Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis
Abstract
1. Introduction
2. Results
2.1. Biochemical and Functional Comparison of CjDsbA1 and CjDsbA2
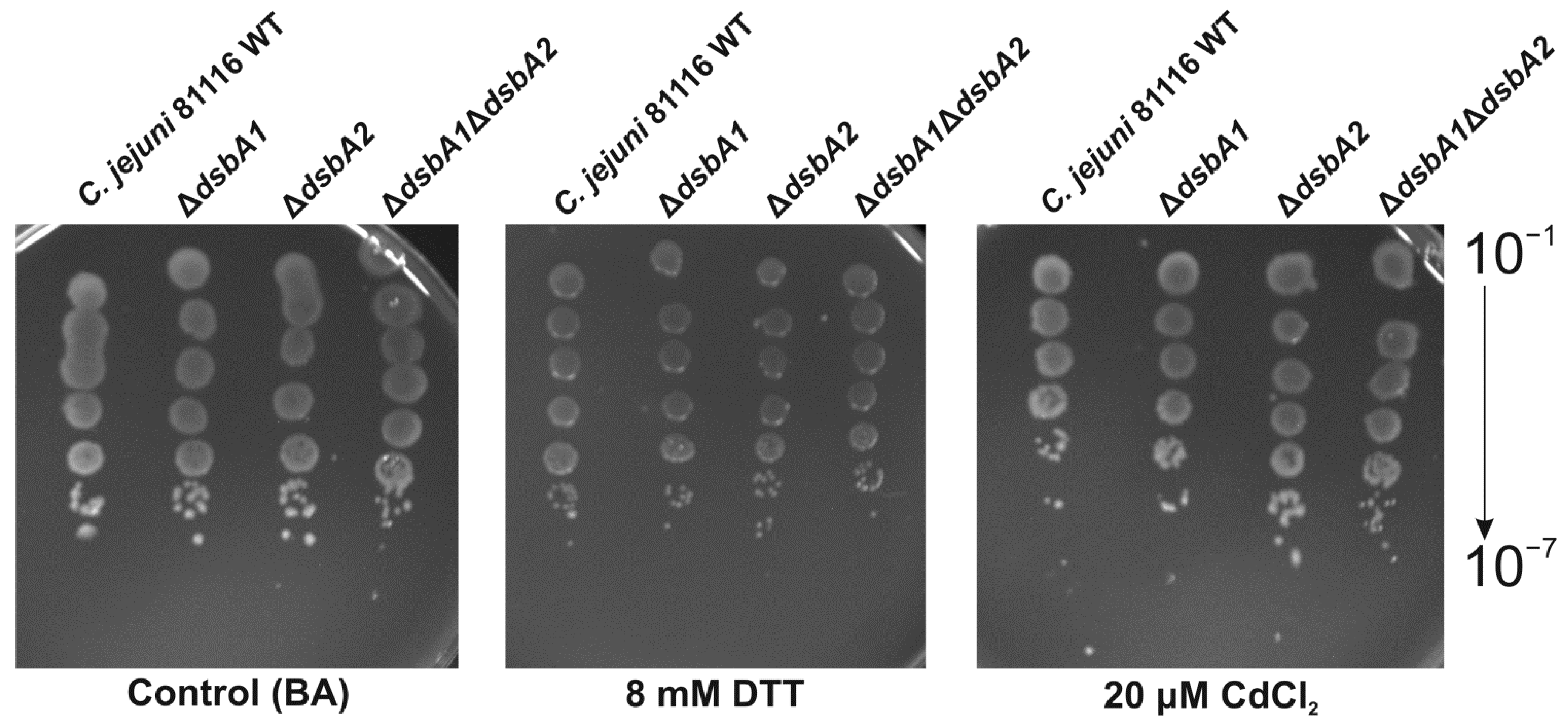
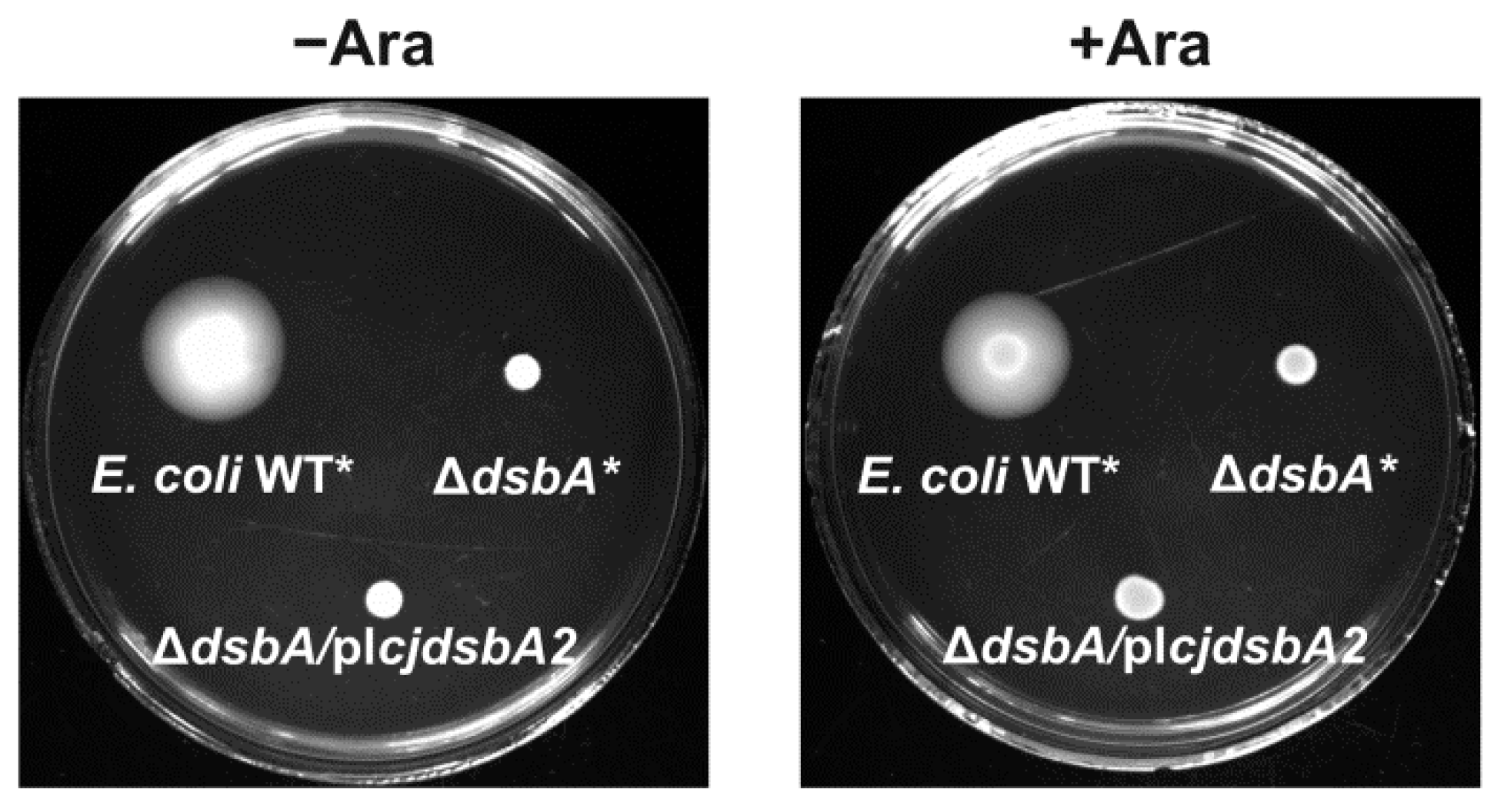
2.2. Functional Studies
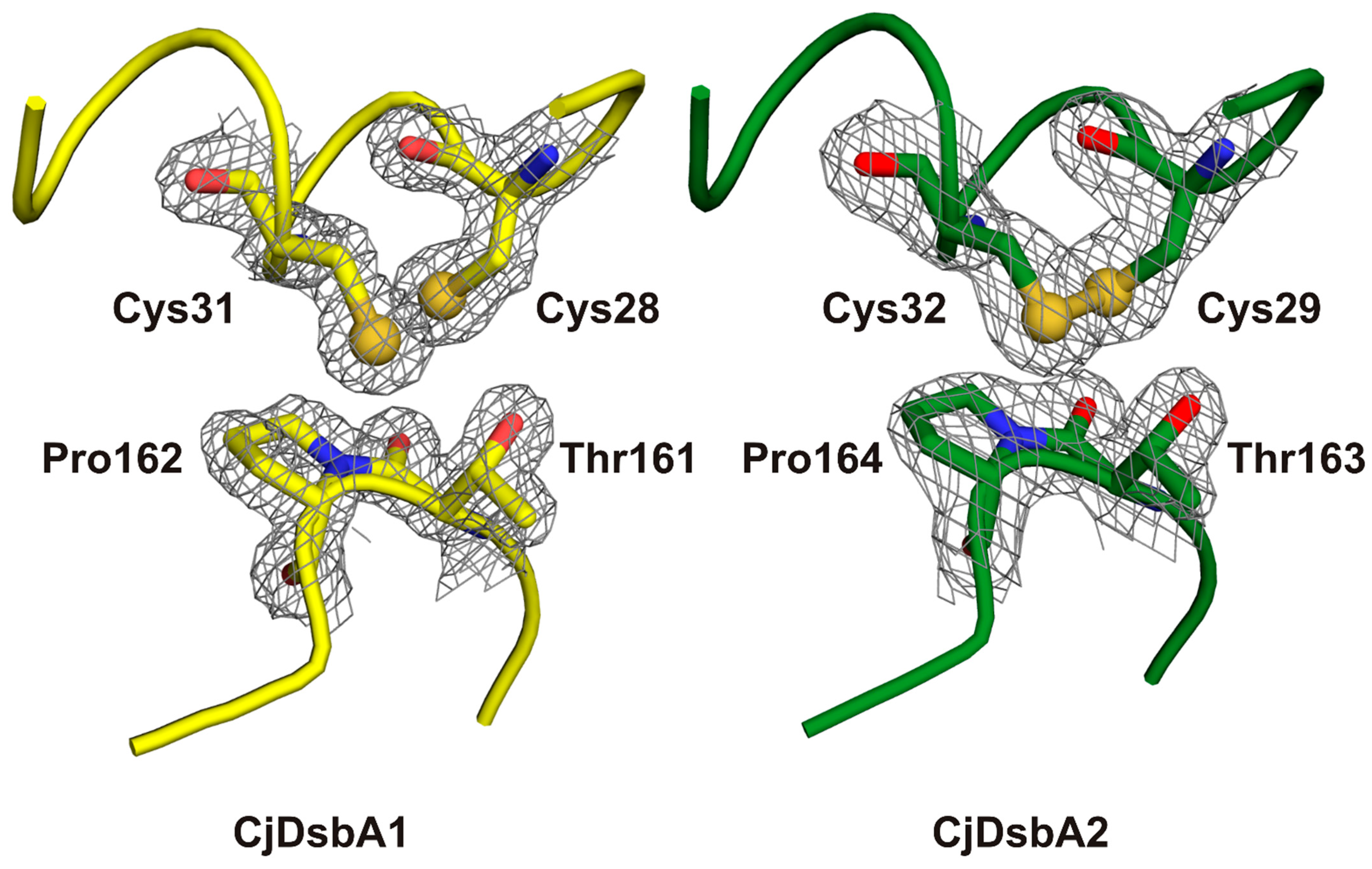
2.3. Structure of C. jejuni DsbA Proteins
2.4. Cooperation of CjDsbB with CjDsbA1 and CjDsbA2
2.5. Identification of Dsb System Targets
2.6. Virulence Tests Using the Galleria mellonella Larvae Infection Model
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains, Primers, Plasmids, Media and Growth Conditions
5.2. DNA Manipulations
5.2.1. General DNA Manipulations
5.2.2. Construction of CjdsbA2+ Plasmids for Complementation Experiments in E. coli
5.2.3. Construction of Recombinant Plasmids for Dsb Protein Overexpression
5.3. Protein Analysis and Biochemical Assays
5.3.1. In Vivo Redox State
5.3.2. Overexpression and Purification
Overexpression and Purification of CjDsbA1, CjDsbA2 and EcDsbA
Overexpression and Purification of CjDsbB Protein
5.3.3. Determination the Redox Potential of CjDsbA1 and CjDsbA2 Proteins
5.3.4. Oxidative Folding of RNaseA
5.3.5. Redox State Analysis of CjDsbA1 and CjDsbA2 in the Presence of CjDsbB
5.3.6. Assessment of CjDsbA1/CjDsbA2 Interactions with CjDsbB Using Microscale Thermophoresis (MST)
5.4. Peptide Synthesis and Purification
5.5. Crystallization, Data Collection and Structure Determination
5.6. Dsb Substrate Identification—Comparison of Periplasmic C. jejuni 81116 Strains Subproteomes
5.6.1. Preparation of Periplasmic Protein Extracts from C. jejuni 81116
5.6.2. Mass Spectrometry
5.6.3. Analysis of Mass Spectrometry Data
5.6.4. Quantification
5.7. Galleria mellonella Virulence Model
5.8. Phenotype Assays
5.8.1. Motility Assay
5.8.2. Spot Titers for Cadmium and DTT Resistance
5.9. Bioinformatic Analysis
5.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Landeta, C.; Boyd, D.; Beckwith, J. Disulfide bond formation in prokaryotes. Nat. Microbiol. 2018, 3, 270–280. [Google Scholar] [CrossRef]
- Hatahet, F.; Boyd, D.; Beckwith, J. Disulfide bond formation in prokaryotes: History, diversity and design. Biochim. Biophys. Acta 2014, 1844, 1402–1414. [Google Scholar] [CrossRef]
- Manta, B.; Boyd, D.; Berkmen, M. Disulfide bond formation in the periplasm of Escherichia coli. EcoSal Plus 2019, 8. [Google Scholar] [CrossRef]
- Shouldice, S.R.; Heras, B.; Walden, P.M.; Totsika, M.; Schembri, M.A.; Martin, J.L. Structure and function of DsbA, a key bacterial oxidative folding catalyst. Antioxid. Redox Signal. 2011, 14, 1729–1760. [Google Scholar] [CrossRef] [PubMed]
- McMahon, R.M.; Premkumar, L.; Martin, J.L. Four structural subclasses of the antivirulence drug target disulfide oxidoreductase DsbA provide a platform for design of subclass-specific inhibitors. Biochim. Biophys. Acta 2014, 1844, 1391–1401. [Google Scholar] [CrossRef]
- Totsika, M.; Vagenas, D.; Paxman, J.J.; Wang, G.; Dhouib, R.; Sharma, P.; Martin, J.L.; Scanlon, M.J.; Heras, B. Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens. Antioxid. Redox Signal. 2018, 29, 653–666. [Google Scholar] [CrossRef]
- Yu, J. Inactivation of DsbA, but not DsbC and DsbD, affects the intracellular survival and virulence of Shigella flexneri. Infect. Immun. 1998, 66, 3909–3917. [Google Scholar] [CrossRef]
- Kurth, F.; Rimmer, K.; Premkumar, L.; Mohanty, B.; Duprez, W.; Halili, M.A.; Shouldice, S.R.; Heras, B.; Fairlie, D.P.; Scanlon, M.J.; et al. Comparative sequence, structure and redox analyses of Klebsiella pneumoniae DsbA show that anti-virulence target DsbA enzymes fall into distinct classes. PLoS ONE 2013, 8, e80210. [Google Scholar] [CrossRef] [PubMed]
- Kurth, F.; Duprez, W.; Premkumar, L.; Schembri, M.A.; Fairlie, D.P.; Martin, J.L. Crystal structure of the dithiol oxidase DsbA enzyme from Proteus mirabilis bound non-covalently to an active site peptide ligand. J. Biol. Chem. 2014, 289, 19810–19822. [Google Scholar] [CrossRef]
