Mapping Resistance to Argentinean Fusarium (Graminearum) Head Blight Isolates in Wheat
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Variation
2.2. QTL Analysis
2.2.1. Additive QTLs
2.2.2. Epistasis QTLs
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Source and Maintenance of F. graminearum Isolates
4.3. Inoculation Method
4.4. Phenotypic Assessments
4.5. Construction of Genetic Linkage Map and QTL Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bai, G.; Shaner, G. Management and resistance in wheat and barley to fusarium head blight. Annu. Rev. Phytopathol. 2004, 42, 135–161. [Google Scholar] [CrossRef] [PubMed]
- Windels, C.E. Economic and Social Impacts of Fusarium Head Blight: Changing Farms and Rural Communities in the Northern Great Plains. Phytopathology 2000, 90, 17–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMullen, M.; Jones, R.; Gallenberg, D. Scab of Wheat and Barley: A Re-emerging Disease of Devastating Impact. Plant Dis. 1997, 81, 1340–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Annone, J. Resistencia Genética a Agentes Fitopatógenos: Conceptos Sobre Hospedante, Patógeno, Relación Hospedante-patógeno y Manejo de la Resistencia. In Curso de Manejo de Enfermedades del Trigo; Kohli, M.M., Annone, J.G., y García, R., Eds.; Centro Internacional de Capacitación INTA-CIMMYT: Pergamino, Argentina, 1995; p. 237. [Google Scholar]
- Wang, Y.; Miller, J. Screening Techniques and Sources of Resistance to Fusarium Head Blight. In Proceedings of the Wheat Production Constraints in Tropical Environments, Chiang Mai, Thailand, 19–23 January 1987. [Google Scholar]
- Jiang, G.-L.; Dong, Y.; Shi, J.; Ward, R.W. QTL analysis of resistance to Fusarium head blight in the novel wheat germplasm CJ 9306. II. Resistance to deoxynivalenol accumulation and grain yield loss. Theor. Appl. Genet. 2007, 115, 1043–1052. [Google Scholar] [CrossRef]
- Buerstmayr, M.; Lemmens, M.; Steiner, B.; Buerstmayr, H. Advanced backcross QTL mapping of resistance to Fusarium head blight and plant morphological traits in a Triticum macha× T. aestivum population. Theor. Appl. Genet. 2011, 123, 293. [Google Scholar] [CrossRef] [Green Version]
- Jiang, G.-L.; Shi, J.; Ward, R.W. QTL analysis of resistance to Fusarium head blight in the novel wheat germplasm CJ 9306. I. Resistance to fungal spread. Theor. Appl. Genet. 2007, 116, 3–13. [Google Scholar] [CrossRef]
- McKendry, A.; Liu, S.; Abate, Z.A. Inheritance of Fusarium Head Blight Resistance in the US Wheat Cultivar Ernie. In Proceedings of the 2nd International Symposium on Fusarium Head Blight Incorporating the 8th European Fusarium Seminar, Orlando, FL, USA, 11–15 December 2004. [Google Scholar]
- Mesterhazy, A. Types and components of resistance to Fusarium head blight of wheat. Plant Breed. 1995, 114, 377–386. [Google Scholar] [CrossRef]
- Schroeder, H.; Christensen, J. Factors affecting resistance of wheat to scab caused by Gibberella zeae. Phytopathology 1963, 53, 831–838. [Google Scholar]
- Mesterházy, Á.; Bartók, T.; Mirocha, C.G.; Komoróczy, R. Nature of wheat resistance to Fusarium head blight and the role of deoxynivalenol for breeding. Plant Breed. 1999, 118, 97–110. [Google Scholar] [CrossRef]
- Miller, J.D.; Young, J.C.; Sampson, D.R. Deoxynivalenol and Fusarium Head Blight Resistance in Spring Cereals. J. Phytopathol. 1985, 113, 359–367. [Google Scholar] [CrossRef]
- Pariyar, S.R.; Erginbas-Orakci, G.; Dadshani, S.; Chijioke, O.B.; Léon, J.; Dababat, A.A.; Grundler, F.M.W. Dissecting the Genetic Complexity of Fusarium Crown Rot Resistance in Wheat. Sci. Rep. 2020, 10, 3200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kollers, S.; Rodemann, B.; Ling, J.; Korzun, V.; Ebmeyer, E.; Argillier, O.; Hinze, M.; Plieske, J.; Kulosa, D.; Ganal, M.W.; et al. Whole Genome Association Mapping of Fusarium Head Blight Resistance in European Winter Wheat (Triticum aestivum L.). PLoS ONE 2013, 8, e57500. [Google Scholar] [CrossRef] [Green Version]
