Hormonal Regulatory Patterns of LaKNOXs and LaBEL1 Transcription Factors Reveal Their Potential Role in Stem Bulblet Formation in LA Hybrid Lily
Abstract
:1. Introduction
2. Results
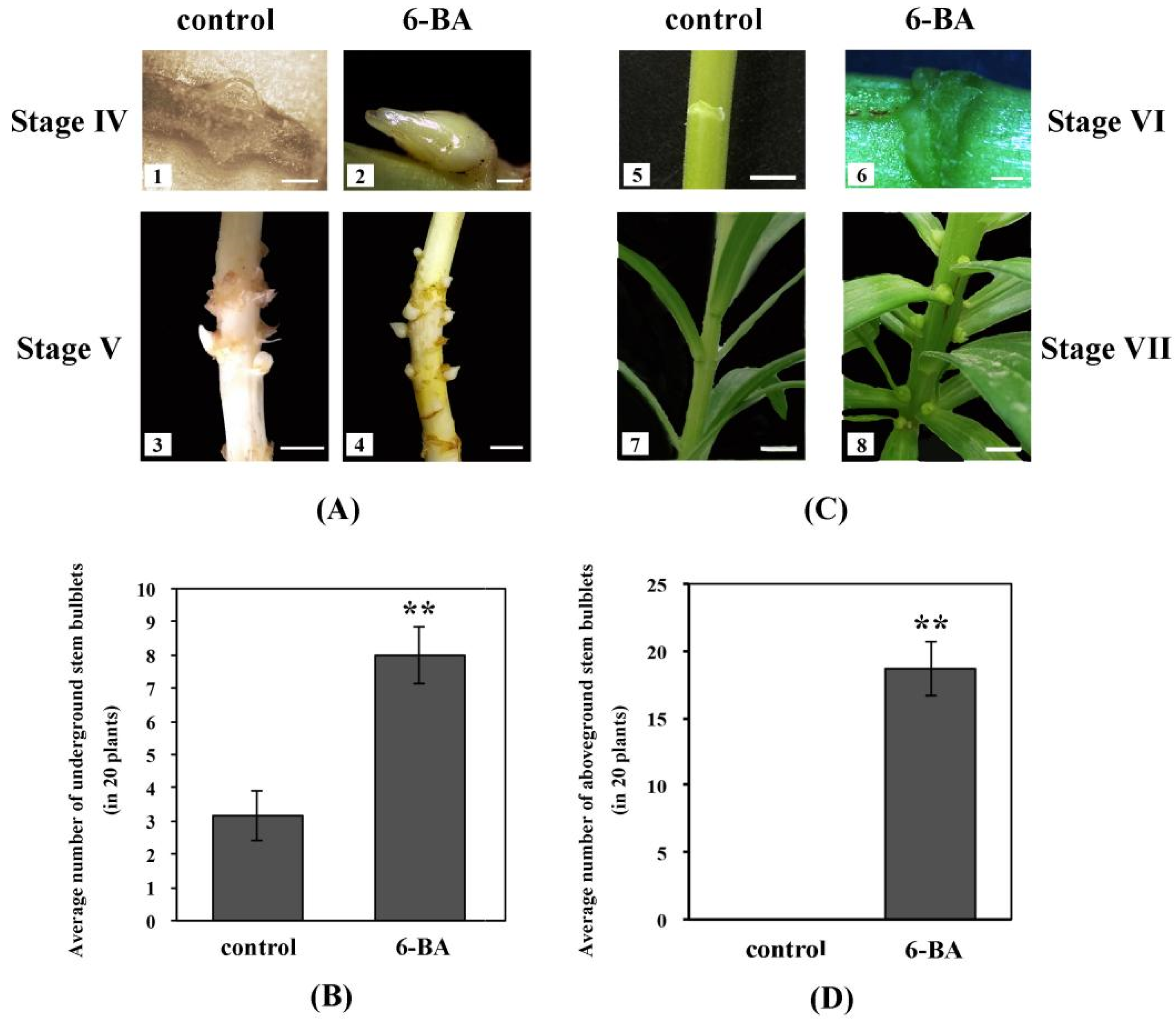
2.1. Effects of Exogenous 6-BA on Stem Bulblet Formation in L. ‘Aladdin’
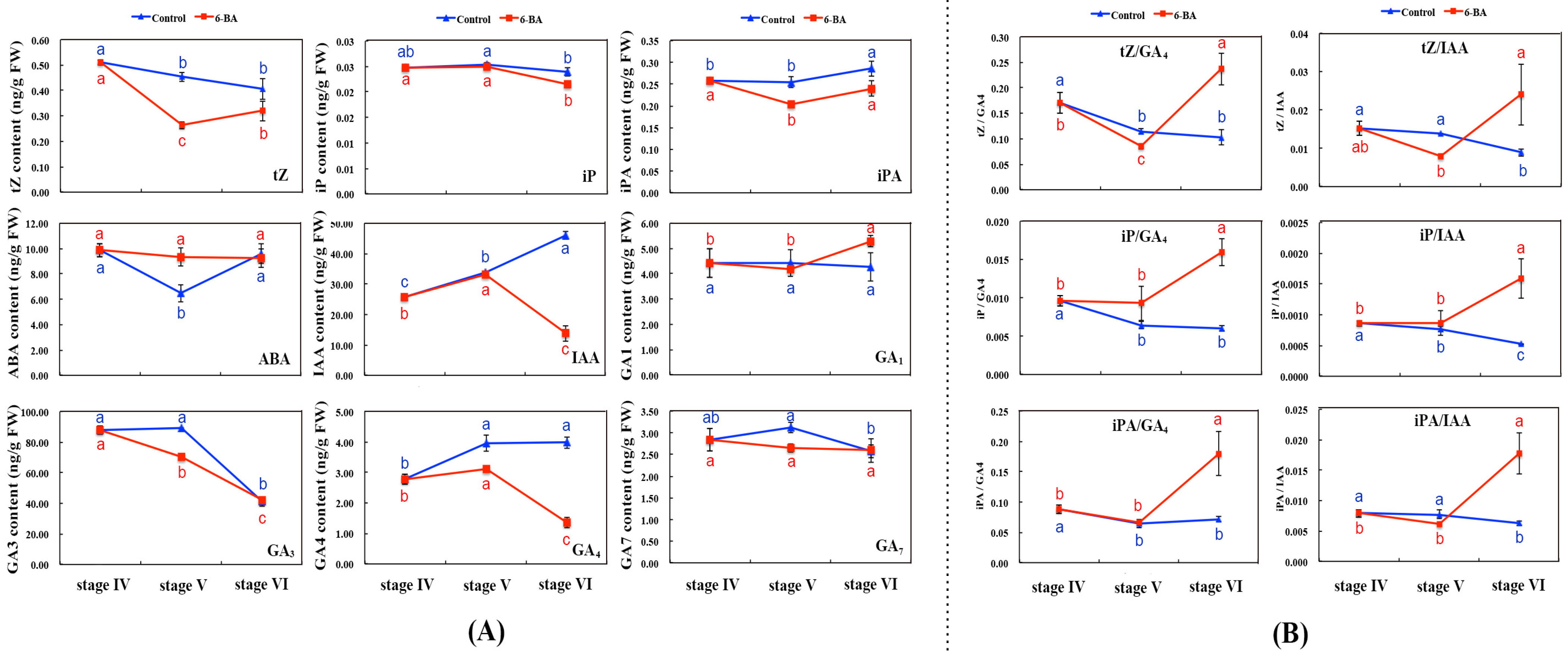
2.2. Determination of Endogenous Hormone Content in Axils of Aboveground Stem after 6-BA Application
2.3. Expression Patterns of LaKNOX1, LaKNOX2 and LaBEL1 Genes during Stem Bulblet Natural Formation under the Ground
2.4. Cloning the Full Length of One LaBELL Family Gene from L. ‘Aladdin’
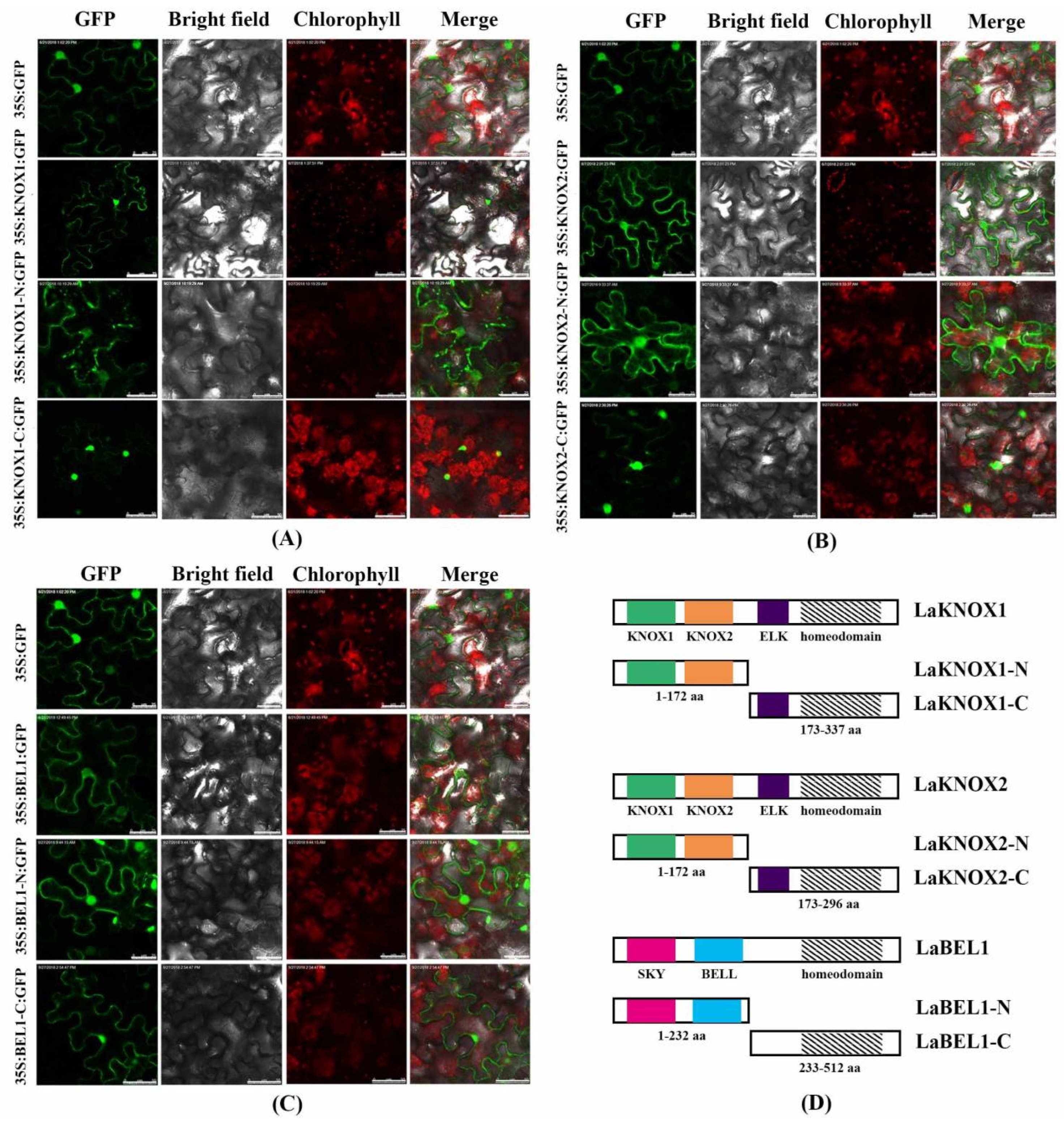
2.5. Subcellular Localization and Transactivation Assay of LaBEL1 and Two LaKNOX Proteins
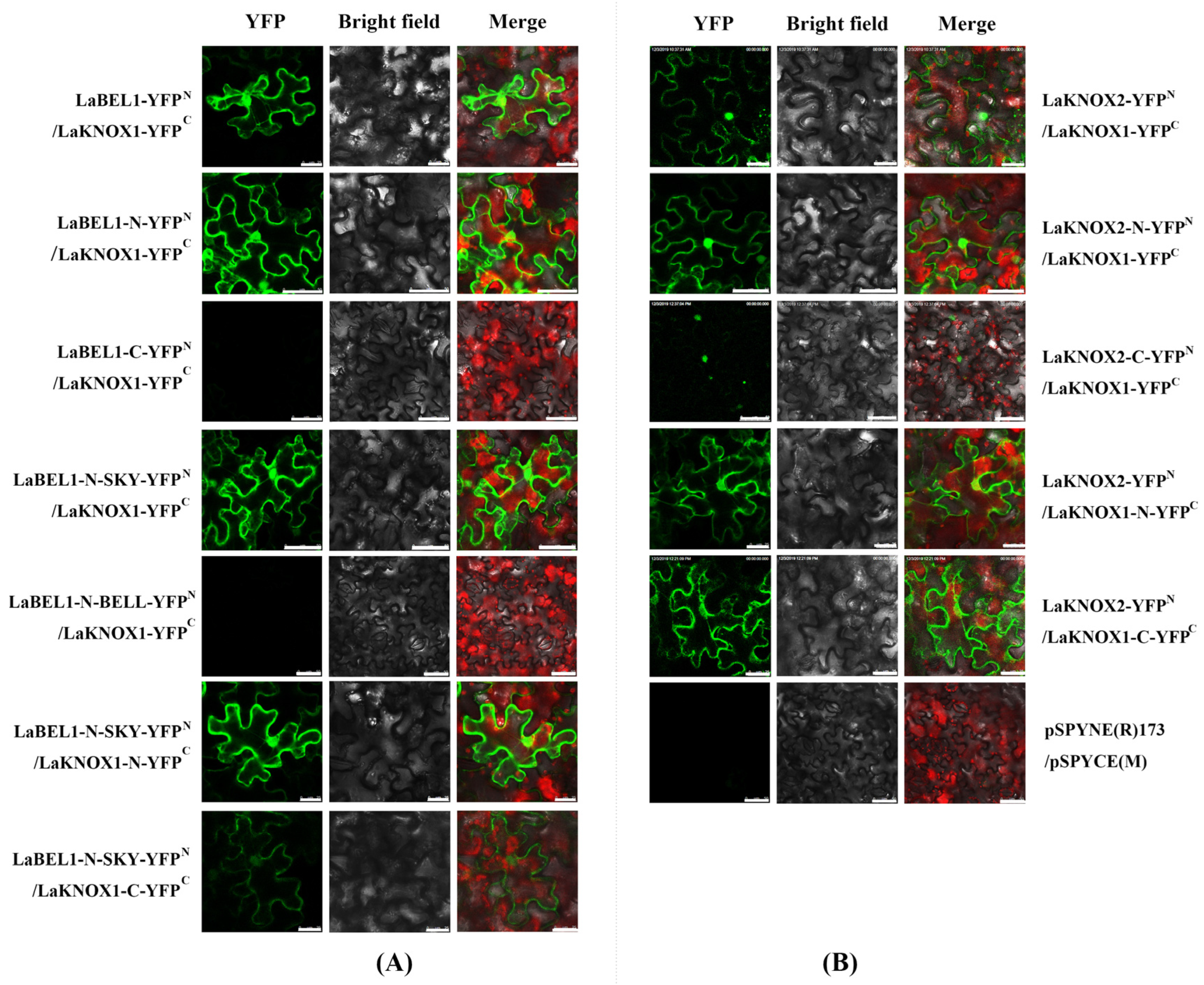
2.6. LaKNOX1 Interacts with LaBEL1 and LaKNOX2
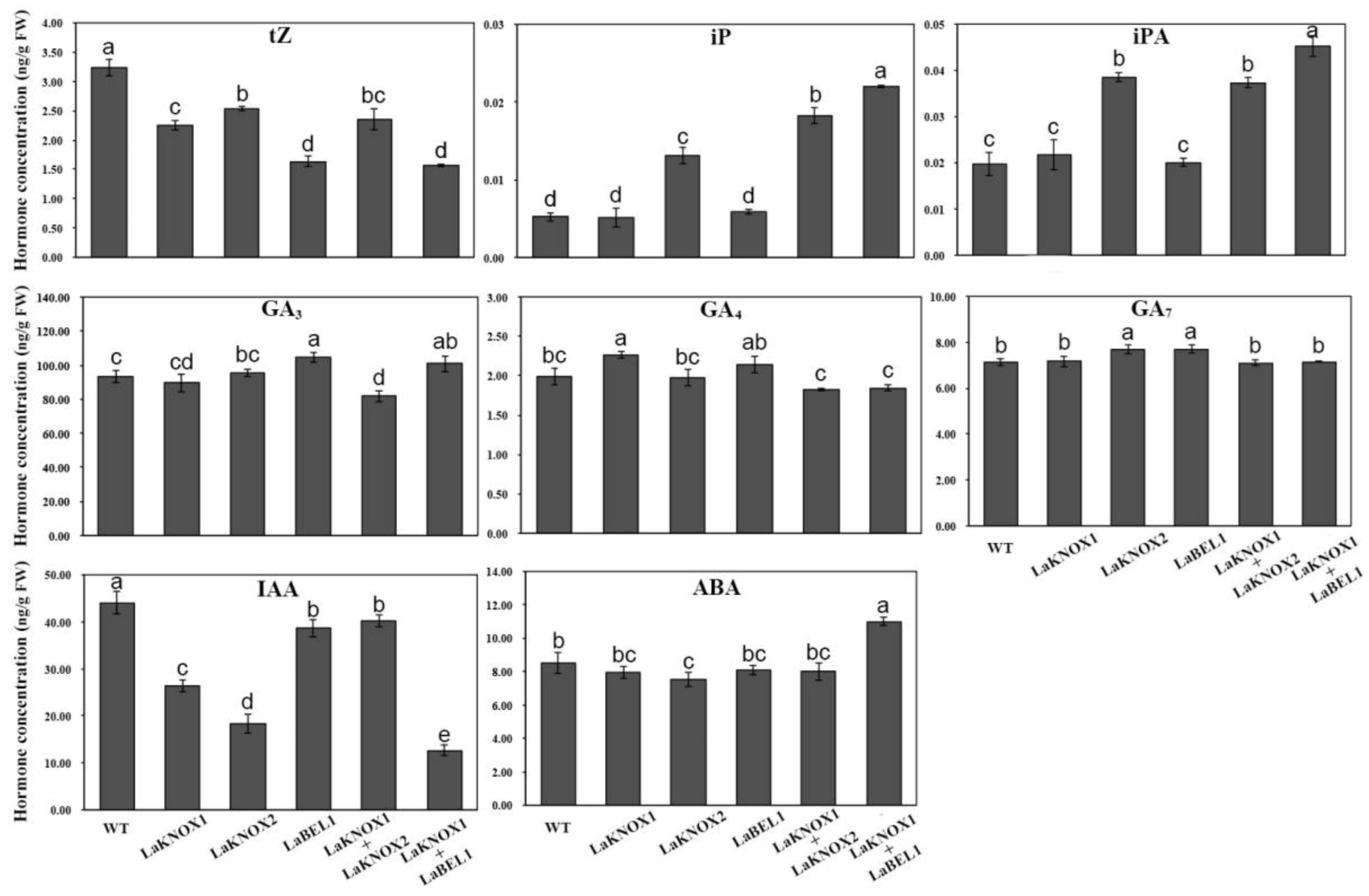
2.7. Altered Phytohormone Levels in Transgenic Arabidopsis Plants
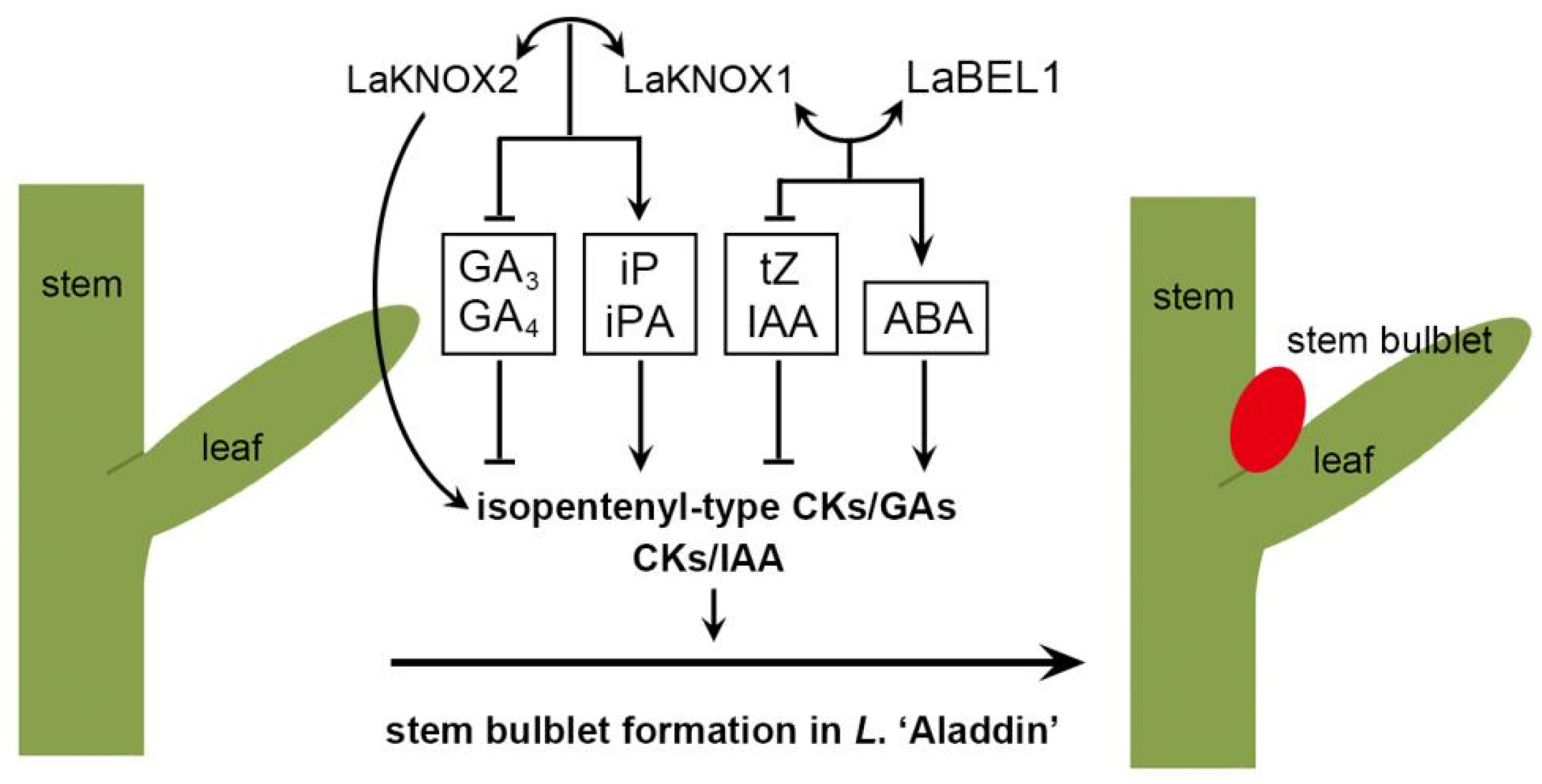
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Exogenous Hormone Treatment
4.2. Effects of Exogenous 6-BA Application on Under- and Above-Ground Stem Bulblet Formation in L. ‘Aladdin’
4.3. Quantitative Real-Time Reverse Transcription-PCR Analysis
4.4. Isolation and Sequencing of LaBEL1 from L. ‘Aladdin’
4.5. Subcellular Localization and Transactivation Assay
4.6. Bimolecular Fluorescence Complementation (BiFC) Analysis
4.7. Yeast Two-Hybrid (Y2H) Assay
4.8. Generation of Transgenic Arabidopsis
4.9. Phytohormone Content Measurement
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yang, P.; Xu, L.; Xu, H.; Tang, Y.; He, G.; Cao, Y.; Feng, Y.; Yuan, S.; Ming, J. Histological and Transcriptomic Analysis during Bulbil Formation in Lilium lancifolium. Front. Plant Sci. 2017, 8, 1508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, L. The Role of Auxin Signaling Pathway Related Genes in the Development of Bulbil for Two Species of Plant; Guizhou University: Guiyang, China, 2019. [Google Scholar]
- Zhang, Y.; Yong, Y.B.; Wang, Q.; Lu, Y.M. Physiological and Molecular Changes during Lily Underground Stem Axillary Bulbils Formation. Russ. J. Plant Physiol. 2018, 65, 372–383. [Google Scholar] [CrossRef]
