energy (∆EVDW) | Electrostatic Energy (∆Eelec) | EPB (∆Epolar) | Enpolar (∆Enon polar) | Binding Energy (∆Gbind) |
|---|---|---|---|---|---|
| Palbociclib | −55.52 ± 2.62 | −30.85 ± 7.84 | 56.3 ± 6.26 | −4.61 ± 0.12 | −34.68 ± 3.46 |
| ZINC585292724 | −57.47 ± 4.24 | −118.27 ± 13.53 | 141.94 ± 11.17 | −3.89 ± 0.11 | −37.69 ± 4.32 |
| ZINC585292614 | −41.67 ± 4.11 | −129.2 ± 12.56 | 145.89 ± 11.60 | −3.33 ± 0.18 | −28.31 ± 4.30 |
| ZINC585292587 | −56.19 ± 2.71 | −25.84 ± 5.43 | 51.88 ± 3.86 | −4.67 ± 0.12 | −34.81 ± 5.07 |
| ZINC585291674 | −55.29 ± 3.24 | −27.40 ± 4.71 | 48.14 ± 4.38 | −3.86 ± 0.07 | −38.41 ± 3.84 |
| ZINC585291474 | −57.22 ± 3.15 | −43.69 ± 5.69 | 68.55 ± 4.91 | −5.17 ± 0.11 | −37.53 ± 4.61 |
| ZINC257310160 | −44.57 ± 4.27 | −27.85 ± 5.67 | 49.33 ± 5.08 | −4.66 ± 0.25 | −27.75 ± 4.19 |
| ZINC257203083 | −47.82 ± 4.23 | −27.91 ± 8.12 | 49.92 ± 7.81 | −4.26 ± 0.21 | −30.08 ± 4.35 |
| ZINC73096242 | −50.94 ± 3.18 | −24.37 ± 45.70 | 50.20 ± 6.73 | −4.65 ± 0.19 | −29.75 ± 4.54 |
| Ligands | Van der Waals Energy (∆EVDW) | Electrostatic Energy (∆Eelec) | EPB (∆Epolar) | Enpolar (∆Enon polar) | Binding Energy (∆Gbind) |
|---|---|---|---|---|---|
| Palbociclib | −55.13 ± 3.21 | −29.12 ± 4.87 | 56.98 ± 4.30 | −4.81 ± 0.12 | −32.08 ± 4.90 |
| ZINC585292724 | −51.50 ± 4.18 | −99.09 ± 11.01 | 118.10 ± 10.01 | −4.49 ± 0.14 | −36.99 ± 5.31 |
| ZINC585292614 | −54.04 ± 3.13 | −28.46 ± 3.96 | 57.23 ± 4.09 | −4.59 ± 0.11 | −29.87 ± 4.56 |
| ZINC585292587 | −54.58 ± 2.75 | −23.03 ± 5.02 | 48.21 ± 4.51 | −4.90 ± 0.12 | −34.30 ± 4.74 |
| ZINC585291674 | −53.01 ± 2.98 | −5.26 ± 3.75 | 23.72 ± 3.58 | −3.79 ± 2.75 | −38.33 ± 2.95 |
| ZINC585291474 | −52.10 ± 3.54 | −14.85 ±4.76 | 34.63 ± 6.13 | −3.92 ± 0.14 | −36.24 ± 3.69 |
| ZINC257310160 | −43.61 ± 4.06 | −95.53 ± 12.22 | 108.53 ± 18.52 | −3.67 ± 0.13 | −34.28 ± 5.41 |
| ZINC257203083 | −41.51 ± 3.84 | −88.11 ±11.45 | 101.01 ± 12.82 | −3.76 ± 0.22 | −32.38 ± 3.77 |
| ZINC73096242 | −54.38 ± 3.20 | −28.07 ± 5.31 | 52.43 ± 5.01 | −3.97 ± 0.15 | −33.99 ± 3.66 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Susanti, N.M.P.; Damayanti, S.; Kartasasmita, R.E.; Tjahjono, D.H. A Search for Cyclin-Dependent Kinase 4/6 Inhibitors by Pharmacophore-Based Virtual Screening, Molecular Docking, and Molecular Dynamic Simulations. Int. J. Mol. Sci. 2021, 22, 13423. https://doi.org/10.3390/ijms222413423
Susanti NMP, Damayanti S, Kartasasmita RE, Tjahjono DH. A Search for Cyclin-Dependent Kinase 4/6 Inhibitors by Pharmacophore-Based Virtual Screening, Molecular Docking, and Molecular Dynamic Simulations. International Journal of Molecular Sciences. 2021; 22(24):13423. https://doi.org/10.3390/ijms222413423
Chicago/Turabian StyleSusanti, Ni Made Pitri, Sophi Damayanti, Rahmana Emran Kartasasmita, and Daryono Hadi Tjahjono. 2021. "A Search for Cyclin-Dependent Kinase 4/6 Inhibitors by Pharmacophore-Based Virtual Screening, Molecular Docking, and Molecular Dynamic Simulations" International Journal of Molecular Sciences 22, no. 24: 13423. https://doi.org/10.3390/ijms222413423
APA StyleSusanti, N. M. P., Damayanti, S., Kartasasmita, R. E., & Tjahjono, D. H. (2021). A Search for Cyclin-Dependent Kinase 4/6 Inhibitors by Pharmacophore-Based Virtual Screening, Molecular Docking, and Molecular Dynamic Simulations. International Journal of Molecular Sciences, 22(24), 13423. https://doi.org/10.3390/ijms222413423