- Löffler, M.; Schön, C.-C.; Miedaner, T. Revealing the genetic architecture of FHB resistance in hexaploid wheat (Triticum aestivum L.) by QTL meta-analysis. Mol. Breed. 2009, 23, 473–488. [Google Scholar] [CrossRef]
- Liu, S.; Hall, M.D.; Griffey, C.A.; McKendry, A.L. Meta-Analysis of QTL Associated with Fusarium Head Blight Resistance in Wheat. Crop Sci. 2009, 49, 1955–1968. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Mardi, M.; Buerstmayr, H.; Ghareyazie, B.; Lemmens, M.; Mohammadi, S.A.; Nolz, R.; Ruckenbauer, P. QTL analysis of resistance to Fusarium head blight in wheat using a ‘Wangshuibai’-derived population. Plant Breed. 2005, 124, 329–333. [Google Scholar] [CrossRef]
- Klahr, A.; Mohler, V.; Herz, M.; Wenzel, G.; Schwarz, G. Enhanced power of QTL detection for Fusarium head blight resistance in wheat by means of codominant scoring of hemizygous molecular markers. Mol. Breed. 2004, 13, 289–300. [Google Scholar] [CrossRef]
- Somers, D.J.; Fedak, G.; Savard, M. Molecular mapping of novel genes controlling Fusarium head blight resistance and deoxynivalenol accumulation in spring wheat. Genome 2003, 46, 555–564. [Google Scholar] [CrossRef]
- Shen, X.; Ittu, M.; Ohm, H.W. Quantitative trait loci conditioning resistance to Fusarium head blight in wheat line F201R. Crop Sci. 2003, 43, 850–857. [Google Scholar] [CrossRef]
- Gervais, L.; Dedryver, F.; Morlais, J.-Y.; Bodusseau, V.; Negre, S.; Bilous, M.; Groos, C.; Trottet, M. Mapping of quantitative trait loci for field resistance to Fusarium head blight in an European winter wheat. Theor. Appl. Genet. 2002, 106, 961–970. [Google Scholar] [CrossRef]
- Lin, F.; Kong, Z.X.; Zhu, H.L.; Xue, S.L.; Wu, J.Z.; Tian, D.G.; Wei, J.B.; Zhang, C.Q.; Ma, Z.Q. Mapping QTL associated with resistance to Fusarium head blight in the Nanda2419× Wangshuibai population. II: Type I resistance. Theor. Appl. Genet. 2006, 112, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Bai, G.; Shaner, G.E. Novel quantitative trait loci (QTL) for Fusarium head blight resistance in wheat cultivar Chokwang. Theor. Appl. Genet. 2005, 111, 1571–1579. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Chen, P.; Qin, G.; Bai, G.; Wang, X.; Wang, S.; Zhou, B.; Zhang, S.; Liu, D. QTLs for Fusarium head blight response in a wheat DH population of Wangshuibai/Alondra‘s’. Euphytica 2005, 146, 183–191. [Google Scholar] [CrossRef]
- Zhou, W.; Kolb, F.L.; Yu, J.; Bai, G.; Boze, L.K.; Domier, L.L. Molecular characterization of Fusarium head blight resistance in Wangshuibai with simple sequence repeat and amplified fragment length polymorphism markers. Genome 2004, 47, 1137–1143. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhou, M.; Ren, L.; Bai, G.; Ma, H.; Scholten, E.O.; Guo, P.; Lu, W. Molecular characterization of Fusarium head blight resistance from wheat variety Wangshuibai. Euphytica 2004, 139, 59–64. [Google Scholar] [CrossRef]
- Zhou, W.-C.; Kolb, F.L.; Bai, G.-H.; Domier, L.L.; Yao, J.-B. Effect of individual Sumai 3 chromosomes on resistance to scab spread within spikes and deoxynivalenol accumulation within kernels in wheat. Hereditas 2002, 137, 81–89. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Lemmens, M.; Hartl, L.; Doldi, L.; Steiner, B.; Stierschneider, M.; Ruckenbauer, P. Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. Resistance to fungal spread (Type II resistance). Theor. Appl. Genet. 2002, 104, 84–91. [Google Scholar] [CrossRef]
- Anderson, J.A.; Stack, R.W.; Liu, S.; Waldron, B.L.; Fjeld, A.D.; Coyne, C.; Moreno-Sevilla, B.; Fetch, J.M.; Song, Q.J.; Cregan, P.B.; et al. DNA markers for Fusarium head blight resistance QTLs in two wheat populations. Theor. Appl. Genet. 2001, 102, 1164–1168. [Google Scholar] [CrossRef]