- Wu, Y. The Mechanisms of Lily Bulblet Initiation and Development Based on the In Vitro Model System; Zhejiang University: Hangzhou, China, 2016. [Google Scholar]
- Borzenkova, R.A.; Sobyanina, E.A.; Pozdeeva, A.A.; Yashkov, M.Y. Effect of phytohormones on starch-synthesizing capacity in growing potato tubers. Russ. J. Plant Physiol. 1998, 45, 472–480. [Google Scholar]
- Wang, Q.; Zhang, L.; Wang, Z. Formation and Thickening of Tuberous Roots in Relation to the Endogenous Hormone Concentrations in Sweetpotato. Sci. Agric. Sin. 2005, 38, 2414–2420. [Google Scholar]
- Cheng, L.; Wang, Y.; Liu, Y.; Zhang, Q.; Gao, H.; Zhang, F. Comparative proteomics illustrates the molecular mechanism of potato (Solanum tuberosum L.) tuberization inhibited by exogenous gibberellins in vitro. Physiol. Plant. 2018, 163, 103–123. [Google Scholar] [CrossRef]
- Moreno-Pachón, N.M. Mechanisms of Vegetative Propagation in Bulbs: A Molecular Approach; Wageningen University: Wageningen, The Netherlands, 2017. [Google Scholar]
- Hao, J. Study on Morphogenesis and Physiological Mechanism of L. formolongi Bulblet; China Agricultural University: Beijing, China, 2004. [Google Scholar]
- Roodbarkelari, F.; Du, F.; Truernit, E.; Laux, T. ZLL/AGO10 maintains shoot meristem stem cells during Arabidopsis embryogenesis by down-regulating ARF2-mediated auxin response. BMC Biol. 2015, 13, 74. [Google Scholar] [CrossRef] [Green Version]
- Rosspopoff, O.; Chelysheva, L.; Saffar, J.; Lecorgne, L.; Gey, D.; Caillieux, E.; Colot, V.; Roudier, F.; Hilson, P.; Berthomé, R.; et al. Direct conversion of root primordium into shoot meristem relies on timing of stem cell niche development. Development 2017, 144, 1187–1200. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Li, T.; Li, Y. Physiological Mechanism of Metabolism of Carbohydrate, Phenols, Free Amino Acid and Endogenous Hormones in Middle Scales of Lilium davidii var. unicolor Bulbs Stored at Low Temperature for Dormancy Release. Sci. Agric. Sin. 2005, 38, 376–382. [Google Scholar]
