Multi-Omics Reveal the Immunological Role and the Theragnostic Value of miR-216a/GDF15 Axis in Human Colon Adenocarcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Multi-Omic Analysis
2.2. In Vitro Cell Experiment
2.3. Immunological Databases
2.4. Construction of PPI Network and Enriched Analysis
2.5. Statistical Analysis
3. Results
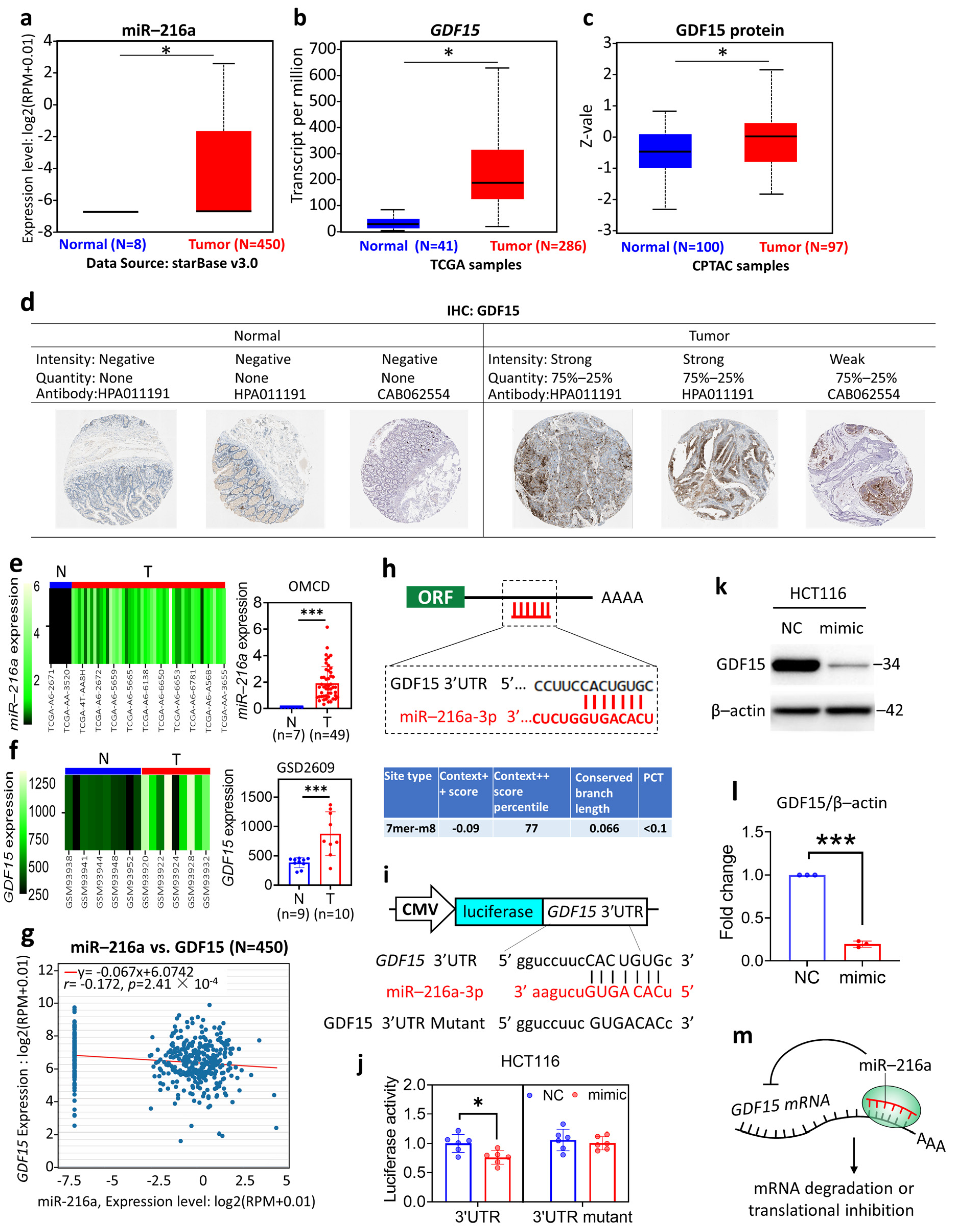
3.1. Differential Expression Profile and the Molecular Interaction of miR-216a and GDF15
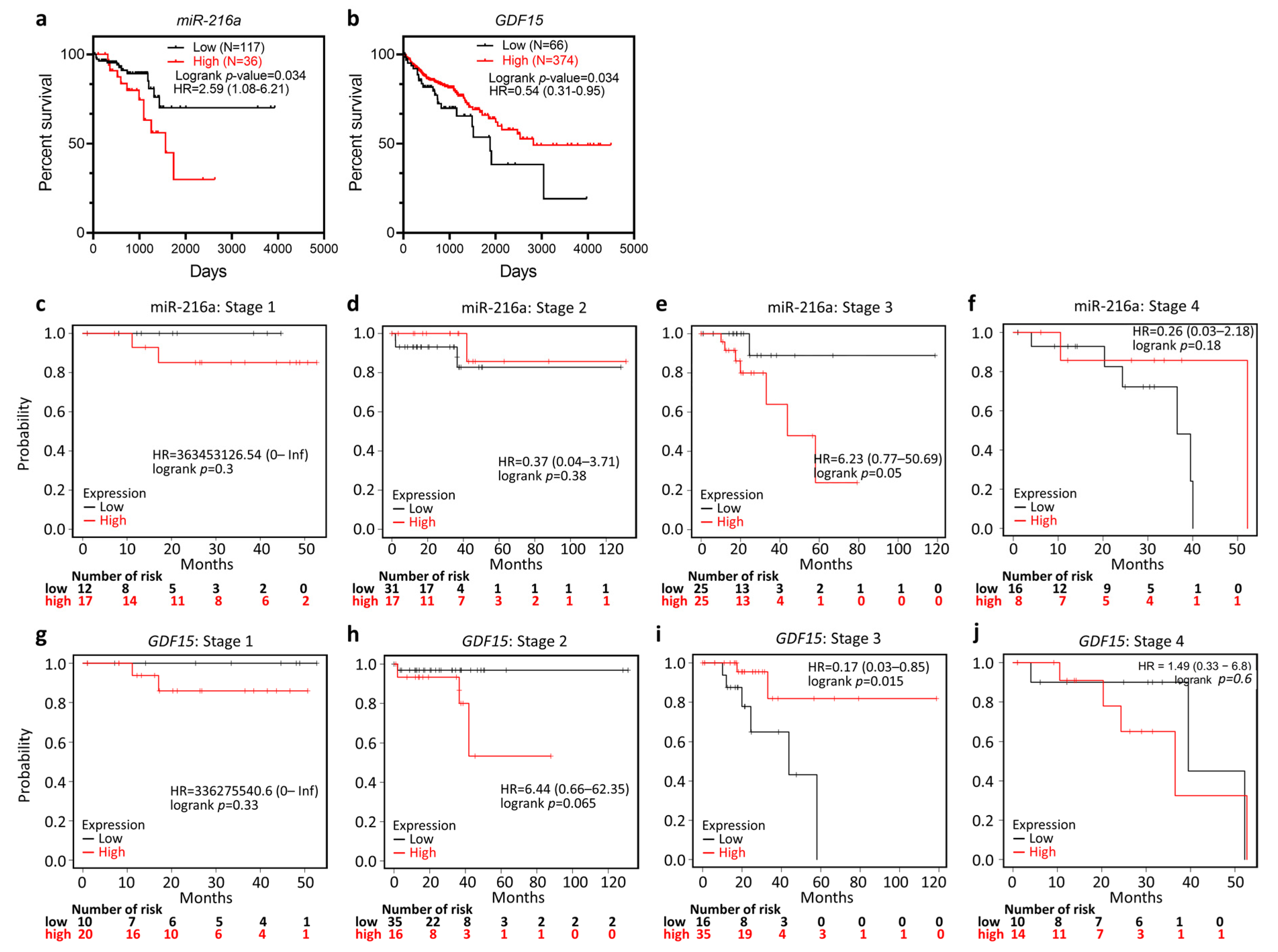
3.2. Prognostic Value of miR-216a and GDF15
3.3. Prognostic Value of miR-216a and GDF15 in the Context of Different Immune Cell Contents
3.4. Mutation Landscape of miR-216a and GDF15
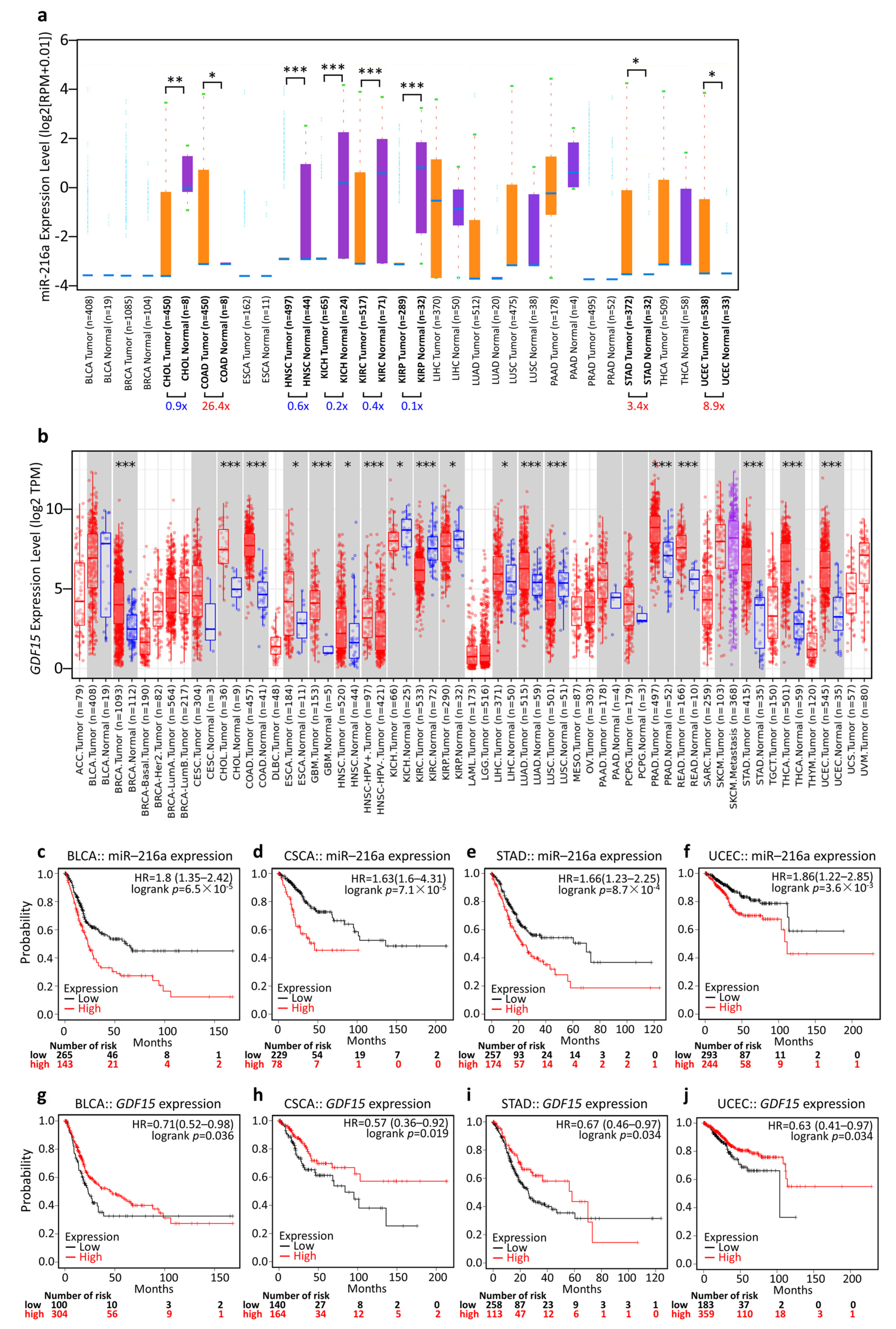
3.5. Generalization Value of the miR-216a/GDF15 Axis in Pan-Cancer
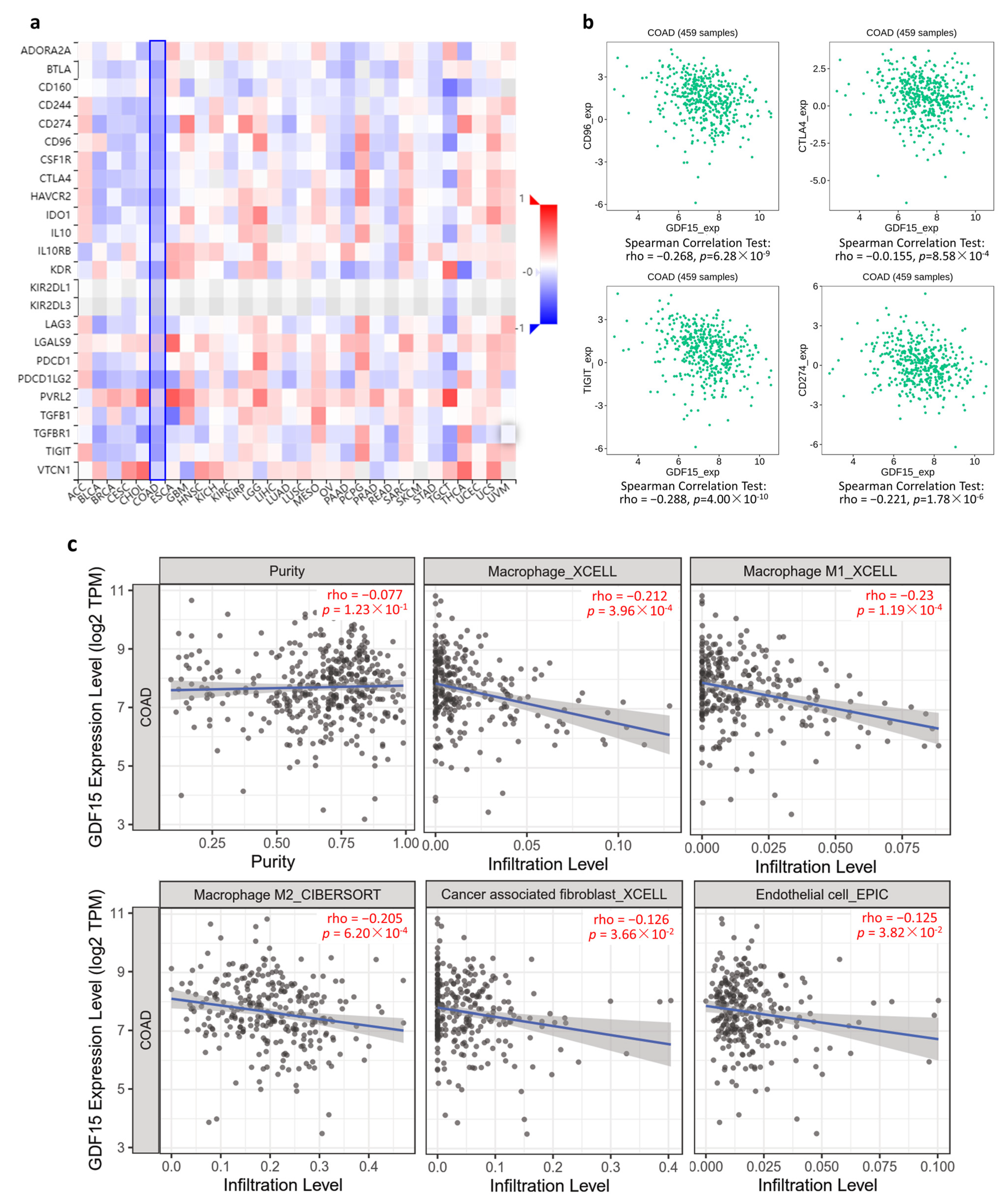
3.6. GDF15 Negatively Correlated with Immunoinhibitory Genes and Immune Infiltration
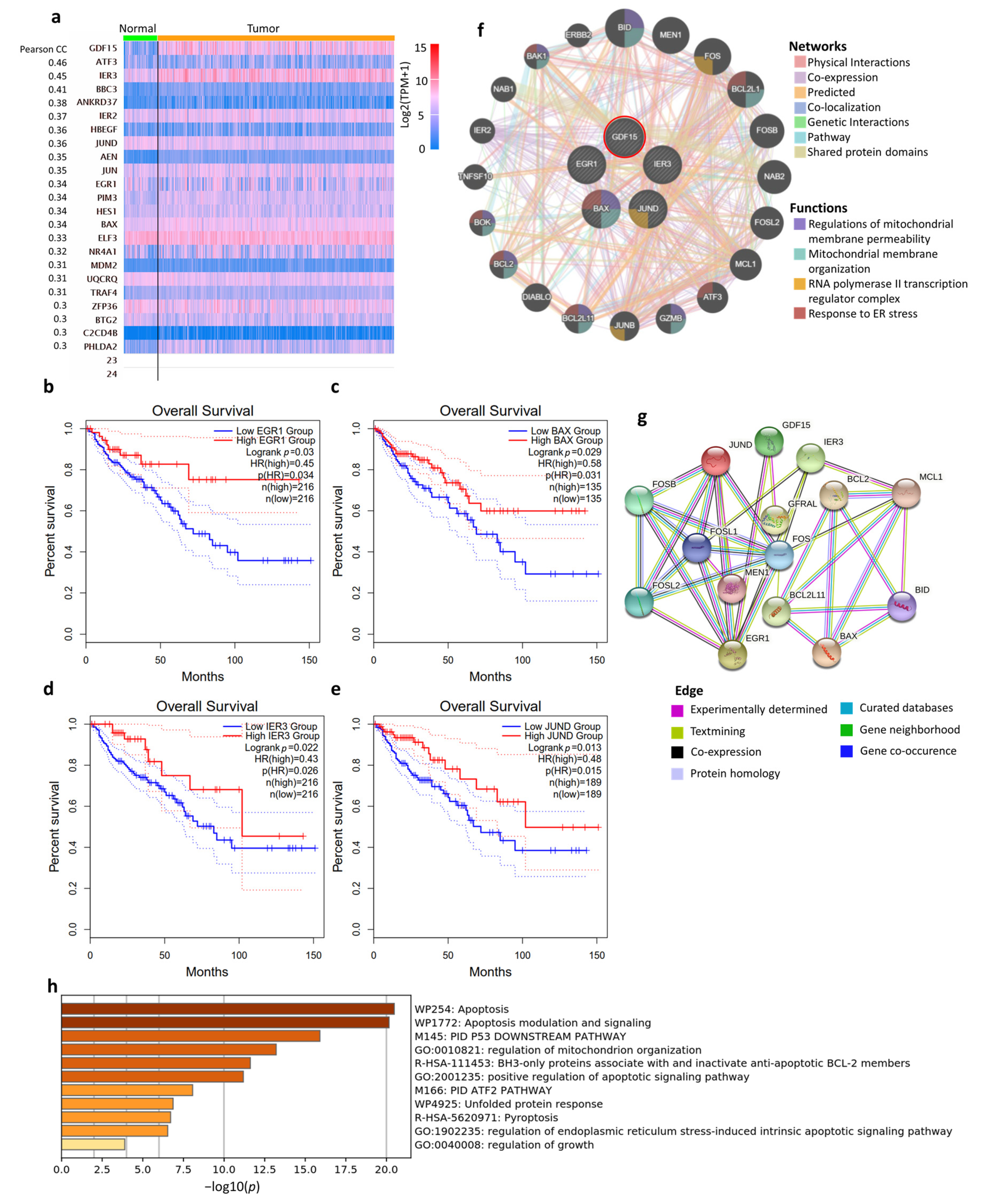
3.7. Co-Expression Network and Biological Functions of GDF15
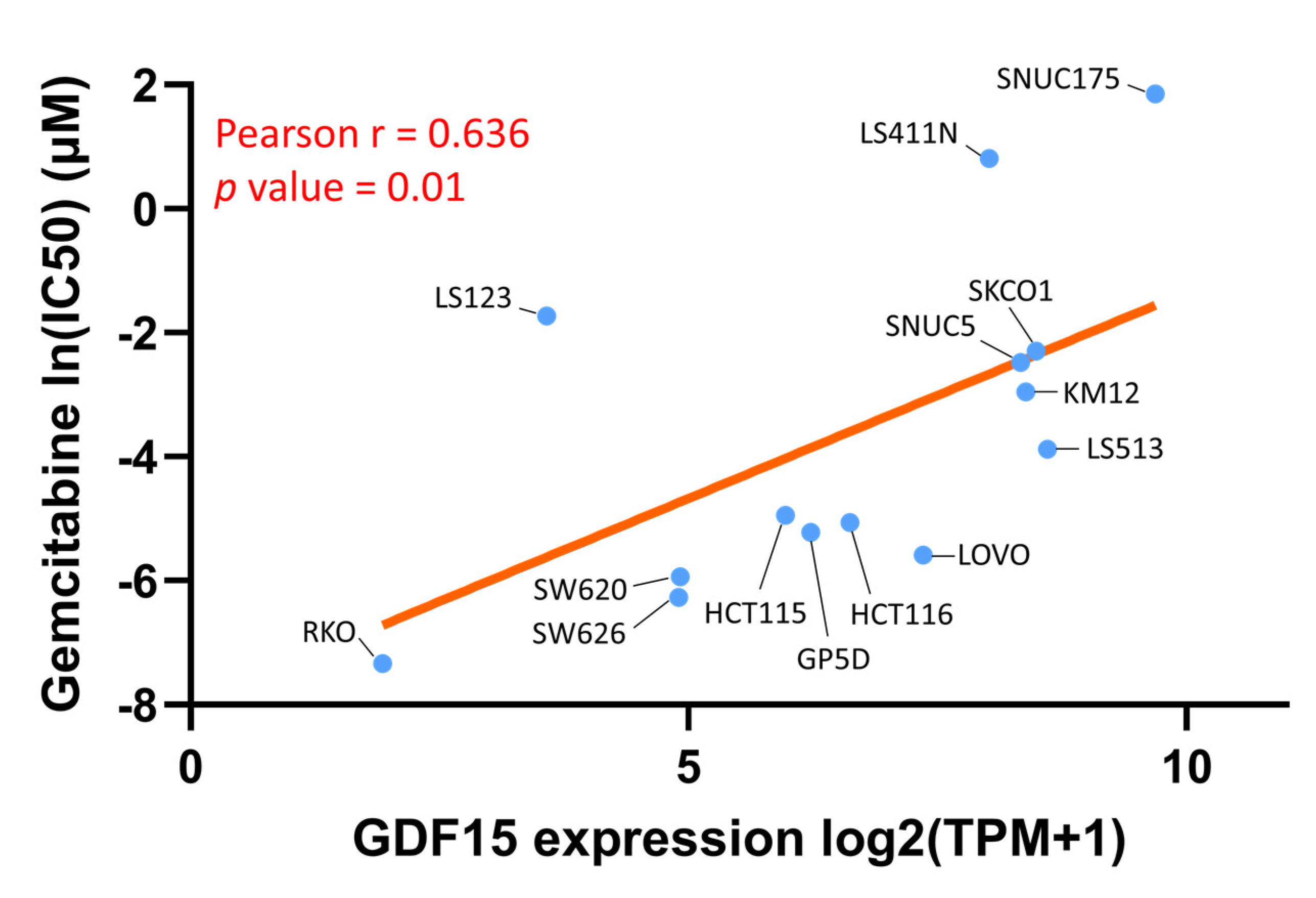
3.8. Gemcitabine as a Therapeutic Option in the Context of Low GDF15
4. Discussion
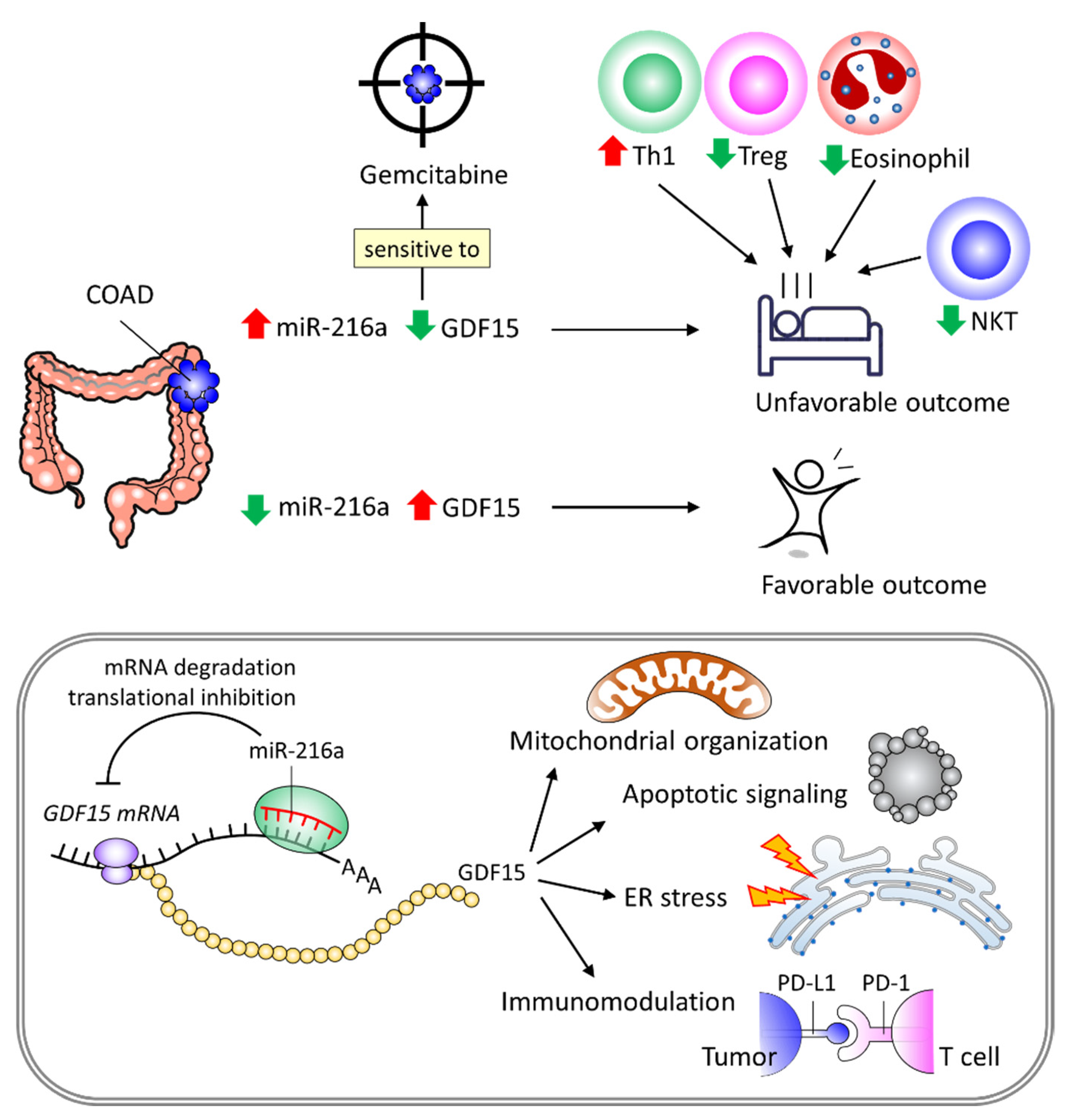
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, M.; Zhou, Y.; Yang, C.; Wei, S.; Bian, X.; Christopher, O.; Xie, L. The Prognostic and Clinicopathological Roles of PD-L1 Expression in Colorectal Cancer: A Systematic Review and Meta-Analysis. Front. Pharmacol. 2019, 10, 139. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klaus, S.; Igual Gil, C.; Ost, M. Regulation of diurnal energy balance by mitokines. Cell. Mol. Life Sci. 2021, 78, 3369–3384. [Google Scholar] [CrossRef]