- Turcot, I.; Ponnampalam, T.V.; Bouwman, C.W.; Martin, N.L. Isolation and characterization of a chromosomally encoded disulphide oxidoreductase from Salmonella enterica serovar Typhimurium. Can. J. Microbiol. 2001, 47, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Heras, B.; Totsika, M.; Jarrott, R.; Shouldice, S.R.; Guncar, G.; Achard, M.E.; Wells, T.J.; Argente, M.P.; McEwan, A.G.; Schembri, M.A. Structural and functional characterization of three DsbA paralogues from Salmonella enterica serovar typhimurium. J. Biol. Chem. 2010, 285, 18423–18432. [Google Scholar] [CrossRef] [PubMed]
- Banas, A.M.; Bocian-Ostrzycka, K.M.; Jagusztyn-Krynicka, E.K. Engineering of the Dsb (disulfide bond) proteins—Contribution towards understanding their mechanism of action and their applications in biotechnology and medicine. Crit. Rev. Microbiol. 2019, 45, 433–450. [Google Scholar] [CrossRef] [PubMed]
- McMahon, R.M.; Coincon, M.; Tay, S.; Heras, B.; Morton, C.J.; Scanlon, M.J.; Martin, J.L. Sent packing: Protein engineering generates a new crystal form of Pseudomonas aeruginosa DsbA1 with increased catalytic surface accessibility. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 2386–2395. [Google Scholar] [CrossRef] [PubMed]
- Lafaye, C.; Iwema, T.; Carpentier, P.; Jullian-Binard, C.; Kroll, J.S.; Collet, J.F.; Serre, L. Biochemical and structural study of the homologues of the thiol-disulfide oxidoreductase DsbA in Neisseria meningitidis. J. Mol. Biol. 2009, 392, 952–966. [Google Scholar] [CrossRef]
- Vivian, J.P.; Scoullar, J.; Robertson, A.L.; Bottomley, S.P.; Horne, J.; Chin, Y.; Wielens, J.; Thompson, P.E.; Velkov, T.; Piek, S.; et al. Structural and biochemical characterization of the oxidoreductase NmDsbA3 from Neisseria meningitidis. J. Biol. Chem. 2008, 283, 32452–32461. [Google Scholar] [CrossRef] [PubMed]
- Vivian, J.P.; Scoullar, J.; Rimmer, K.; Bushell, S.R.; Beddoe, T.; Wilce, M.C.; Byres, E.; Boyle, T.P.; Doak, B.; Simpson, J.S.; et al. Structure and function of the oxidoreductase DsbA1 from Neisseria meningitidis. J. Mol. Biol. 2009, 394, 931–943. [Google Scholar] [CrossRef]
- Ireland, P.M.; McMahon, R.M.; Marshall, L.E.; Halili, M.; Furlong, E.; Tay, S.; Martin, J.L.; Sarkar-Tyson, M. Disarming Burkholderia pseudomallei: Structural and functional characterization of a disulfide oxidoreductase (DsbA) required for virulence in vivo. Antioxid. Redox Signal. 2014, 20, 606–617. [Google Scholar] [CrossRef]
- Kurz, M.; Iturbe-Ormaetxe, I.; Jarrott, R.; Shouldice, S.R.; Wouters, M.A.; Frei, P.; Glockshuber, R.; O’Neill, S.L.; Heras, B.; Martin, J.L. Structural and functional characterization of the oxidoreductase alpha-DsbA1 from Wolbachia pipientis. Antioxid. Redox Signal. 2009, 11, 1485–1500. [Google Scholar] [CrossRef]
- Arts, I.S.; Ball, G.; Leverrier, P.; Garvis, S.; Nicolaes, V.; Vertommen, D.; Ize, B.; Tamu Dufe, V.; Messens, J.; Voulhoux, R.; et al. Dissecting the machinery that introduces disulfide bonds in Pseudomonas aeruginosa. mBio 2013, 4, e00912–e00913. [Google Scholar] [CrossRef]
- Christensen, S.; Groftehauge, M.K.; Byriel, K.; Huston, W.M.; Furlong, E.; Heras, B.; Martin, J.L.; McMahon, R.M. Structural and biochemical characterization of Chlamydia trachomatis DsbA reveals a cysteine-rich and weakly oxidising oxidoreductase. PLoS ONE 2016, 11, e0168485. [Google Scholar] [CrossRef]
- Daniels, R.; Mellroth, P.; Bernsel, A.; Neiers, F.; Normark, S.; von Heijne, G.; Henriques-Normark, B. Disulfide bond formation and cysteine exclusion in Gram-positive bacteria. J. Biol. Chem. 2010, 285, 3300–3309. [Google Scholar] [CrossRef]
- Zhou, Y.; Cierpicki, T.; Jimenez, R.H.; Lukasik, S.M.; Ellena, J.F.; Cafiso, D.S.; Kadokura, H.; Beckwith, J.; Bushweller, J.H. NMR solution structure of the integral membrane enzyme DsbB: Functional insights into DsbB-catalyzed disulfide bond formation. Mol. Cell 2008, 31, 896–908. [Google Scholar] [CrossRef]
- Inaba, K.; Murakami, S.; Suzuki, M.; Nakagawa, A.; Yamashita, E.; Okada, K.; Ito, K. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell 2006, 127, 789–801. [Google Scholar] [CrossRef]
- Grimshaw, J.P.; Stirnimann, C.U.; Brozzo, M.S.; Malojcic, G.; Grutter, M.G.; Capitani, G.; Glockshuber, R. DsbL and DsbI form a specific dithiol oxidase system for periplasmic arylsulfate sulfotransferase in uropathogenic Escherichia coli. J. Mol. Biol. 2008, 380, 667–680. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Kim, B.; Slauch, J.M. DsbL and DsbI contribute to periplasmic disulfide bond formation in Salmonella enterica serovar Typhimurium. Microbiology 2009, 155, 4014–4024. [Google Scholar] [CrossRef] [PubMed]