- Jasinski, S.; Piazza, P.; Craft, J.; Hay, A.; Woolley, L.; Rieu, I.; Phillips, A.; Hedden, P.; Tsiantis, M. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 2005, 15, 1560–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, L. The Research of Developmental Mechanism during the Axillary Buds Regeneration for the In Vitro Culture of Lycoris chinensis; Nanjing Forestry University: Nanjing, China, 2012. [Google Scholar]
- Burglin, T.R. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997, 25, 4173–4180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerstetter, R.; Vollbrecht, E.; Lowe, B.; Veit, B.; Yamaguchi, J.; Hake, S. Sequence Analysis and Expression Patterns Divide the Maize knottedl-like Homeobox Genes into Two Classes. Plant Cell 1994, 6, 1877–1887. [Google Scholar]
- Sinha, N.R.; Williams, R.E.; Hake, S. Overexpression of the maize homeobox gene, KNOTTED-I, causes a switch from determinate to indeterminate cell fates. Genes Dev. 1993, 7, 787–795. [Google Scholar] [CrossRef] [Green Version]
- Clark, S.E.; Jacobsen, S.E.; Levin, J.Z.; Meyerowitz, E.M. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 1996, 122, 1567–1575. [Google Scholar] [CrossRef] [PubMed]
- Kerstetter, R.A.; Laudencia-Chingcuanco, D.; Smith, L.G.; Hake, S. Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 1997, 124, 3045–3054. [Google Scholar] [CrossRef] [PubMed]
- Chan, R.L.; Gago, G.M.; Palena, C.M.; Gonzalez, D.H. Homeoboxes in plant development. Biochim. et Biophys. Acta 1998, 1442, 1–19. [Google Scholar] [CrossRef]
- Robinson-Beers, K.; Pruitt, R.E.; Gasser, C.S. Ovule Development in Wild-Type Arabidopsis Two Female-Sterile Mutants. Plant Cell 1992, 4, 1237–1249. [Google Scholar] [CrossRef]
- Modrusan, Z.; Reiser, L.; Feldmann, K.A.; Fischer, R.L.; Haughnai, G.W. Homeotic Transformation of Ovules into Carpel-like Structures in Arabidopsis. Plant Cell 1994, 6, 333–349. [Google Scholar] [CrossRef]
- Rosin, F.M.; Hart, J.K.; Horner, H.T.; Davies, P.J.; Hannapel, D.J. Overexpression of a knotted-like homeobox gene of potato alters vegetative development by decreasing gibberellin accumulation. Plant Physiol. 2003, 132, 106–117. [Google Scholar] [PubMed] [Green Version]