- Baek, S.J.; Eling, T. Growth differentiation factor 15 (GDF15): A survival protein with therapeutic potential in metabolic diseases. Pharmacol. Ther. 2019, 198, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Emmerson, P.J.; Wang, F.; Du, Y.; Liu, Q.; Pickard, R.T.; Gonciarz, M.D.; Coskun, T.; Hamang, M.J.; Sindelar, D.K.; Ballman, K.K.; et al. The metabolic effects of GDF15 are mediated by the orphan receptor GFRAL. Nat. Med. 2017, 23, 1215–1219. [Google Scholar] [CrossRef]
- Emmerson, P.J.; Duffin, K.L.; Chintharlapalli, S.; Wu, X. GDF15 and Growth Control. Front. Physiol. 2018, 9, 1712. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Jiang, M.; Nouraie, M.; Roth, M.G.; Tabib, T.; Winters, S.; Chen, X.; Sembrat, J.; Chu, Y.; Cardenes, N.; et al. GDF15 is an epithelial-derived biomarker of idiopathic pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2019, 317, L510–L521. [Google Scholar] [CrossRef]
- Li, C.; Wang, X.; Casal, I.; Wang, J.; Li, P.; Zhang, W.; Xu, E.; Lai, M.; Zhang, H. Growth differentiation factor 15 is a promising diagnostic and prognostic biomarker in colorectal cancer. J. Cell. Mol. Med. 2016, 20, 1420–1426. [Google Scholar] [CrossRef]