- Totsika, M.; Heras, B.; Wurpel, D.J.; Schembri, M.A. Characterization of two homologous disulfide bond systems involved in virulence factor biogenesis in uropathogenic Escherichia coli CFT073. J. Bacteriol. 2009, 191, 3901–3908. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority, and European Centre for Disease Prevention and Control. The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef]
- Burnham, P.M.; Hendrixson, D.R. Campylobacter jejuni: Collective components promoting a successful enteric lifestyle. Nat. Rev. Microbiol. 2018, 16, 551–565. [Google Scholar] [CrossRef]
- Tresse, O.; Alvarez-Ordonez, A.; Connerton, I.F. Editorial: About the foodborne pathogen campylobacter. Front. Microbiol. 2017, 8, 1908. [Google Scholar] [CrossRef]
- Kaakoush, N.O.; Castano-Rodriguez, N.; Mitchell, H.M.; Man, S.M. Global epidemiology of campylobacter infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef]
- Bocian-Ostrzycka, K.M.; Grzeszczuk, M.J.; Dziewit, L.; Jagusztyn-Krynicka, E.K. Diversity of the Epsilonproteobacteria Dsb (disulfide bond) systems. Front. Microbiol. 2015, 6, 570. [Google Scholar] [CrossRef] [PubMed]
- Grabowska, A.D.; Wywial, E.; Dunin-Horkawicz, S.; Lasica, A.M.; Wosten, M.M.; Nagy-Staron, A.; Godlewska, R.; Bocian-Ostrzycka, K.; Pienkowska, K.; Laniewski, P.; et al. Functional and bioinformatics analysis of two Campylobacter jejuni homologs of the thiol-disulfide oxidoreductase, DsbA. PLoS ONE 2014, 9, e106247. [Google Scholar] [CrossRef] [PubMed]
- Roszczenko, P.; Radomska, K.A.; Wywial, E.; Collet, J.F.; Jagusztyn-Krynicka, E.K. A novel insight into the oxidoreductase activity of Helicobacter pylori HP0231 protein. PLoS ONE 2012, 7, e46563. [Google Scholar] [CrossRef]
- Denoncin, K.; Nicolaes, V.; Cho, S.H.; Leverrier, P.; Collet, J.F. Protein disulfide bond formation in the periplasm: Determination of the in vivo redox state of cysteine residues. Methods Mol. Biol. 2013, 966, 325–336. [Google Scholar] [CrossRef]
- Bocian-Ostrzycka, K.M.; Lasica, A.M.; Dunin-Horkawicz, S.; Grzeszczuk, M.J.; Drabik, K.; Dobosz, A.M.; Godlewska, R.; Nowak, E.; Collet, J.F.; Jagusztyn-Krynicka, E.K. Functional and evolutionary analyses of Helicobacter pylori HP0231 (DsbK) protein with strong oxidative and chaperone activity characterized by a highly diverged dimerization domain. Front. Microbiol. 2015, 6, 1065. [Google Scholar] [CrossRef]
- Banas, A.M.; Bocian-Ostrzycka, K.M.; Plichta, M.; Dunin-Horkawicz, S.; Ludwiczak, J.; Placzkiewicz, J.; Jagusztyn-Krynicka, E.K. C8J_1298, a bifunctional thiol oxidoreductase of Campylobacter jejuni, affects Dsb (disulfide bond) network functioning. PLoS ONE 2020, 15, e0230366. [Google Scholar] [CrossRef]
- Mayer, M.P. A new set of useful cloning and expression vectors derived from pBlueScript. Gene 1995, 163, 41–46. [Google Scholar] [CrossRef]
- Dailey, F.E.; Berg, H.C. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA 1993, 90, 1043–1047. [Google Scholar] [CrossRef]
- Wunderlich, M.; Otto, A.; Maskos, K.; Mucke, M.; Seckler, R.; Glockshuber, R. Efficient catalysis of disulfide formation during protein folding with a single active-site cysteine. J. Mol. Biol. 1995, 247, 28–33. [Google Scholar] [CrossRef]
- Ondo-Mbele, E.; Vives, C.; Kone, A.; Serre, L. Intriguing conformation changes associated with the trans/cis isomerization of a prolyl residue in the active site of the DsbA C33A mutant. J. Mol. Biol. 2005, 347, 555–563. [Google Scholar] [CrossRef]
- Duprez, W.; Premkumar, L.; Halili, M.A.; Lindahl, F.; Reid, R.C.; Fairlie, D.P.; Martin, J.L. Peptide inhibitors of the Escherichia coli DsbA oxidative machinery essential for bacterial virulence. J. Med. Chem. 2015, 58, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Paxman, J.J.; Borg, N.A.; Horne, J.; Thompson, P.E.; Chin, Y.; Sharma, P.; Simpson, J.S.; Wielens, J.; Piek, S.; Kahler, C.M.; et al. The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes. J. Biol. Chem. 2009, 284, 17835–17845. [Google Scholar] [CrossRef]