- Abraham-Juarez, M.J.; Martinez-Hernandez, A.; Leyva-Gonzalez, M.A.; Herrera-Estrella, L.; Simpson, J. Class I KNOX genes are associated with organogenesis during bulbil formation in Agave tequilana. J. Exp. Bot. 2010, 61, 4055–4067. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Lin, T.; Hannapel, D.J. Targets of the StBEL5 Transcription Factor Include the FT Ortholog StSP6A. Plant Physiol. 2016, 170, 310–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, F.; Hu, G.; Ren, Z.; Deng, W.; Li, Z. Ectopic expression a tomato KNOX Gene Tkn4 affects the formation and the differentiation of meristems and vasculature. Plant Mol. Biol. 2015, 89, 589–605. [Google Scholar] [CrossRef]
- Yanai, O.; Shani, E.; Dolezal, K.; Tarkowski, P.; Sablowski, R.; Sandberg, G.; Samach, A.; Ori, N. Arabidopsis KNOXI Proteins Activate Cytokinin Biosynthesis. Curr. Biol. 2005, 15, 1566–1571. [Google Scholar] [CrossRef] [Green Version]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef]
- Sakamoto, T.; Kamiya, N.; Ueguchi-Tanaka, M.; Iwahori, S.; Matsuoka, M. KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 2001, 15, 581–590. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Rosin, F.M.; Prat, S.; Hannapel, D.J. Interacting transcription factors from the three-amino acid loop extension superclass regulate tuber formation. Plant Physiol. 2003, 132, 1391–1404. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Banerjee, A.K.; Hannapel, D.J. The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J. 2004, 38, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, N.; Hake, S. The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 2009, 21, 1647–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolduc, N.; Yilmaz, A.; Mejia-Guerra, M.K.; Morohashi, K.; O’Connor, D.; Grotewold, E.; Hake, S. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 2012, 26, 1685–1690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hay, A.; Tsiantis, M. KNOX genes: Versatile regulators of plant development and diversity. Development 2010, 137, 3153–3165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Cho, Y.-H.; Ryu, H.; Kim, Y.; Kim, T.-H.; Hwang, I. BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J. 2013, 75, 755–766. [Google Scholar] [CrossRef]