- Wallin, U.; Glimelius, B.; Jirstrom, K.; Darmanis, S.; Nong, R.Y.; Ponten, F.; Johansson, C.; Pahlman, L.; Birgisson, H. Growth differentiation factor 15: A prognostic marker for recurrence in colorectal cancer. Br. J. Cancer 2011, 104, 1619–1627. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Yu, S.; Zhu, C.; Guo, T.; Liu, F.; Xu, Y. HIF1alpha promotes tumor chemoresistance via recruiting GDF15-producing TAMs in colorectal cancer. Exp. Cell Res. 2021, 398, 112394. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Guo, C.; Fang, L.; Sang, T.; Wang, Y.; Wu, K.; Guo, C.; Wang, Y.; Pan, H.; Chen, R.; et al. Qizhen capsule inhibits colorectal cancer by inducing NAG-1/GDF15 expression that mediated via MAPK/ERK activation. J. EthnoPharmacol. 2021, 273, 113964. [Google Scholar] [CrossRef]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Xiao, M.; Li, J.; Li, W.; Wang, Y.; Wu, F.; Xi, Y.; Zhang, L.; Ding, C.; Luo, H.; Li, Y.; et al. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017, 14, 1326–1334. [Google Scholar] [CrossRef]
- Lee, I.; Ajay, S.S.; Yook, J.I.; Kim, H.S.; Hong, S.H.; Kim, N.H.; Dhanasekaran, S.M.; Chinnaiyan, A.M.; Athey, B.D. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009, 19, 1175–1183. [Google Scholar] [CrossRef] [Green Version]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Croce, C.M. A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol. 2013, 10, 169–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Bedja, D.; Campbell, N.; Dunkerly, B.; Chenna, V.; Maitra, A.; Steenbergen, C. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS ONE 2014, 9, e96820. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Li, Y.; Zhang, C.; Li, X.; Yu, J. MiR-216a-3p inhibits colorectal cancer cell proliferation through direct targeting COX-2 and ALOX5. J. Cell. Biochem. 2018, 119, 1755–1766. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S.; Dang, S.; Fang, X.; Liu, M. Overexpression of microRNA-216a inhibits autophagy by targeting regulated MAP1S in colorectal cancer. Onco Targets Ther. 2019, 12, 4621–4629. [Google Scholar] [CrossRef] [Green Version]
- Zeng, X.; Liu, Y.; Zhu, H.; Chen, D.; Hu, W. Downregulation of miR-216a-5p by long noncoding RNA PVT1 suppresses colorectal cancer progression via modulation of YBX1 expression. Cancer Manag Res. 2019, 11, 6981–6993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [Green Version]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thul, P.J.; Akesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Bjork, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, 806. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Sarver, A.L.; Sarver, A.E.; Yuan, C.; Subramanian, S. OMCD: OncomiR Cancer Database. BMC Cancer 2018, 18, 1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagy, A.; Munkacsy, G.; Gyorffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Jeong, E.; Lee, Y.; Kim, Y.; Lee, J.; Yoon, S. Analysis of Cross-Association between mRNA Expression and RNAi Efficacy for Predictive Target Discovery in Colon Cancers. Cancers 2020, 12, 3091. [Google Scholar] [CrossRef]