- Grauschopf, U.; Winther, J.R.; Korber, P.; Zander, T.; Dallinger, P.; Bardwell, J.C. Why is DsbA such an oxidizing disulfide catalyst? Cell 1995, 83, 947–955. [Google Scholar] [CrossRef]
- Grzeszczuk, M.J.; Bocian-Ostrzycka, K.M.; Banas, A.M.; Roszczenko-Jasinska, P.; Malinowska, A.; Stralova, H.; Haas, R.; Meyer, T.F.; Jagusztyn-Krynicka, E.K. Thioloxidoreductase HP0231 of Helicobacter pylori impacts HopQ-dependent CagA translocation. Int. J. Med. Microbiol. 2018, 308, 977–985. [Google Scholar] [CrossRef]
- Hiniker, A.; Bardwell, J.C. In vivo substrate specificity of periplasmic disulfide oxidoreductases. J. Biol. Chem. 2004, 279, 12967–12973. [Google Scholar] [CrossRef] [PubMed]
- Andreesen, J.R.; Makdessi, K. Tungsten, the surprisingly positively acting heavy metal element for prokaryotes. Ann. N. Y. Acad. Sci. 2008, 1125, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Smart, J.P.; Cliff, M.J.; Kelly, D.J. A role for tungsten in the biology of Campylobacter jejuni: Tungstate stimulates formate dehydrogenase activity and is transported via an ultra-high affinity ABC system distinct from the molybdate transporter. Mol. Microbiol. 2009, 74, 742–757. [Google Scholar] [CrossRef]
- Rathbun, K.M.; Thompson, S.A. Mutation of PEB4 alters the outer membrane protein profile of Campylobacter jejuni. FEMS Microbiol. Lett. 2009, 300, 188–194. [Google Scholar] [CrossRef]
- Rathbun, K.M.; Hall, J.E.; Thompson, S.A. Cj0596 is a periplasmic peptidyl prolyl cis-trans isomerase involved in Campylobacter jejuni motility, invasion, and colonization. BMC Microbiol. 2009, 9, 160. [Google Scholar] [CrossRef]
- Atack, J.M.; Kelly, D.J. Oxidative stress in Campylobacter jejuni: Responses, resistance and regulation. Future Microbiol. 2009, 4, 677–690. [Google Scholar] [CrossRef]
- Boschi-Muller, S.; Branlant, G. Methionine sulfoxide reductase: Chemistry, substrate binding, recycling process and oxidase activity. Bioorg. Chem. 2014, 57, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Oakland, M.; Jeon, B.; Sahin, O.; Shen, Z.; Zhang, Q. Functional characterization of a lipoprotein-encoding operon in Campylobacter jejuni. PLoS ONE 2011, 6, e20084. [Google Scholar] [CrossRef]
- Bleumink-Pluym, N.M.C.; Verschoor, F.; Gaastra, W.; van der Zeijst, B.A.M.; Fry, B.N. A novel approach for the construction of a Campylobacter mutant library. Microbiology (Reading) 1999, 145 Pt 8, 2145–2151. [Google Scholar] [CrossRef][Green Version]
- Beeby, M.; Ribardo, D.A.; Brennan, C.A.; Ruby, E.G.; Jensen, G.J.; Hendrixson, D.R. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc. Natl. Acad. Sci. USA 2016, 113, E1917–E1926. [Google Scholar] [CrossRef] [PubMed]
- Gundogdu, O.; da Silva, D.T.; Mohammad, B.; Elmi, A.; Mills, D.C.; Wren, B.W.; Dorrell, N. The Campylobacter jejuni MarR-like transcriptional regulators RrpA and RrpB both influence bacterial responses to oxidative and aerobic stresses. Front. Microbiol. 2015, 6, 724. [Google Scholar] [CrossRef]
- Gundogdu, O.; da Silva, D.T.; Mohammad, B.; Elmi, A.; Wren, B.W.; van Vliet, A.H.; Dorrell, N. The Campylobacter jejuni oxidative stress regulator RrpB is associated with a genomic hypervariable region and rltered oxidative stress resistance. Front. Microbiol. 2016, 7, 2117. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Tinsley, C.R.; Voulhoux, R.; Beretti, J.L.; Tommassen, J.; Nassif, X. Three homologues, including two membrane-bound proteins, of the disulfide oxidoreductase DsbA in Neisseria meningitidis: Effects on bacterial growth and biogenesis of functional type IV pili. J. Biol. Chem. 2004, 279, 27078–27087. [Google Scholar] [CrossRef]
- Taylor, A.J.; Kelly, D.J. The function, biogenesis and regulation of the electron transport chains in Campylobacter jejuni: New insights into the bioenergetics of a major food-borne pathogen. Adv. Microb. Physiol. 2019, 74, 239–329. [Google Scholar] [CrossRef]
- Howlett, R.M.; Hughes, B.M.; Hitchcock, A.; Kelly, D.J. Hydrogenase activity in the foodborne pathogen Campylobacter jejuni depends upon a novel ABC-type nickel transporter (NikZYXWV) and is SlyD-independent. Microbiology (Reading) 2012, 158, 1645–1655. [Google Scholar] [CrossRef] [PubMed]