- Bellaoui, M.; Pidkowich, M.S.; Samach, A.; Kushalappa, K.; Kohalmi, S.E.; Modrusan, Z.; Crosby, W.L.; Haughn, G.W. The Arabidopsis BELL1 and KNOX TALE Homeodomain Proteins Interact through a Domain Conserved between Plants and Animals. Plant Cell 2001, 13, 2455–2470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiser, L.; Modrusan, Z.; Margossian, L.; Samach, A.; Ohad, N.; Haughn, G.W.; Fischer, R.L. The BELL1 Gene Encodes a Homeodomain Protein Involved in Pattern Formation in the Arabidopsis Ovule Primordium. Cell 1995, 83, 735–742. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Awasthi, V.; Kanwar, J.K. Influence of growth regulators and nitrogenous compounds on in vitro bulblet formation and growth in oriental lily. HortScience 2007, 34, 77–83. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Zhao, J.; An, X.; Jin, X. Effect of plant growth regulators on scale cutting propagation of Lilium davidii var. unicolor. North. Hortic. 2014, 20, 68–71. [Google Scholar]
- Di Giacomo, E.; Iannelli, M.A.; Frugis, G. TALE and Shape: How to Make a Leaf Different. Plants 2013, 2, 317–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brambilla, V.; Battaglia, R.; Colombo, M.; Masiero, S.; Bencivenga, S.; Kater, M.M.; Colombo, L. Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell 2007, 19, 2544–2556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huh, W.-K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef]
- Pesch, M.; Schultheiss, I.; Digiuni, S.; Uhrig, J.F.; Hulskamp, M. Mutual control of intracellular localisation of the patterning proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development 2013, 140, 3456–3467. [Google Scholar] [CrossRef] [Green Version]
- Ding, K.; Ma, P.; Jia, Y.; Pei, T.; Bai, Z.; Liang, Z. Subcellular localization and transactivation analysis of three R2R3-MYB in Salvia miltiorrhiza Bunge. Acta Agric. Boreali Occident. Sin. 2017, 27, 586–594. [Google Scholar]
- Muller, J.; Wang, Y.; Franzen, R.; Santi, L.; Salamini, F.; Rohde, W. In Vitro interactions between barley TALE homeodomain proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J. 2001, 27, 13–23. [Google Scholar] [CrossRef]