- Liu, C.J.; Hu, F.F.; Xia, M.X.; Han, L.; Zhang, Q.; Guo, A.Y. GSCALite: A web server for gene set cancer analysis. Bioinformatics 2018, 34, 3771–3772. [Google Scholar] [CrossRef]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.S.W.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef]
- Zuberi, K.; Franz, M.; Rodriguez, H.; Montojo, J.; Lopes, C.T.; Bader, G.D.; Morris, Q. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013, 41, W115–W122. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.A.; Lindmark, F.; Stattin, P.; Balter, K.; Adami, H.O.; Zheng, S.L.; Xu, J.; Isaacs, W.B.; Gronberg, H.; Breit, S.N.; et al. Macrophage inhibitory cytokine 1: A new prognostic marker in prostate cancer. Clin. Cancer Res. 2009, 15, 6658–6664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koopmann, J.; Buckhaults, P.; Brown, D.A.; Zahurak, M.L.; Sato, N.; Fukushima, N.; Sokoll, L.J.; Chan, D.W.; Yeo, C.J.; Hruban, R.H.; et al. Serum macrophage inhibitory cytokine 1 as a marker of pancreatic and other periampullary cancers. Clin. Cancer Res. 2004, 10, 2386–2392. [Google Scholar] [CrossRef] [Green Version]
- Welsh, J.B.; Sapinoso, L.M.; Kern, S.G.; Brown, D.A.; Liu, T.; Bauskin, A.R.; Ward, R.L.; Hawkins, N.J.; Quinn, D.I.; Russell, P.J.; et al. Large-scale delineation of secreted protein biomarkers overexpressed in cancer tissue and serum. Proc. Natl. Acad. Sci. USA 2003, 100, 3410–3415. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.A.; Ward, R.L.; Buckhaults, P.; Liu, T.; Romans, K.E.; Hawkins, N.J.; Bauskin, A.R.; Kinzler, K.W.; Vogelstein, B.; Breit, S.N. MIC-1 serum level and genotype: Associations with progress and prognosis of colorectal carcinoma. Clin. Cancer Res. 2003, 9, 2642–2650. [Google Scholar]