- Waldron, B.L.; Moreno-Sevilla, B.; Anderson, J.A.; Stack, R.W.; Frohberg, R.C. RFLP Mapping of QTL for Fusarium Head Blight Resistance in Wheat. Crop Sci. 1999, 39, 805–811. [Google Scholar] [CrossRef]
- Goddard, R.; Steed, A.; Scheeren, P.L.; Maciel, J.L.N.; Caierão, E.; Torres, G.A.M.; Consoli, L.; Santana, F.M.; Fernandes, J.M.C.; Simmonds, J.; et al. Identification of Fusarium head blight resistance loci in two Brazilian wheat mapping populations. PLoS ONE 2021, 16, e0248184. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.; Tekauz, A. Review: Recent developments in research on fusarium head blight of wheat in Canada. Can. J. Plant Pathol. 2000, 22, 1–8. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Steiner, B.; Hartl, L.; Griesser, M.; Angerer, N.; Lengauer, D.; Miedaner, T.; Schneider, B.; Lemmens, M. Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. II. Resistance to fungal penetration and spread. Theor. Appl. Genet. 2003, 107, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Kolb, F.L.; Shaner, G.; Domier, L.L. Amplified Fragment Length Polymorphism Markers Linked to a Major Quantitative Trait Locus Controlling Scab Resistance in Wheat. Phytopathology 1999, 89, 343–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miedaner, T.; Wilde, F.; Steiner, B.; Buerstmayr, H.; Korzun, V.; Ebmeyer, E. Stacking quantitative trait loci (QTL) for Fusarium head blight resistance from non-adapted sources in an European elite spring wheat background and assessing their effects on deoxynivalenol (DON) content and disease severity. Theor. Appl. Genet. 2005, 112, 562–569. [Google Scholar] [CrossRef]
- Bai, G.; Guo, P.; Kolb, F.L. Genetic Relationships among Head Blight Resistant Cultivars of Wheat Assessed on the Basis of Molecular Markers. Crop Sci. 2003, 43, 498–507. [Google Scholar] [CrossRef]
- Steiner, B.; Lemmens, M.; Griesser, M.; Scholz, U.; Schondelmaier, J.; Buerstmayr, H. Molecular mapping of resistance to Fusarium head blight in the spring wheat cultivar Frontana. Theor. Appl. Genet. 2004, 109, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Schmolke, M.; Zimmermann, G.; Buerstmayr, H.; Schweizer, G.; Miedaner, T.; Korzun, V.; Ebmeyer, E.; Hartl, L. Molecular mapping of Fusarium head blight resistance in the winter wheat population Dream/Lynx. Theor. Appl. Genet. 2005, 111, 747–756. [Google Scholar] [CrossRef]
- Wilde, F.; Korzun, V.; Ebmeyer, E.; Geiger, H.H.; Miedaner, T. Comparison of phenotypic and marker-based selection for Fusarium head blight resistance and DON content in spring wheat. Mol. Breed. 2007, 19, 357–370. [Google Scholar] [CrossRef]
- Wilde, F.; Miedaner, T. Selection for Fusarium head blight resistance in early generations reduces the deoxynivalenol (DON) content in grain of winter and spring wheat. Plant Breed. 2006, 125, 96–98. [Google Scholar] [CrossRef]
- Semagn, K.; Skinnes, H.; Bjørnstad, Å.; Marøy, A.G. Quantitative trait loci controlling Fusarium head blight resistance and low deoxynivalenol content in hexaploid wheat population from ‘Arina’ and NK93604. Crop Sci. 2007, 47, 294–303. [Google Scholar] [CrossRef]
- Dababat, A.; Arif, M.A.R.; Toktay, H.; Atiya, O.; Shokat, S.; E-Orakci, G.; Imren, M.; Singh, S. A GWAS to identify the cereal cyst nematode (Heterodera filipjevi) resistance loci in diverse wheat prebreeding lines. J. Appl. Genet. 2021, 62, 93–98. [Google Scholar] [CrossRef]
- Arif, M.A.R.; Shokat, S.; Plieske, J.; Ganal, M.; Lohwasser, U.; Chesnokov, Y.V.; Kocherina, N.V.; Kulwal, P.; Kumar, N.; McGuire, P.E.; et al. A SNP-based genetic dissection of versatile traits in bread wheat (Triticum aestivum L.). Plant J. 2021, 108, 960–976. [Google Scholar] [CrossRef]