- Cole, M.; Nolte, C.; Werr, W. Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res. 2006, 34, 1281–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meisel, L.; Lam, E. The conserved ELK-homeodomain of KNOTTED-I contains two regions that signal nuclear localization. Plant Mol. Biol. 1996, 30, 1–14. [Google Scholar] [CrossRef]
- Smith, H.M.S.; Boschke, I.; Hake, S. Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. USA 2002, 99, 9579–9584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, Y.; Chen, M.; Shu, Y.; Zhu, Y.; Wang, S.; Huang, L.; Yu, X.; Wang, Z.; Qian, P.; Gu, W.; et al. Identification and functional characterization of a novel BEL1-LIKE homeobox transcription factor GmBLH4 in soybean. Plant Cell Tissue Organ Cult. PCTOC 2018, 134, 331–344. [Google Scholar] [CrossRef]
- Nhut, D.T. Micropropagation of lily (Lilium longiflorum) via in vitro stem node and pseudo-bulblet culture. Plant Cell Rep. 1998, 17, 913–916. [Google Scholar] [CrossRef]
- Li, E.; Bhargava, A.; Qiang, W.; Friedmann, M.C.; Forneris, N.; Savidge, R.A.; Douglas, C.J.; Johnson, L.A.; Mansfield, S.D.; Ellis, B.E. The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol. 2012, 194, 102–115. [Google Scholar] [CrossRef]
- Liu, Y.; You, S.; Taylor-Teeples, M.; Li, W.L.; Schuetz, M.; Brady, S.M.; Douglas, C.J. BEL1-LIKE HOMEODOMAIN6 and KNOTTED Arabidopsis THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell 2014, 26, 4843–4861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frugis, G.; Giannino, D.; Mele, G.; Nicolodi, C.; Chiappetta, A.; Bitonti, M.B.; Innocenti, A.M.; Dewitte, W.; van Onckelen, H.; Mariotti, D. Overexpression of KNAT1 in Lettuce Shifts Leaf Determinate Growth to a Shoot-Like Indeterminate Growth Associated with an Accumulation of Isopentenyl-Type Cytokinins. Plant Physiol. 2001, 126, 1370–1380. [Google Scholar] [CrossRef] [Green Version]
- Marcel, L.N.; Christine, D.; Gwenaëlle, W.; Noëlle, D.; Claude, B.; Casenave, M.; Leboeuf, J. Observations sur la multiplication in vitro de la tulipe (Tulipa gesneriaha L.) à partir de hampes florales prélevées chez des bulbes en cours de conservation. Agronomie 1987, 7, 321–329. [Google Scholar]