- Weerakoon, D.R.; Borden, N.J.; Goodson, C.M.; Grimes, J.; Olson, J.W. The role of respiratory donor enzymes in Campylobacter jejuni host colonization and physiology. Microb. Pathog. 2009, 47, 8–15. [Google Scholar] [CrossRef]
- van der Stel, A.X.; van de Lest, C.H.A.; Huynh, S.; Parker, C.T.; van Putten, J.P.M.; Wosten, M. Catabolite repression in Campylobacter jejuni correlates with intracellular succinate levels. Environ. Microbiol. 2018, 20, 1374–1388. [Google Scholar] [CrossRef]
- Kassem, I.I.; Candelero-Rueda, R.A.; Esseili, K.A.; Rajashekara, G. Formate simultaneously reduces oxidase activity and enhances respiration in Campylobacter jejuni. Sci. Rep. 2017, 7, 40117. [Google Scholar] [CrossRef] [PubMed]
- Sparacino-Watkins, C.; Stolz, J.F.; Basu, P. Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 2014, 43, 676–706. [Google Scholar] [CrossRef]
- Rothery, R.A.; Workun, G.J.; Weiner, J.H. The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim. Biophys. Acta 2008, 1778, 1897–1929. [Google Scholar] [CrossRef] [PubMed]
- Garg, N.; Taylor, A.J.; Kelly, D.J. Bacterial periplasmic nitrate and trimethylamine-N-oxide respiration coupled to menaquinol-cytochrome c reductase (Qcr): Implications for electrogenic reduction of alternative electron acceptors. Sci. Rep. 2018, 8, 15478. [Google Scholar] [CrossRef]
- Mintmier, B.; McGarry, J.M.; Sparacino-Watkins, C.E.; Sallmen, J.; Fischer-Schrader, K.; Magalon, A.; McCormick, J.R.; Stolz, J.F.; Schwarz, G.; Bain, D.J.; et al. Molecular cloning, expression and biochemical characterization of periplasmic nitrate reductase from Campylobacter jejuni. FEMS Microbiol. Lett. 2018, 365, fny151. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A. Where in the world do bacteria experience oxidative stress? Environ. Microbiol. 2019, 21, 521–530. [Google Scholar] [CrossRef]
- Liu, Y.W.; Kelly, D.J. Cytochrome c biogenesis in Campylobacter jejuni requires cytochrome c6 (CccA; Cj1153) to maintain apocytochrome cysteine thiols in a reduced state for haem attachment. Mol. Microbiol. 2015, 96, 1298–1317. [Google Scholar] [CrossRef] [PubMed]
- Champion, O.L.; Karlyshev, A.V.; Senior, N.J.; Woodward, M.; La Ragione, R.; Howard, S.L.; Wren, B.W.; Titball, R.W. Insect infection model for Campylobacter jejuni reveals that O-methyl phosphoramidate has insecticidal activity. J. Infect. Dis. 2010, 201, 776–782. [Google Scholar] [CrossRef]
- Senior, N.J.; Bagnall, M.C.; Champion, O.L.; Reynolds, S.E.; La Ragione, R.M.; Woodward, M.J.; Salguero, F.J.; Titball, R.W. Galleria mellonella as an infection model for Campylobacter jejuni virulence. J. Med. Microbiol. 2011, 60, 661–669. [Google Scholar] [CrossRef]
- Mehat, J.W.; Park, S.F.; van Vliet, A.H.M.; La Ragione, R.M. CapC, a novel autotransporter and virulence factor of Campylobacter jejuni. Appl. Environ. Microbiol. 2018, 84, e01032-18. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Cawthraw, S.; Bagnall, M.C.; Gielbert, A.J.; Woodward, M.J.; Petrovska, L. Identification of temperature regulated factors of Campylobacter jejuni and their potential roles in virulence. AIMS Microbiol. 2017, 3, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Hermans, D.; Van Deun, K.; Martel, A.; Van Immerseel, F.; Messens, W.; Heyndrickx, M.; Haesebrouck, F.; Pasmans, F. Colonization factors of Campylobacter jejuni in the chicken gut. Vet. Res. 2011, 42, 82. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Miroux, B.; Walker, J.E. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996, 260, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Bardwell, J.C.; McGovern, K.; Beckwith, J. Identification of a protein required for disulfide bond formation in vivo. Cell 1991, 67, 581–589. [Google Scholar] [CrossRef]
- Palmer, S.R.; Gully, P.R.; White, J.M.; Pearson, A.D.; Suckling, W.G.; Jones, D.M.; Rawes, J.C.; Penner, J.L. Water-borne outbreak of Campylobacter gastroenteritis. Lancet 1983, 1, 287–290. [Google Scholar] [CrossRef]
- Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Wunderlich, M.; Glockshuber, R. Redox properties of protein disulfide isomerase (DsbA) from Escherichia coli. Protein Sci. 1993, 2, 717–726. [Google Scholar] [CrossRef]