- Agacka-Mołdoch, M.; Arif, M.A.R.; Lohwasser, U.; Doroszewska, T.; Qualset, C.O.; Börner, A. The inheritance of wheat grain longevity: A comparison between induced and natural ageing. J. Appl. Genet. 2016, 57, 477–481. [Google Scholar] [CrossRef]
- Arif, M.A.R.; Nagel, M.; Neumann, K.; Kobiljski, B.; Lohwasser, U.; Börner, A. Genetic studies of seed longevity in hexaploid wheat using segregation and association mapping approaches. Euphytica 2011, 186, 1–13. [Google Scholar] [CrossRef]
- Miedaner, T.; Würschum, T.; Maurer, H.P.; Korzun, V.; Ebmeyer, E.; Reif, J.C. Association mapping for Fusarium head blight resistance in European soft winter wheat. Mol. Breed. 2010, 28, 647–655. [Google Scholar] [CrossRef]
- Somers, D.; McCartney, C.; Depauw, R.; Thomas, J.; Fox, S.; Fedak, G.; Humphreys, G.; Gilbert, J.; McCallum, B.; Banks, T. Molecular Breeding for Multiple Pest Resistance in Wheat. Dev. Plant Breed. 2007, 12, 667–676. [Google Scholar] [CrossRef]
- Abedi, T.; Khalil, M.F.M.; Koike, K.; Hagura, Y.; Tazoe, Y.; Ishida, N.; Kitamura, K.; Tanaka, N. Expression of the human UDP-galactose transporter gene hUGT1 in tobacco plants’ enhanced plant hardness. J. Biosci. Bioeng. 2018, 126, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Bird, D.A. The role of ABC transporters in cuticular lipid secretion. Plant Sci. 2008, 174, 563–569. [Google Scholar] [CrossRef]
- He, W.-J.; Zhang, L.; Yi, S.-Y.; Tang, X.-L.; Yuan, Q.-S.; Guo, M.-W.; Wu, A.-B.; Qu, B.; Li, H.-P.; Liao, Y.-C. An aldo-keto reductase is responsible for Fusarium toxin-degrading activity in a soil Sphingomonas strain. Sci. Rep. 2017, 7, 9549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Sun, H.; Zhang, J.; Hou, Y.; Zhang, T.; Kang, J.; Wang, Z.; Yang, Q.; Long, R. Analysis of aldo–keto reductase gene family and their responses to salt, drought, and abscisic acid stresses in Medicago truncatula. Int. J. Mol. Sci. 2020, 21, 754. [Google Scholar] [CrossRef] [Green Version]
- Walter, S.; Doohan, F. Transcript profiling of the phytotoxic response of wheat to the Fusarium mycotoxin deoxynivalenol. Mycotoxin Res. 2011, 27, 221–230. [Google Scholar] [CrossRef]
- Forde, B.G.; Roberts, M.R. Glutamate receptor-like channels in plants: A role as amino acid sensors in plant defence? F1000 Prime Rep. 2014, 6, 37. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jing, Y.; Zhang, X.; Li, L.; Wang, P.; Zhang, S.; Zhou, H.; Wu, J. Evolutionary and Expression Analysis Provides Evidence for the Plant Glutamate-like Receptors Family is Involved in Woody Growth-related Function. Sci. Rep. 2016, 6, 32013. [Google Scholar] [CrossRef] [Green Version]
- Kawashima, C.G.; Berkowitz, O.; Hell, R.; Noji, M.; Saito, K. Characterization and Expression Analysis of a Serine Acetyltransferase Gene Family Involved in a Key Step of the Sulfur Assimilation Pathway in Arabidopsis. Plant Physiol. 2005, 137, 220–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walter, S.; Kahla, A.; Arunachalam, C.; Perochon, A.; Khan, M.R.; Scofield, S.R.; Doohan, F.M. A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J. Exp. Bot. 2015, 66, 2583–2593. [Google Scholar] [CrossRef] [Green Version]
- Theivendren, P.; Kunjiappan, S.; Hegde, Y.M.; Vellaichamy, S.; Gopal, M.; Dhramalingam, S.R.; Kumar, S. Importance of Protein Kinase and Its Inhibitor: A Review. Protein Kinase-New Oppor. Chall. Future Perspect. 2021. [Google Scholar] [CrossRef]
- Guo, C.; Guo, L.; Li, X.; Gu, J.; Zhao, M.; Duan, W.; Ma, C.; Lu, W.; Xiao, K. TaPT2, a high-affinity phosphate transporter gene in wheat (Triticum aestivum L.), is crucial in plant Pi uptake under phosphorus deprivation. Acta Physiol. Plant. 2014, 36, 1373–1384. [Google Scholar] [CrossRef]
- Raffan, S.; Halford, N.G. Cereal asparagine synthetase genes. Ann. Appl. Biol. 2020, 178, 6–22. [Google Scholar] [CrossRef]