- Danta, M.; Barber, D.A.; Zhang, H.P.; Lee-Ng, M.; Baumgart, S.W.L.; Tsai, V.W.W.; Husaini, Y.; Saxena, M.; Marquis, C.P.; Errington, W.; et al. Macrophage inhibitory cytokine-1/growth differentiation factor-15 as a predictor of colonic neoplasia. Aliment. Pharmacol. Ther. 2017, 46, 347–354. [Google Scholar] [CrossRef] [Green Version]
- Klaus, S.; Ost, M. Mitochondrial uncoupling and longevity—A role for mitokines? Exp. Gerontol. 2020, 130, 110796. [Google Scholar] [CrossRef]
- Kang, Y.E.; Kim, J.M.; Lim, M.A.; Lee, S.E.; Yi, S.; Kim, J.T.; Oh, C.; Liu, L.; Jin, Y.; Jung, S.N.; et al. Growth Differentiation Factor 15 Is a Cancer Cell-Induced Mitokine That Primes Thyroid Cancer Cells for Invasiveness. Thyroid 2021, 31, 772–786. [Google Scholar] [CrossRef]
- Guo, Y.; Ayers, J.L.; Carter, K.T.; Wang, T.; Maden, S.K.; Edmond, D.; Newcomb, P.P.; Li, C.; Ulrich, C.; Yu, M.; et al. Senescence-associated tissue microenvironment promotes colon cancer formation through the secretory factor GDF15. Aging Cell 2019, 18, e13013. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.Y.; Kim, J.H.; Lee, J.H.; Lim, Y.; Lee, Y.H. 2′-Hydroxyflavanone induces apoptosis through Egr-1 involving expression of Bax, p21, and NAG-1 in colon cancer cells. Mol. Nutr. Food Res. 2012, 56, 761–774. [Google Scholar] [CrossRef]