- Roszczenko, P.; Grzeszczuk, M.; Kobierecka, P.; Wywial, E.; Urbanowicz, P.; Wincek, P.; Nowak, E.; Jagusztyn-Krynicka, E.K. Helicobacter pylori HP0377, a member of the Dsb family, is an untypical multifunctional CcmG that cooperates with dimeric thioldisulfide oxidase HP0231. BMC Microbiol. 2015, 15, 135. [Google Scholar] [CrossRef]
- AAT Bioquest, I. Quest Calculate™ Protein Concentration Calculator. Available online: https://www.aatbio.com/tools/calculate-protein-concentration (accessed on 30 July 2021).
- Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Krug, M.; Weiss, M.S.; Heinemann, U.; Mueller, U. XDSAPP: A graphical user interface for the convenient processing of diffraction data using XDS. J. Appl. Crystallogr. 2012, 45, 568–572. [Google Scholar] [CrossRef]
- Moriarty, N.W.; Grosse-Kunstleve, R.W.; Adams, P.D. Electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 2009, 65, 1074–1080. [Google Scholar] [CrossRef]
- Bunkoczi, G.; Echols, N.; McCoy, A.J.; Oeffner, R.D.; Adams, P.D.; Read, R.J. Phaser.MRage: Automated molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 2276–2286. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Moriarty, N.W.; Zwart, P.H.; Hung, L.W.; Read, R.J.; Adams, P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 2008, 64, 61–69. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubak, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Malinowska, A.; Kistowski, M.; Bakun, M.; Rubel, T.; Tkaczyk, M.; Mierzejewska, J.; Dadlez, M. Diffprot—Software for non-parametric statistical analysis of differential proteomics data. J. Proteomics 2012, 75, 4062–4073. [Google Scholar] [CrossRef]
- Elias, J.E.; Haas, W.; Faherty, B.K.; Gygi, S.P. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods 2005, 2, 667–675. [Google Scholar] [CrossRef]
- proteom.pl. MScan. Available online: http://proteom.ibb.waw.pl/mscan/ (accessed on 12 January 2021).
- Bakun, M.; Karczmarski, J.; Poznanski, J.; Rubel, T.; Rozga, M.; Malinowska, A.; Sands, D.; Hennig, E.; Oledzki, J.; Ostrowski, J.; et al. An integrated LC-ESI-MS platform for quantitation of serum peptide ladders. Application for colon carcinoma study. Proteomics Clin. Appl. 2009, 3, 932–946. [Google Scholar] [CrossRef]
- Askoura, M.; Stintzi, A. Using Galleria mellonella as an infection model for Campylobacter jejuni pathogenesis. Methods Mol. Biol. 2017, 1512, 163–169. [Google Scholar] [CrossRef]
- Ren, G.; Stephan, D.; Xu, Z.; Zheng, Y.; Tang, D.; Harrison, R.S.; Kurz, M.; Jarrott, R.; Shouldice, S.R.; Hiniker, A.; et al. Properties of the thioredoxin fold superfamily are modulated by a single amino acid residue. J. Biol. Chem. 2009, 284, 10150–10159. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Armenteros, J.J.A.; Johansen, A.R.; Gislason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 6.0 achieves signal peptide prediction across all types using protein language models. bioRxiv 2021. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]












| Feature | CjDsbA1 | CjDsbA2 | EcDsbA | EcDsbL |
|---|---|---|---|---|
| Catalytic motif | ||||
| CXXC | CIHC | CTHC | CPHC | CPFC |
| cis-Pro | TcisP | TcisP | VcisP | VcisP |
| Biochemical characteristics | ||||
| Redox potential | −60 mV | −116 mV | −122 mV a | −95 mV a |
| Insulin reduction activity assay | – | – | + a | – a |
| RNaseA oxidation activity assay | + | ++ | ++ a | – a |
| EcDsbA (4TKY) | EcDsbL (3C7M) | CjDsbA1 (7PQ7) | CjDsbA2 (7PQF) | |||||
|---|---|---|---|---|---|---|---|---|
| Seq. ID (%) | RMSD [Å] (Aligned Residues) | Seq. ID (%) | RMSD (Å) (Aligned Residues) | Seq. ID (%) | RMSD (Å) (Aligned Residues) | Seq. ID (%) | RMSD (Å) (Aligned Residues) | |
| EcDsbA (4TKY) | X | X | 22.5 | 2.22 (173) | 24.2 | 2.36 (131) | 23.5 | 2.56 (136) |
| EcDsbL (3C7M) | X | X | 28.3 | 1.78 (152) | 39.8 | 1.26 (186) | ||
| CjDsbA1 (7PQ7) | X | X | 52.4 | 1.09 (187) | ||||
| CjDsbA2 (7PQF) | X | X | ||||||
| L1 | L2 | L3 | CXXC | |
|---|---|---|---|---|
| EcDsbA | 61VNFMGG66 | 147LRGVPAM153 | 162NPQGMDTSNMDVFVQ176 | 30 CPHC33 |
| EcDsbL | 58LETKGE63 | 158AKIQGVPAY166 | 175YTKSIKSID183 | 29CPFC32 |
| CjDsbA1 | 57VSLMNG62 | 155IAKTYGTPAF164 | 173NPSAINSMQ181 | 28CIHC31 |