- Brauer, E.K.; Rocheleau, H.; Balcerzak, M.; Pan, Y.; Fauteux, F.; Liu, Z.; Wang, L.; Zheng, W.; Ouellet, T. Transcriptional and hormonal profiling of Fusarium graminearum-infected wheat reveals an association between auxin and susceptibility. Physiol. Mol. Plant Pathol. 2019, 107, 33–39. [Google Scholar] [CrossRef]
- Ohmiya, A. Carotenoid cleavage dioxygenases and their apocarotenoid products in plants. Plant Biotechnol. 2009, 26, 351–358. [Google Scholar] [CrossRef] [Green Version]
- Rampino, P.; De Pascali, M.; Perrotta, C.; Gullì, M. New gene functions are involved in the thermotolerance of the wild wheat relative Aegilops umbellulata. Plant Physiol. Biochem. 2020, 156, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Michaels, S.D. SUPPRESSOR OF FRI 4 encodes a nuclear-localized protein that is required for delayed flowering in winter-annual Arabidopsis. Development 2006, 133, 4699–4707. [Google Scholar] [CrossRef] [Green Version]
- Chi, Q.; Guo, L.; Ma, M.; Zhang, L.; Mao, H.; Wu, B.; Liu, X.; Ramirez-Gonzalez, R.H.; Uauy, C.; Appels, R.; et al. Global transcriptome analysis uncovers the gene co-expression regulation network and key genes involved in grain development of wheat (Triticum aestivum L.). Funct. Integr. Genom. 2019, 19, 853–866. [Google Scholar] [CrossRef] [Green Version]
- Curtis, T.Y.; Bo, V.; Tucker, A.; Halford, N.G. Construction of a network describing asparagine metabolism in plants and its application to the identification of genes affecting asparagine metabolism in wheat under drought and nutritional stress. Food Energy Secur. 2018, 7, e00126. [Google Scholar] [CrossRef]
- Kong, X.; Pan, J.; Zhang, D.; Jiang, S.; Cai, G.; Wang, L.; Li, D. Identification of mitogen-activated protein kinase kinase gene family and MKK–MAPK interaction network in maize. Biochem. Biophys. Res. Commun. 2013, 441, 964–969. [Google Scholar] [CrossRef] [PubMed]
- Suliman, S.; Alemu, A.; Abdelmula, A.A.; Badawi, G.H.; Al-Abdallat, A.; Tadesse, W. Genome-wide association analysis uncovers stable QTLs for yield and quality traits of spring bread wheat (Triticum aestivum) across contrasting environments. Plant Gene 2020, 25, 100269. [Google Scholar] [CrossRef]
- Gonzalez, D.; Bowen, A.J.; Carroll, T.S.; Conlan, R.S. The Transcription Corepressor LEUNIG Interacts with the Histone Deacetylase HDA19 and Mediator Components MED14 (SWP) and CDK8 (HEN3) To Repress Transcription. Mol. Cell. Biol. 2007, 27, 5306–5315. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Kaur, G.; Kumar, A.; Meena, V.; Kaur, J.; Pandey, A.K. Overlapping transcriptional expression response of wheat zinc-induced facilitator-like transporters emphasize important role during Fe and Zn stress. BMC Mol. Biol. 2019, 20, 22. [Google Scholar] [CrossRef] [Green Version]
- Feng, L.; Schaefer, A.L.; Hu, M.; Chen, R.; Greenberg, E.P.; Zhou, J. Virulence Factor Identification in the Banana Pathogen Dickeya zeae MS2. Appl. Environ. Microbiol. 2019, 85, e1611–e1619. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.-X.; Bai, G.-H.; Gill, B.S.; Hart, L.P. Deletion of a Chromosome Arm Altered Wheat Resistance to Fusarium Head Blight and Deoxynivalenol Accumulation in Chinese Spring. Plant Dis. 2006, 90, 1545–1549. [Google Scholar] [CrossRef] [PubMed]
- Niwa, S.; Kubo, K.; Lewis, J.; Kikuchi, R.; Alagu, M.; Ban, T. Variations for Fusarium head blight resistance associated with genomic diversity in different sources of the resistant wheat cultivar ‘Sumai 3’. Breed. Sci. 2014, 64, 90–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourdoncle, W.; Ohm, H. Quantitative trait loci for resistance to Fusarium head blight in recombinant inbred wheat lines from the cross Huapei 57-2/Patterson. Euphytica 2003, 131, 131–136. [Google Scholar] [CrossRef]