- Podwyszyńska, M.; Novák, O.; Doležal, K.; Strnad, M. Endogenous cytokinin dynamics in micropropagated tulips during bulb formation process influenced by TDZ and iP pretreatment. Plant Cell Tiss Organ Cult. 2014, 119, 331–346. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Li, T.; Li, Y. Changes of carbohydrate and amylase in lily bulb during bulb development. Bull. Bot. Res. 2005, 25, 59–63. [Google Scholar] [CrossRef] [Green Version]
- Podwyszynska, M. The Mechanisms of in vitro Storage Organ Formation in Ornamental Geophytes. Floric. Ornam. Biotechnol. 2012, 6, 9–23. [Google Scholar]
- Shoub, J.; de Hertogh, A.A. Effects of ancymidol and gibberellins A3 and A4+7 on Tulipa Gesneriana, L. cv. Paul Richter during development in the greenhouse. Sci. Hortic. 1974, 2, 55–67. [Google Scholar] [CrossRef]
- Ranwala, A.P.; Miller, W.B. Ancymidol Drenches, Reversed Greenhouse Temperatures, Postgreenhouse Cold Storage, and Hormone Sprays Affect Postharvest Leaf Chlorosis in Easter Lily. J. Amer. Soc. Hort. Sci. 2000, 125, 248–253. [Google Scholar] [CrossRef] [Green Version]
- Ranwala, A.P.; Miller, W.B. Gibberellin-mediated changes in carbohydrate metabolism during flower stalk elongation in tulips. Plant Growth Regul. 2008, 55, 241–248. [Google Scholar] [CrossRef]
- Liu, Q.; Wei, C.; Zhang, M.F.; Jia, G.X. Evaluation of putative reference genes for quantitative real-time PCR normalization in Lilium regale during development and under stress. PeerJ 2016, 4, e1837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Yong, Y.; Zhang, Y.; Lyu, Y. Transcriptional Regulatory Network of GA Floral Induction Pathway in LA Hybrid Lily. Int. J. Mol. Sci. 2019, 20, 2694. [Google Scholar] [CrossRef] [Green Version]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef] [Green Version]
- Sheludko, Y.V.; Sindarovska, Y.R.; Gerasymenko, I.M.; Bannikova, M.A.; Kuchuk, N.V. Comparison of several Nicotiana species as hosts for high-scale Agrobacterium-mediated transient expression. Biotechnol. Bioeng. 2007, 96, 608–614. [Google Scholar] [CrossRef]