| CjDsbA2 | 58VSSMGD63 | 157ISQNYGTPAF166 | 175IPSAINSPE183 | 29CTHC32 |
| Locus Tag C. jejuni 81116 | Gene | NCBI Protein ID | q-Value | Ratio ΔdsbA1Δc8j1298/WT | Peptide Number | Description | 1 SignalP Prediction | 2 Cysteine Residues Number |
|---|---|---|---|---|---|---|---|---|
| C8J_0082 | WP_002866718.1 | NA | only in WT | 4 | Putative lipoprotein | Sec/SPII | 4 | |
| C8J_1462 | pflA | WP_002866855.1 | NA | only in WT | 2 | Paralyzed flagellum protein | Sec/SPI | 6 |
| C8J_0543 | rppH (nudH) | WP_002854797.1 | NA | only in A1 | 2 | RNA pyrophosphohydrolase | 1 | |
| C8J_0596 | msrA | WP_002866497.1 | NA | only in WT | 2 | Peptide methionine sulfoxide reductase MsrA | 4 | |
| C8J_0412 | mfrA | WP_002867001.1 | 0.00010 | 0.30 | 54 | Succinate dehydrogenase, flavoprotein subunit | Tat/SPI | 12 |
| C8J_0241 | WP_002871921.1 | 0.00010 | 0.37 | 62 | Trimethylamine-N-oxide reductase | Tat/SPI | 10 | |
| C8J_0413 | mfrB | WP_002854950.1 | 0.00010 | 0.23 | 45 | Succinate dehydrogenase, iron–sulfur protein subunit | 13 | |
| C8J_1439 | tupA | WP_002877290.1 | 0.00010 | 0.27 | 29 | Tungstate-binding protein TupA | Sec/SPI | 2 |
| C8J_0558 | peb4 (ppiC) | WP_002876664.1 | 0.00010 | 0.38 | 32 | Putative peptidyl-prolyl cis-trans isomerase Cbf2 | Sec/SPI | 0 |
| C8J_0731 | napA | WP_002866908.1 | 0.00010 | 0.52 | 91 | Periplasmic nitrate reductase | Tat/SPI | 12 |
| C8J_0040 | cccC | WP_002859743.1 | 0.00010 | 0.34 | 16 | Putative cytochrome C | Sec/SPI | 5 |
| C8J_0028 | WP_072238642.1 | 0.00017 | 0.35 | 27 | Cytoplasmic L-asparaginase | Sec/SPII | 0 | |
| C8J_0079 | aspA | WP_002854440.1 | 0.00083 | 0.51 | 67 | Aspartate ammonia-lyase | 7 | |
| C8J_1282 | fumC | WP_002877339.1 | 0.00296 | 0.51 | 55 | Fumarate hydratase class II | 5 | |
| C8J_0955 | livK | WP_002853905.1 | 0.00715 | 0.25 | 18 | Branched-chain amino-acid ABC transport system, periplasmic binding protein | Sec/SPI | 3 |
| C8J_0335 | WP_012006644.1 | 0.01006 | 0.45 | 26 | Cytochrome c551 peroxidase | Sec/SPI | 4 | |
| C8J_1099 | cccA | WP_002852762.1 | 0.02261 | 0.48 | 15 | Cytochrome c553 | Sec/SPI | 3 |
| Name | Relevant Characteristics | Source/Ref. | |
|---|---|---|---|
| E. coli STRAINS | |||
| TG1 | supE44 hsdΔ 5 thi Δ(lac− proAB) F’ [traD36 proAB+ lacIq lacZΔM15] | [75] | |
| Rosetta(DE3)pLysS | F− ompT hsdSB(rB−mB−) gal dcm (DE3) pLysS RARE (CmR) | Novagen | |
| BL21 (DE3) | F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | [76] | |
| BL21C43 (DE3) | F− ompT gal dcm lon hsdSB(rB−mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) and two uncharacterized mutations | [77] | |
| JCB816 | MC1000 phoR λ102 | [78] | |
| JCB816/pMPM-A6 | MC1000 phoR λ102 carrying pMPM-A6; ApR | DBG Collection* | |
| JCB817 | MC1000 phoR λ102 dsbA::kan1; KmR | [78] | |
| JCB817/pMPM-A6 | MC1000 phoR λ102 dsbA::kan1 carrying pMPM-A6; KmR ApR | DBG Collection* | |
| JCB818 | MC1000 phoR λ102 dsbA::kan1, dsbB::kan; KmR | [78] | |
| JCB818/pMPM-A6 | MC1000 phoR λ102 dsbA::kan1, dsbB::Kan carrying pMPM-A6; KmR ApR | DBG Collection* | |
| BL21/EcdsbA | BL21 carrying EcdsbA+ in pET28a KmR | J.F. Collet Collection | |
| KBO1436 | Rosetta(DE3)pLysS carrying pUWM1430 (CjdsbA1+ in pET28a) KmR CmR | [36] | |
| KBO1441 | Rosetta(DE3)pLysS carrying pUWM1432 (CjdsbA2+ in pET24a) KmR CmR | [36] | |
| C43(DE3)/pUWM1469 | Rosetta(DE3)pLysS carrying pUWM1469 (CjdsbB+ in pET24a) KmR CmR | This study | |
| AG1254 | JCB817 carrying ss’pelB-cjdsbA1+ in pMPM-A6 KmR ApR | [32] | |
| AG1256 | JCB818 carrying ss’pelB-cjdsbA1+ in pMPM-A6 KmR ApR | [32] | |
| AB1525 | JCB817 carrying pUWM1523 (ss’pelB- dsbA2+ in pMPM-A6) KmR ApR | This study | |
| AB1566 | JCB818 carrying pUWM1523 (ss’pelB- dsbA2+ in pMPM-A6) KmR ApR | This study | |
| Campylobacter jejuni 81116 STRAINS | |||
| wild-type—81116 (NCTC11828) | Parental strain | [79] | |
| KBO1 | CjdsbA1::cat | [36] | |
| KBO2 | CjdsbA1::aph3, dsbA2::cat | [36] | |
| AG2 | CjdsbA2::cat | [32] | |
| AG3 | CjdsbB::kan | [32] | |
| AG4 | CjdsbI::cat | [32] | |
| AB4 | CjdsbA1::cat, c8j_1298:aph3 | [36] | |
| General cloning and expression vectors | |||
| pJet 1.2 blunt | ApR, CloneJET PCR Cloning Kit | Thermo Fisher Scientific | |
| pET22b | ApR; ss’pelB, IPTG inducible | Novagen | |