- Börner, A.; Schumann, E.; Fürste, A.; Cöster, H.; Leithold, B.; Röder, M.; Weber, W. Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor. Appl. Genet. 2002, 105, 921–936. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Plattner, R.; Desjardins, A.; Kolb, F.; McIntosh, R.A. Resistance to Fusarium head blight and deoxynivalenol accumulation in wheat. Plant Breed. 2001, 120, 1–6. [Google Scholar] [CrossRef]
- Malbrán, I.; Mourelos, C.; Girotti, J.R.; Aulicino, M.; Balatti, P.; Lori, G. Aggressiveness variation of Fusarium graminearum isolates from Argentina following point inoculation of field grown wheat spikes. Crop Prot. 2012, 42, 234–243. [Google Scholar] [CrossRef]
- Cappellini, R.; Peterson, J. Macroconidium formation in submerged cultures by a nonsporulating strain of Gibberella zeae. Mycologia 1965, 57, 962–966. [Google Scholar] [CrossRef]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Stack, R.W.; McMullen, M.P. A Visual Scale to Estimate Severity of Fusarium Head Blight in Wheat; North Dakota State University Extension Service: Fargo, ND, USA, 1995; p. 1095. [Google Scholar]
- Soleimani, B.; Lehnert, H.; Keilwagen, J.; Plieske, J.; Ordon, F.; Rad, S.N.; Ganal, M.; Beier, S.; Perovic, D. Comparison Between Core Set Selection Methods Using Different Illumina Marker Platforms: A Case Study of Assessment of Diversity in Wheat. Front. Plant Sci. 2020, 11, 1040. [Google Scholar] [CrossRef]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [Green Version]
- Allen, A.M.; Winfield, M.O.; Burridge, A.J.; Downie, R.C.; Benbow, H.R.; Barker, G.L.A.; Wilkinson, P.A.; Coghill, J.; Waterfall, C.; Davassi, A.; et al. Characterization of a Wheat Breeders’ Array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum). Plant Biotechnol. J. 2017, 15, 390–401. [Google Scholar] [CrossRef] [Green Version]
- Ooijen, J.W.; Voorrips, R. JoinMap: Version 3.0: Software for the Calculation of Genetic Linkage Maps; University and Research Center: Wageningen, The Netherlands, 2002. [Google Scholar]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef] [Green Version]
- Arif, M.A.R.; Börner, A. Mapping of QTL associated with seed longevity in durum wheat (Triticum durum Desf.). J. Appl. Genet. 2018, 60, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
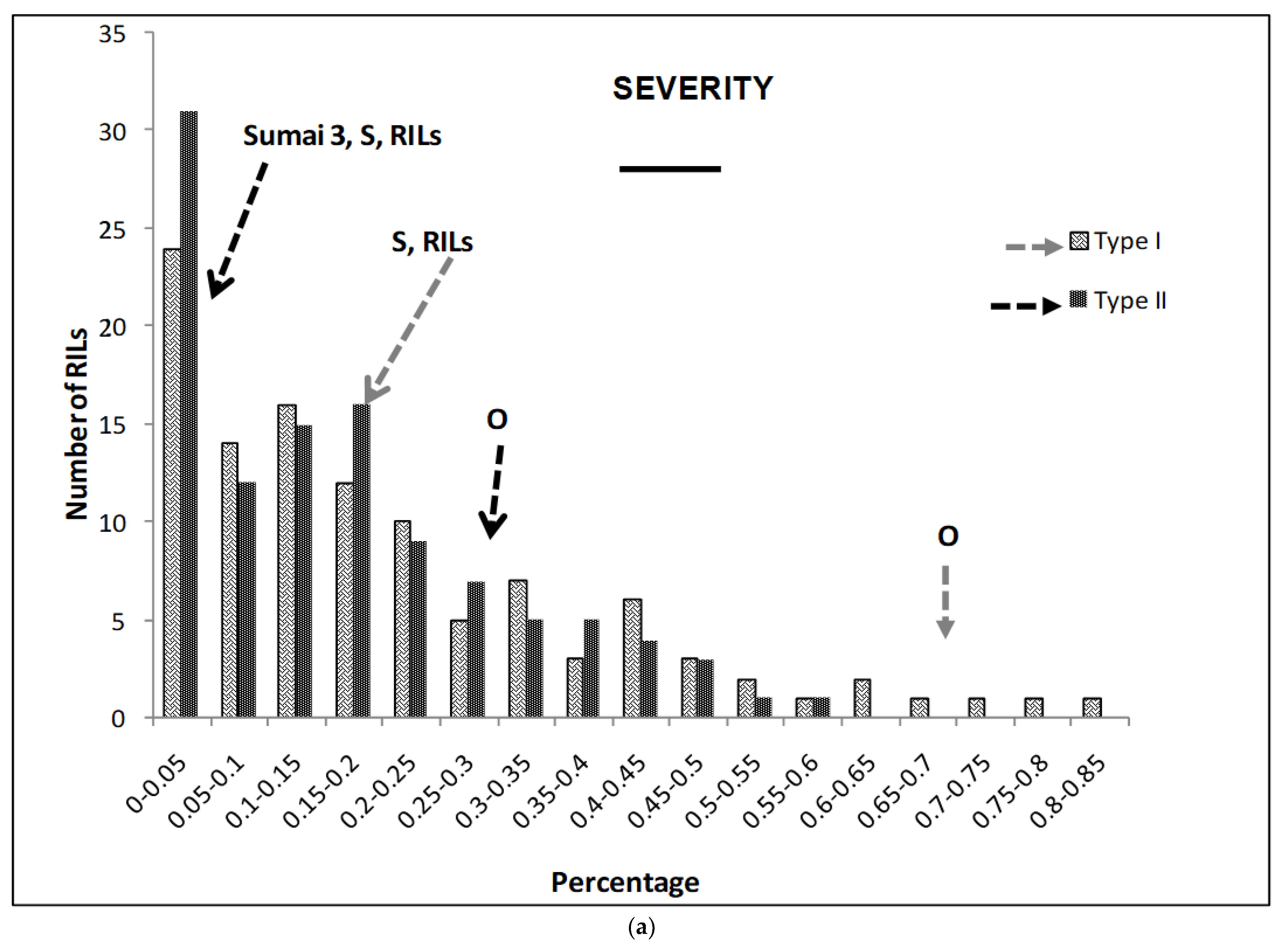





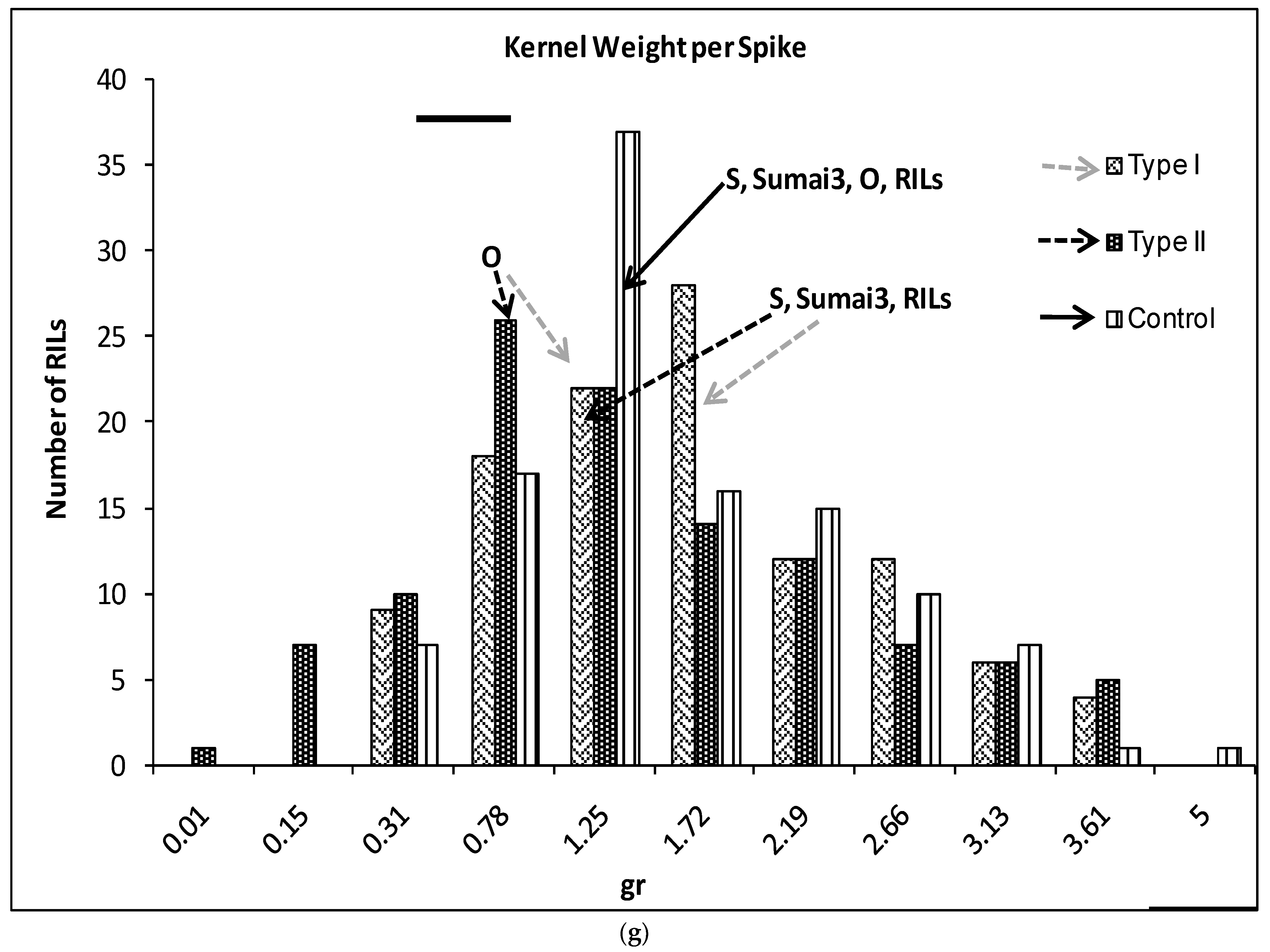
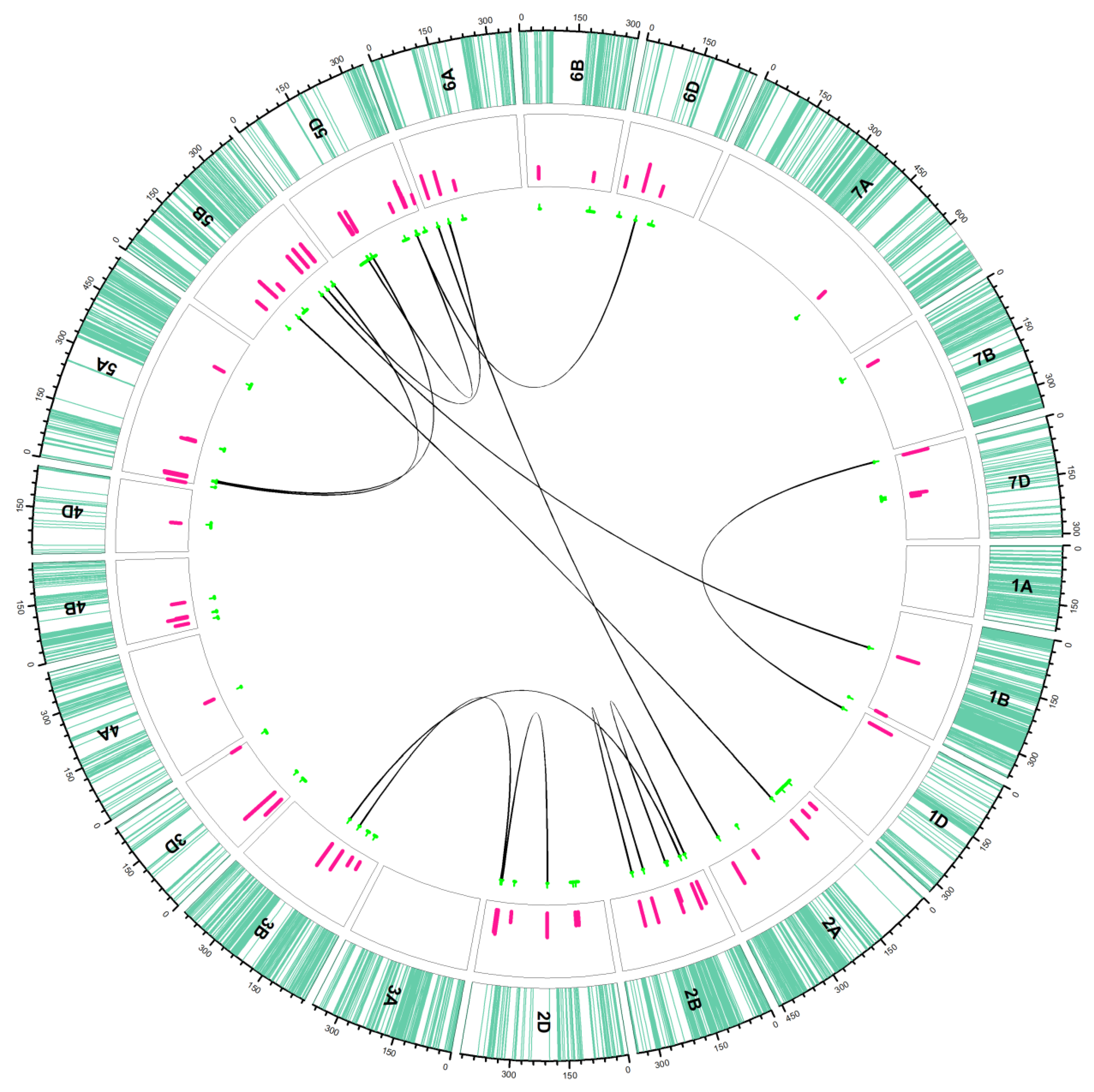
| Sources | DF | Mean Squares | ||||||
|---|---|---|---|---|---|---|---|---|
| Severity | Fusarium Index | TKW | FDK | Number of Spikes | Kernel Weight per Plant | Kernel Weight per Spike | ||
| Genotypes | 112 | 0.119 *** | 0.054 *** | 135.69 ** | 0.0078732ns | 25.89 ** | 74.78 ** | 0.55 * |
| Treatment | 2 | 1.476 *** | 0.157 *** | 920.91 *** | 0.1735337 *** | 35.68ns | 535.02 *** | 4.32 *** |
| G × T | 224 | 0.101 *** | 0.030 ** | 93.96 ** | 0.008631ns | 12.57 * | 46.80 ** | 0.45 ** |
| Error | 1029 | 0.000 | 0.005 | 73.09 | 0.0075382 | 12.09 | 32.81 | 0.44 |
| Trait | Specific Trait | Chr. | Pos. | Left Marker | Right Marker | LOD | PVE (%) | Add | LeftCI | RightCI |
|---|---|---|---|---|---|---|---|---|---|---|
| Type I resistance | FDK_Type_I_2009 | 2B | 114 | AX-94381628 | BobWhite_c8253_397 | 2.8 | 13.63 | −4.13 | 111.5 | 116.5 |
| Sev_Type_I_2009 + | 2D | 97 | CAP12_c1503_76 | D_GBUVHFX02GV41H_67 | 3 | 9.22 | 0.07 | 85.5 | 115.5 | |
| FI_Type I_2008 | 3D | 15 | BS00033229_51 | D_contig00455_358 | 5.14 | 16.61 | −0.06 | 1.5 | 18.5 | |
| FI_Type I_2008 | 3D | 50 | D_GDS7LZN02IJRXZ_309 | CAP12_c2615_128 | 9.14 | 47.74 | 0.10 | 49.5 | 55.5 | |
| FI_Type I_2008 | 4B | 23 | wsnp_Ex_c4148_7494801 | Kukri_rep_c71670_163 | 3.04 | 7.84 | −0.04 | 17.5 | 25.5 | |
| Sev_Type_I_2009 | 4B | 45 | tplb0027f12_503 | BS00035426_51 | 4.51 | 9.85 | 0.07 | 43.5 | 45.5 | |
| Sev_Type_I_2009 | 4D | 131.5 | AX-94838884 | AX-95126745 | 2.35 | 6.49 | 0.06 | 119.98 | 139.98 | |
| FI_Type_I_2009 | 5A | 0 | wsnp_Ex_c905_1748920 | AX-95114232 | 4.2 | 15.15 | 0.06 | 0 | 0.5 | |