- Wei, H.T.; Guo, E.N.; Liao, X.W.; Chen, L.S.; Wang, J.L.; Ni, M.; Liang, C. Genomescale analysis to identify potential prognostic microRNA biomarkers for predicting overall survival in patients with colon adenocarcinoma. Oncol. Rep. 2018, 40, 1947–1958. [Google Scholar] [CrossRef]
- Chen, J.; Pitmon, E.; Wang, K. Microbiome, inflammation and colorectal cancer. Semin. Immunol. 2017, 32, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Allen, T.D.; May, R.J.; Lightfoot, S.; Houchen, C.W.; Huycke, M.M. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008, 68, 9909–9917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Huycke, M.M. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology 2007, 132, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Mangerich, A.; Knutson, C.G.; Parry, N.M.; Muthupalani, S.; Ye, W.; Prestwich, E.; Cui, L.; McFaline, J.L.; Mobley, M.; Ge, Z.; et al. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl. Acad. Sci. USA 2012, 109, E1820–E1829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, S.J.; Zheng, S.P.; Huang, F.Y.; Jiao, C.L.; Wu, R.L. Synergistic anti-tumor effect of recombinant chicken fibroblast growth factor receptor-1-mediated anti-angiogenesis and low-dose gemcitabine in a mouse colon adenocarcinoma model. World J. Gastroenterol. 2007, 13, 2484–2489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merl, M.; Hoimes, C.; Pham, T.; Saif, M.W. Is there a palliative benefit of gemcitabine plus fluoropyrimidines in patients with refractory colorectal cancer? A review of the literature previously presented: Poster at the 2008 Gastrointestinal Cancer Symposium (Abstract No. 512). Expert Opin. Investig. Drugs 2009, 18, 1257–1264. [Google Scholar] [CrossRef]