- Bent, A. Arabidopsis thaliana Floral Dip Transformation Method. Agrobacterium Protoc. 2006, 343, 87–104. [Google Scholar]
- Pan, X.Q.; Welti, W.X.M. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 986–992. [Google Scholar] [CrossRef] [PubMed]



| Diameter, cm | 6-BA | Control | ||||
|---|---|---|---|---|---|---|
| Number | Average Weight 1, g | Percentage % | Number | Average Weight 1, g | Percentage % | |
| >1.8 | 6 | 1.85 ± 0.31 a | 3.75 | 1 | 2.56 a | 1.59 |
| 1.6–1.8 | 13 | 1.41 ± 0.21 b | 8.12 | 12 | 1.34 ± 0.22 b | 19.05 |
| 1.4–1.6 | 35 | 1.00 ± 0.15 c | 21.88 | 13 | 0.97 ± 0.14 c | 20.63 |
| 1.2–1.4 | 44 | 0.68 ± 0.09 d | 27.50 | 14 | 0.81 ± 0.11 cd | 22.22 |
| 1.0–1.2 | 16 | 0.52 ± 0.12 de | 10.00 | 9 | 0.59 ± 0.02 de | 14.29 |
| 0.8–1.0 | 18 | 0.36 ± 0.12 ef | 11.25 | 9 | 0.34 ± 0.01 ef | 14.29 |
| <0.8 | 28 | 0.17 ± 0.01 f | 17.50 | 5 | 0.17 ± 0.01 f | 7.93 |
| Diameter, cm | 6-BA | ||
|---|---|---|---|
| Number | Average Weight 1, g | Percentage, % | |
| >1.8 | 0 | 0 | 0 |
| 1.6–1.8 | 4 | 1.25 ± 0.18 a | 1.07 |
| 1.4–1.6 | 6 | 0.86 ± 0.12 b | 1.61 |
| 1.2–1.4 | 4 | 0.54 ± 0.08 c | 1.07 |
| 1.0–1.2 | 40 | 0.31 ± 0.02 d | 10.72 |
| 0.8–1.0 | 43 | 0.21 ± 0.03 de | 11.53 |
| <0.8 | 276 | 0.08 ± 0.01 f | 74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zeng, Z.; Yong, Y.; Lyu, Y. Hormonal Regulatory Patterns of LaKNOXs and LaBEL1 Transcription Factors Reveal Their Potential Role in Stem Bulblet Formation in LA Hybrid Lily. Int. J. Mol. Sci. 2021, 22, 13502. https://doi.org/10.3390/ijms222413502
Zhang Y, Zeng Z, Yong Y, Lyu Y. Hormonal Regulatory Patterns of LaKNOXs and LaBEL1 Transcription Factors Reveal Their Potential Role in Stem Bulblet Formation in LA Hybrid Lily. International Journal of Molecular Sciences. 2021; 22(24):13502. https://doi.org/10.3390/ijms222413502
Chicago/Turabian StyleZhang, Yue, Zhen Zeng, Yubing Yong, and Yingmin Lyu. 2021. "Hormonal Regulatory Patterns of LaKNOXs and LaBEL1 Transcription Factors Reveal Their Potential Role in Stem Bulblet Formation in LA Hybrid Lily" International Journal of Molecular Sciences 22, no. 24: 13502. https://doi.org/10.3390/ijms222413502