| FI_Type I_2008 | 5B | 114 | BS00065390_51 | AX-95145462 | 2.88 | 8.03 | −0.04 | 112.5 | 115.5 | |
| FI_Type_I_2009 | 5B | 356 | RAC875_c278_1801 | Excalibur_c48387_58 | 2.8 | 10.11 | 0.05 | 349.5 | 356.5 | |
| Sev_Type_I_2009 * | 5D | 109 | Kukri_c13045_302 | IAAV6265 | 2.12 | 15.09 | 0.09 | 70.5 | 139.5 | |
| FI_Type I_2008 * | 5D | 124 | Kukri_c13045_302 | IAAV6265 | 2.7 | 7.95 | −0.04 | 87.5 | 137.5 | |
| Sev_Type_I_2009 | 5D | 318 | AX-95190974 | Kukri_rep_c106820_591 | 3.35 | 7.39 | 0.06 | 313.5 | 320.5 | |
| Type II resistance | FI_Type_II_2009 | 2A | 291 | BobWhite_c4743_63 | Excalibur_c47535_389 | 2.16 | 5.57 | 0.032 | 288.5 | 291.5 |
| Sev_Type_II_2009 | 2A | 30 | RAC875_c24364_307 | wsnp_Ex_rep_c66358_64543089 | 2.13 | 9.89 | 0.13 | 20.5 | 38.5 | |
| FI_Type_II_2009 | 3B | 84.2 | RAC875_c60169_200 | BS00084607_51 | 2.02 | 5.27 | 0.03 | 78.679 | 89.679 | |
| FI_Type_II_2009 | 3D | 249 | TA020105-1083 | AX-94637066 | 2.38 | 6.13 | −0.03 | 238.5 | 252 | |
| FI_Type_II_2009 | 4B | 48 | Kukri_c32958_390 | Excalibur_c19547_1012 | 2.56 | 8.03 | 0.03 | 47.5 | 49.5 | |
| Sev_Type_II_2009 | 4B | 47 | wsnp_Ku_c12503_20174234 | Kukri_c28022_54 | 2.06 | 6.55 | 0.04 | 43.5 | 48.5 | |
| FI_Type II_2008 | 4B | 99 | AX-94899864 | AX-94465680 | 2.97 | 6.39 | 0.14 | 96.5 | 104.5 | |
| FI_Type II_2008 | 5A | 148 | wsnp_RFL_Contig4307_5006558 | Kukri_c12738_882 | 3.35 | 6.39 | 0.14 | 144.5 | 151.5 | |
| Sev_Type_II_2009 | 5D | 270 | IACX3123 | TA015368-0126 | 2.24 | 10.09 | 0.13 | 258.5 | 279.5 | |
| FI_Type_II_2009 ** | 5D | 317 | AX-95190974 | Kukri_rep_c106820_591 | 4.8 | 15.99 | 0.05 | 314.5 | 320.5 | |
| Sev_Type_II_2009 ** | 5D | 319 | AX-95190974 | Kukri_rep_c106820_591 | 2.25 | 3.74 | 0.05 | 312.5 | 323.5 | |
| Sev_Type_II_2009 | 5D | 352 | RAC875_c16419_585 | BS00078603_51 | 2.07 | 5.6 | 0.04 | 344.5 | 355 | |
| FI_Type_II_2009 | 6B | 233 | GENE-0221_350 | Kukri_c32307_481 | 2.06 | 5.36 | −0.03 | 224.5 | 250.5 | |
| Sev_Type_II_2009 | 6D | 153 | AX-95107291 | Excalibur_rep_c99143_422 | 2.36 | 7.35 | 0.05 | 142.5 | 162.5 | |
| TKW | TKW_C_2010 | 1B | 366 | AX-95154820 | AX-94621372 | 2.72 | 16.27 | 5.39 | 364.5 | 366 |
| TKW_C_2008 | 2D | 332 | TA021271-0482 | Excalibur_c1451_660 | 2.48 | 18.04 | −4.34 | 328.5 | 334.5 | |
| TKW_Type II_2010 | 6A | 126 | Excalibur_c23748_452 | AX-94978875 | 2.27 | 13.6 | −6.03 | 122.5 | 134.5 | |
| TKW_C_2010 | 6D | 24 | AX-94633926 | IACX10982 | 2.36 | 19.9 | 6.17 | 19.5 | 31.5 | |
| TKW_C_2010 *** | 7D | 145 | BS00066128_51 | Ku_c32426_324 | 3.6 | 19.9 | −6.17 | 140.5 | 149.5 | |
| TKW_Type II_2010 | 7D | 155 | Kukri_c15768_1383 | BS00062644_51 | 2.03 | 14.9 | −4.25 | 146.5 | 158.5 | |
| KWS | KWS_Type II_2010 | 2A | 67 | wsnp_Ex_rep_c66358_64543089 | Kukri_c33374_1048 | 2.23 | 15.55 | 0.18 | 38.5 | 92.5 |
| KWS_Type II_2008 + | 2D | 107 | CAP12_c1503_76 | D_GBUVHFX02GV41H_67 | 3.04 | 35 | 0.37 | 90.5 | 117.5 | |
| KWS_C_2010 | 3B | 115.2 | BS00065934_51 | BobWhite_c22370_352 | 2.24 | 19.18 | 31.0 | 111.67 | 121.67 | |
| KWS_Type I_2008 | 4A | 168 | RAC875_c25124_182 | wsnp_Ex_c24474_33721784 | 2.25 | 19.72 | −0.20 | 166.5 | 170.5 | |
| KWS_C_2008 | 5A | 151 | wsnp_RFL_Contig4307_5006558 | Kukri_c12738_882 | 2.03 | 19.28 | 0.80 | 144.5 | 153.5 | |
| KWS_Type I_2008 | 5A | 416 | IAAV1179 | AX-94706027 | 2.5 | 20.95 | 0.21 | 403.5 | 423.5 | |
| KWS_Type II_2010 | 5B | 206 | Kukri_c52_225 | wsnp_Ku_c3102_5810751 | 2.07 | 14.21 | −0.18 | 194.5 | 215.5 | |
| KWS_Type II_2010 | 6B | 40 | BS00068245_51 | Ex_c17379_1431 | 2.68 | 17.3 | −0.19 | 38.5 | 43.5 | |
| KWS_C_2008 | 7A | 512 | AX-94582130 | Tdurum_contig45618_1089 | 2.15 | 13.98 | 0.40 | 509.5 | 514.5 | |
| KWS_Type II_2010 | 7B | 48 | BobWhite_c44404_312 | Ex_c101666_634 | 2.68 | 31.19 | −0.30 | 38.5 | 54.5 | |
| KWS_Type I_2008 *** | 7D | 150 | Ku_c32426_324 | BS00022610_51 | 2.15 | 18.5 | 0.19 | 143.5 | 154.5 |