- Saif, M.W.; Kaley, K.; Penney, R.; Hotchkiss, S.; Syrigos, K.N.; Strimpakos, A.S. The efficacy of gemcitabine as salvage treatment in patients with refractory advanced colorectal cancer (CRC): A single institution experience. Anticancer Res. 2011, 31, 2971–2974. [Google Scholar] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tung, C.-B.; Li, C.-Y.; Lin, H.-Y. Multi-Omics Reveal the Immunological Role and the Theragnostic Value of miR-216a/GDF15 Axis in Human Colon Adenocarcinoma. Int. J. Mol. Sci. 2021, 22, 13636. https://doi.org/10.3390/ijms222413636
Tung C-B, Li C-Y, Lin H-Y. Multi-Omics Reveal the Immunological Role and the Theragnostic Value of miR-216a/GDF15 Axis in Human Colon Adenocarcinoma. International Journal of Molecular Sciences. 2021; 22(24):13636. https://doi.org/10.3390/ijms222413636
Chicago/Turabian StyleTung, Chun-Bin, Chia-Ying Li, and Hung-Yu Lin. 2021. "Multi-Omics Reveal the Immunological Role and the Theragnostic Value of miR-216a/GDF15 Axis in Human Colon Adenocarcinoma" International Journal of Molecular Sciences 22, no. 24: 13636. https://doi.org/10.3390/ijms222413636
APA StyleTung, C.-B., Li, C.-Y., & Lin, H.-Y. (2021). Multi-Omics Reveal the Immunological Role and the Theragnostic Value of miR-216a/GDF15 Axis in Human Colon Adenocarcinoma. International Journal of Molecular Sciences, 22(24), 13636. https://doi.org/10.3390/ijms222413636