| pET24a | KmR, IPTG inducible | Novagen | |
| pMPM-A6 | ApR, SpecR, ParaBAD | [37] | |
| Plasmids for recombinant Dsb overexpression and purification | |||
| pUWM1430 | CjdsbA1-His6 in pET28a | [36] | |
| pUWM1432 | CjdsbA2-His6 in pET24a | [36] | |
| pUWM1459 | CjdsbB in pJet 1.2 blunt (to generate pUWM1469) | This study | |
| pUWM1469 | CjdsbB-His6 in pET24a | This study | |
| pET28a/EcdsbA | EcdsbA in pET28a | J.F. Collet Collection | |
| Plasmid for complementation experiments | |||
| pUWM1522 | CjdsbA2 in pET22b | This study | |
| pUWM1523 | ss’pelB-cjdsbA2 in pMPM-A6 | This study | |
| Primers | |||
| Name | Sequence | Restriction Site | Source |
| Primers for complementation | |||
| c8j0811Ec_BamHI | GTCGGATCCGTTAAGTGAAGGTAAAG | BamHI | This study |
| c8j0811Ec_stopXhoI | GCACTCGAGAACTCATTTTTGTTTGCTAAG | XhoI | This study |
| Primers for recombinant proteins | |||
| dsbB_pur_rev | AGTAAGCTTAACGACTTGATTTAAATGATTTAAACT | HindIII | This study |
| dsbB_pur_for | AGTCATATGAAAGATAATTGCAGAAAATTTTCACT | NdeI | This study |
| CjDsbA1 | CjDsbA2 | ||
|---|---|---|---|
| PDB ID | 7PQ7 | 7PQ8 | 7PQF |
| Xray source | Elettra XRD2 | BESSY 14.1 | SLS PXIII |
| Wavelength (Å) | 0.9789 | 0.9184 | 1.0 |
| Resolution range (Å) | 45.83–1.55 (1.64–1.55) | 48.89–1.33 (1.41–1.33) | 39.26–1.81 (1.92–1.81) |
| Space group | C2 | P212121 | P3221 |
| Unit cell (Å, °) | 120.88 51.73 75.54 90.0 125.14 90.0 | 34.49 57.67 93.78 90.0 90.0 90.0 | 43.49 43.49 196.31 90.0 90.0 120.0 |
| Total reflections | 194386 (30,855) | 307,623 (47,208) | 283,766 (42,669) |
| Unique reflections | 55,179 (8791) | 81,341 (12,965) | 20,471 (3072) |
| Multiplicity | 3.52 | 3.78 | 13.86 |
| Completeness (%) | 98.6 (97.4) | 97.8 (96.4) | 99.2 (95.3) |
| Mean I/sigma(I) | 16.31 (1.53) | 8.94 (1.77) | 14.60 (1.04) |
| Wilson B-factor | 33.05 | 18.69 | 41.48 |
| R-merge (%) | 3.6 (69.3) | 8.2 (67) | 11.6 (271.4) |
| R-meas (%) | 4.3 (81.6) | 9.5 (78.3) | 12.1 (281.7) |
| CC1/2 | 99.9 (77.2) | 99.6 (65.5) | 99.9 (63.2) |
| Reflections used in refinement | 55,153 (5392) | 42145 | 20,349 (1926) |
| R-work | 0.1768 (0.3444) | 0.1759 | 0.1990 (0.3609) |
| R-free | 0.2009 (0.3550) | 0.2192 | 0.2346 (0.3675) |
| Number of non-hydrogen atoms | 3569 | 1942 | 1681 |
| Macromolecules | 3163 | 1631 | 1574 |
| Solvent | 383 | 311 | 107 |
| Protein residues | 379 | 191 | 194 |
| RMS(bonds) | 0.019 | 0.011 | 0.009 |
| RMS(angles) | 1.62 | 1.75 | 1.01 |
| Ramachandran favored (%) | 99.20 | 98.94 | 99.48 |
| Ramachandran allowed (%) | 0.80 | 1.06 | 0.52 |
| Ramachandran outliers (%) | 0 | 0 | 0 |
| Rotamer outliers (%) | 0.56 | 0.55 | 0.58 |
| Clashscore | 4.42 | 4.00 | 3.18 |
| Average B-factor | 37.96 | 14.02 | 41.93 |
| Macromolecules | 37.03 | 13.69 | 41.72 |
| Solvent | 44.59 | 27.04 | 45.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banaś, A.M.; Bocian-Ostrzycka, K.M.; Dunin-Horkawicz, S.; Ludwiczak, J.; Wilk, P.; Orlikowska, M.; Wyszyńska, A.; Dąbrowska, M.; Plichta, M.; Spodzieja, M.; et al. Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis. Int. J. Mol. Sci. 2021, 22, 13451. https://doi.org/10.3390/ijms222413451
Banaś AM, Bocian-Ostrzycka KM, Dunin-Horkawicz S, Ludwiczak J, Wilk P, Orlikowska M, Wyszyńska A, Dąbrowska M, Plichta M, Spodzieja M, et al. Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis. International Journal of Molecular Sciences. 2021; 22(24):13451. https://doi.org/10.3390/ijms222413451
Chicago/Turabian StyleBanaś, Anna M., Katarzyna M. Bocian-Ostrzycka, Stanisław Dunin-Horkawicz, Jan Ludwiczak, Piotr Wilk, Marta Orlikowska, Agnieszka Wyszyńska, Maria Dąbrowska, Maciej Plichta, Marta Spodzieja, and et al. 2021. "Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis" International Journal of Molecular Sciences 22, no. 24: 13451. https://doi.org/10.3390/ijms222413451
APA StyleBanaś, A. M., Bocian-Ostrzycka, K. M., Dunin-Horkawicz, S., Ludwiczak, J., Wilk, P., Orlikowska, M., Wyszyńska, A., Dąbrowska, M., Plichta, M., Spodzieja, M., Polańska, M. A., Malinowska, A., & Jagusztyn-Krynicka, E. K. (2021). Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis. International Journal of Molecular Sciences, 22(24), 13451. https://doi.org/10.3390/ijms222413451