| Trait | Specific Trait | Chr1 | Pos1 | LeftMarker1 | RightMarker1 | Chr2 | Pos2 | LeftMarker2 | RightMarker2 | LOD | PVE (%) | Add1 | Add2 | AddbyAdd |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type I resistance | FI_Type_I_2009 | 1B | 160 | AX-94503785 | BobWhite_c48071_144 | 5B | 295 | BobWhite_c17845_132 | AX-94579117 | 5.04 | 6.15 | 0.0507 | 0.0454 | 0.0735 |
| FDK_Type_I_2009 | 1D | 25 | Excalibur_c27873_266 | RAC875_c51493_471 | 7D | 5 | AX-94741998 | BobWhite_c40479_283 | 5.73 | 10.34 | 5.1292 | −7.0357 | −6.5405 | |
| FI_Type_I_2009 | 2A | 120 | wsnp_Ex_rep_c66358_64543089 | Kukri_c33374_1048 | 5B | 170 | Excalibur_rep_c67473_264 | RAC875_c19099_308 | 5.45 | 10.5 | 0.069 | 0.0477 | 0.0837 | |
| FDK_Type_I_2009 | 2A | 375 | wsnp_Ex_c14953_23104041 | RAC875_rep_c69619_78 | 6A | 25 | BS00098857_51 | Kukri_c14679_1082 | 5.34 | 6.43 | −0.0818 | −1.9157 | −5.325 | |
| FI_Type_I_2009 | 2B | 40 | RAC875_c98387_130 | RAC875_c30797_179 | 2B | 120 | Kukri_c18459_2622 | AX-94588421 | 5.39 | 5.24 | −0.0101 | −0.012 | −0.0714 | |
| Sev_Type_I_2009 | 2B | 60 | RAC875_c27650_216 | BS00010988_51 | 3B | 201.2 | Excalibur_c9001_569 | RAC875_c195_499 | 6.04 | 10.83 | 0.0343 | −0.0503 | −0.121 | |
| FDK_Type_I_2009 | 2B | 210 | Tdurum_contig47202_1699 | RAC875_c41476_217 | 2B | 255 | AX-94430027 | RAC875_c22429_249 | 5.74 | 8.96 | −2.4307 | 3.8205 | −12.4406 | |
| FDK_Type_I_2009 | 2D | 205 | Kukri_c33486_128 | Excalibur_c24307_739 | 2D | 380 | AX-94602542 | AX-95115363 | 5.51 | 11.24 | 5.9524 | 4.5552 | 7.3597 | |
| FDK_Type_I_2009 | 2D | 385 | AX-94702227 | BS00086534_51 | 3B | 156.2 | IACX971 | Kukri_c35146_2094 | 5.08 | 5.82 | −0.2105 | 0.7398 | 5.3925 | |
| FI_Type_I_2009 | 5A | 20 | AX-94694404 | wsnp_Ex_c16551_25061517 | 5B | 355 | RAC875_c278_1801 | Excalibur_c48387_58 | 5.11 | 12.31 | 0.0668 | 0.1015 | 0.0997 | |
| FI_Type_I_2009 | 5A | 25 | AX-94694404 | wsnp_Ex_c16551_25061517 | 5D | 125 | Kukri_c13045_302 | IAAV6265 | 5.26 | 12.25 | 0.0327 | 0.0683 | 0.089 | |
| FI_Type_I_2009 | 5B | 325 | Tdurum_contig45588_730 | Excalibur_c8168_226 | 6A | 70 | AX-95180013 | BS00074992_51 | 5.32 | 10.7 | 0.0559 | 0.0674 | 0.1193 | |
| Type II resistance | FI_Type_II_2009 | 5D | 105 | Kukri_c13045_302 | IAAV6265 | 5D | 315 | AX-95190974 | Kukri_rep_c106820_591 | 5.72 | 6.76 | 0.0475 | 0.0648 | 0.0577 |
| FI_Type_II_2009 | 5D | 315 | AX-95190974 | Kukri_rep_c106820_591 | 6D | 90 | TA001847-0566 | D_GB5Y7FA02FHK0M_407 | 6.31 | 6.56 | 0.0708 | −0.0443 | −0.0622 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sgarbi, C.; Malbrán, I.; Saldúa, L.; Lori, G.A.; Lohwasser, U.; Arif, M.A.R.; Börner, A.; Yanniccari, M.; Castro, A.M. Mapping Resistance to Argentinean Fusarium (Graminearum) Head Blight Isolates in Wheat. Int. J. Mol. Sci. 2021, 22, 13653. https://doi.org/10.3390/ijms222413653
Sgarbi C, Malbrán I, Saldúa L, Lori GA, Lohwasser U, Arif MAR, Börner A, Yanniccari M, Castro AM. Mapping Resistance to Argentinean Fusarium (Graminearum) Head Blight Isolates in Wheat. International Journal of Molecular Sciences. 2021; 22(24):13653. https://doi.org/10.3390/ijms222413653
Chicago/Turabian StyleSgarbi, Carolina, Ismael Malbrán, Luciana Saldúa, Gladys Albina Lori, Ulrike Lohwasser, Mian Abdur Rehman Arif, Andreas Börner, Marcos Yanniccari, and Ana Maria Castro. 2021. "Mapping Resistance to Argentinean Fusarium (Graminearum) Head Blight Isolates in Wheat" International Journal of Molecular Sciences 22, no. 24: 13653. https://doi.org/10.3390/ijms222413653
APA StyleSgarbi, C., Malbrán, I., Saldúa, L., Lori, G. A., Lohwasser, U., Arif, M. A. R., Börner, A., Yanniccari, M., & Castro, A. M. (2021). Mapping Resistance to Argentinean Fusarium (Graminearum) Head Blight Isolates in Wheat. International Journal of Molecular Sciences, 22(24), 13653. https://doi.org/10.3390/ijms222413653








