Molecular Characterization of U-box E3 Ubiquitin Ligases (TaPUB2 and TaPUB3) Involved in the Positive Regulation of Drought Stress Response in Arabidopsis
Abstract
:1. Introduction
2. Results
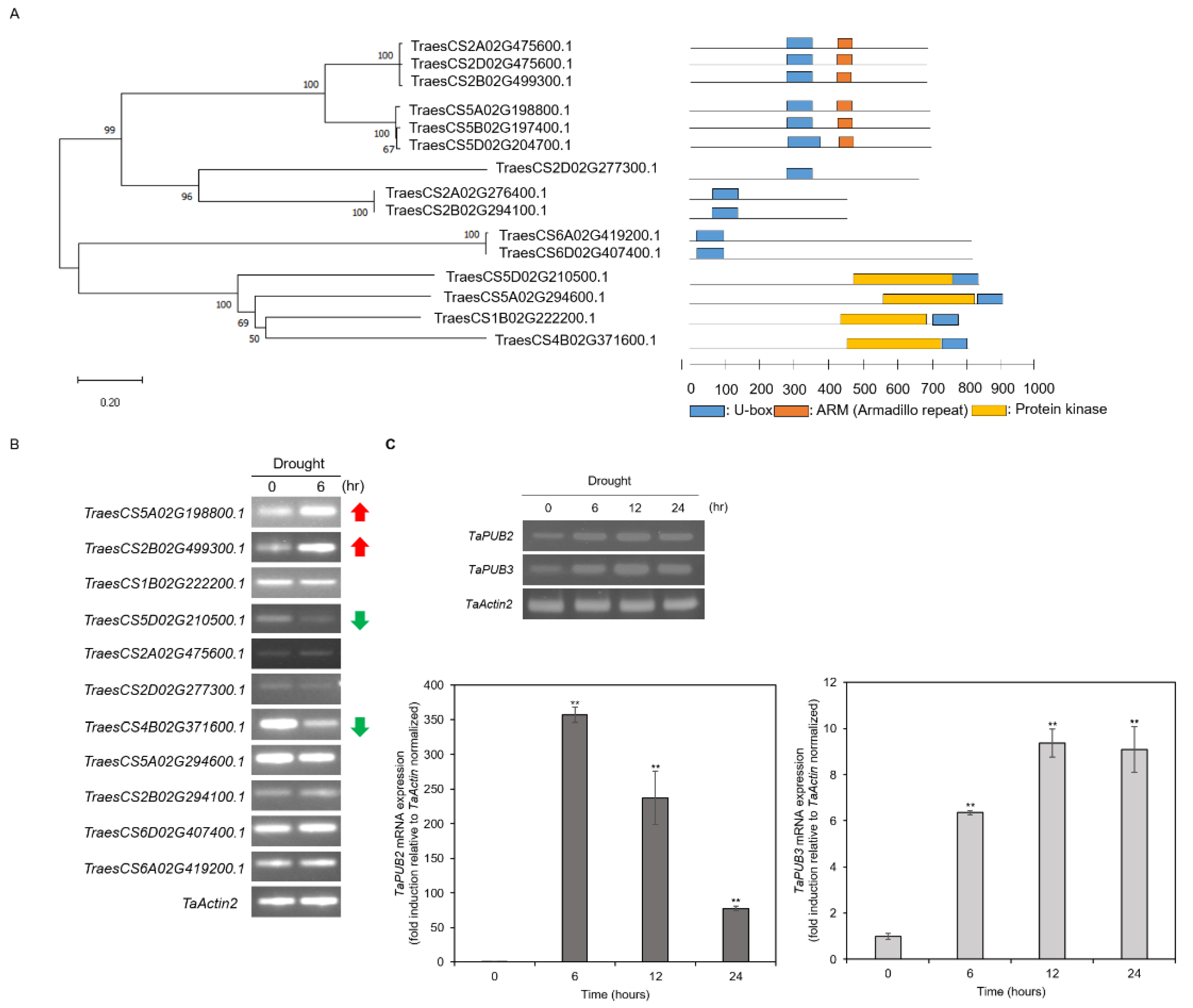
2.1. Isolation of Drought-Induced PUB Genes
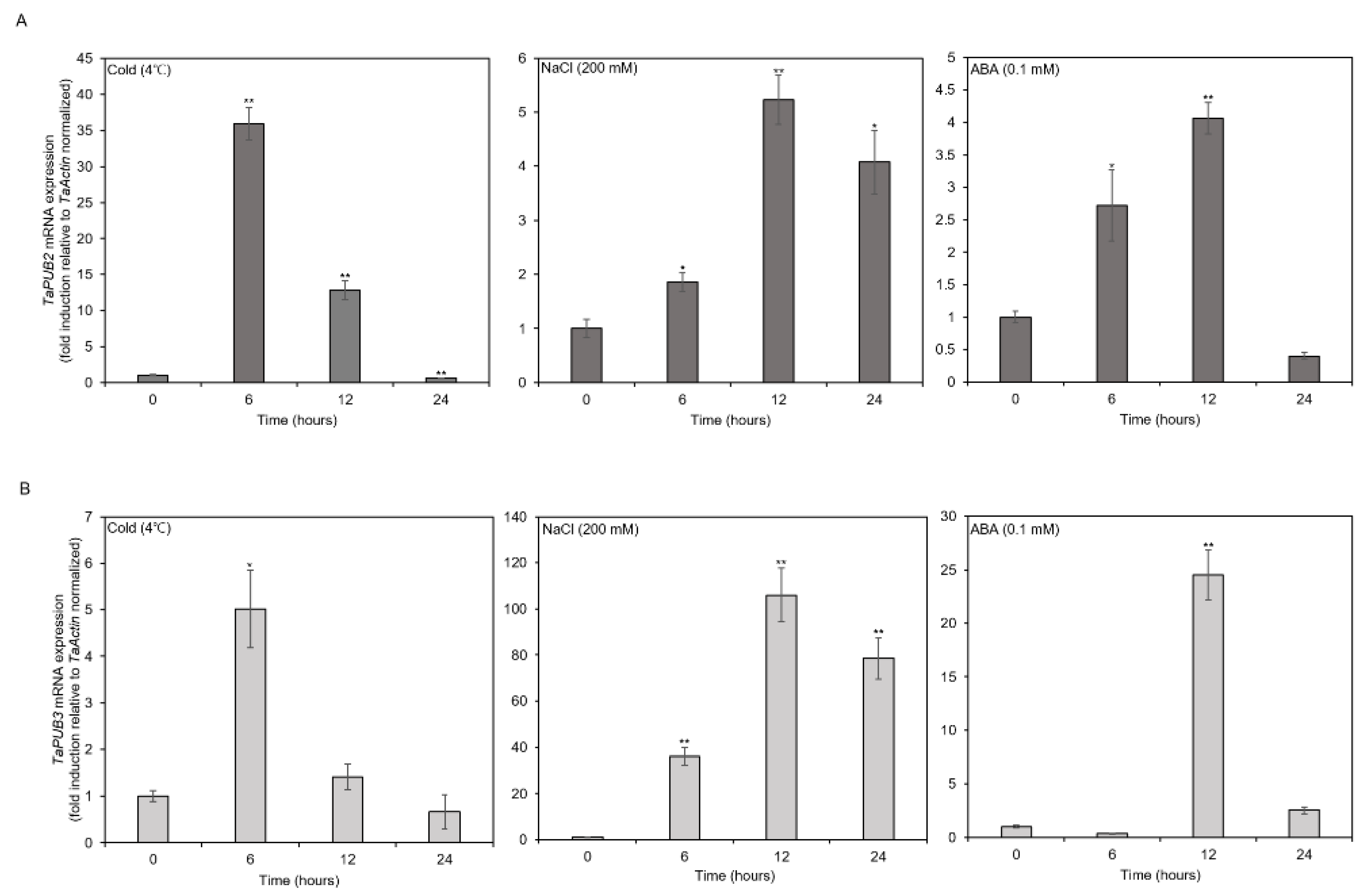
2.2. TaPUB2 and TaPUB3 in Response to Different Abiotic Stresses
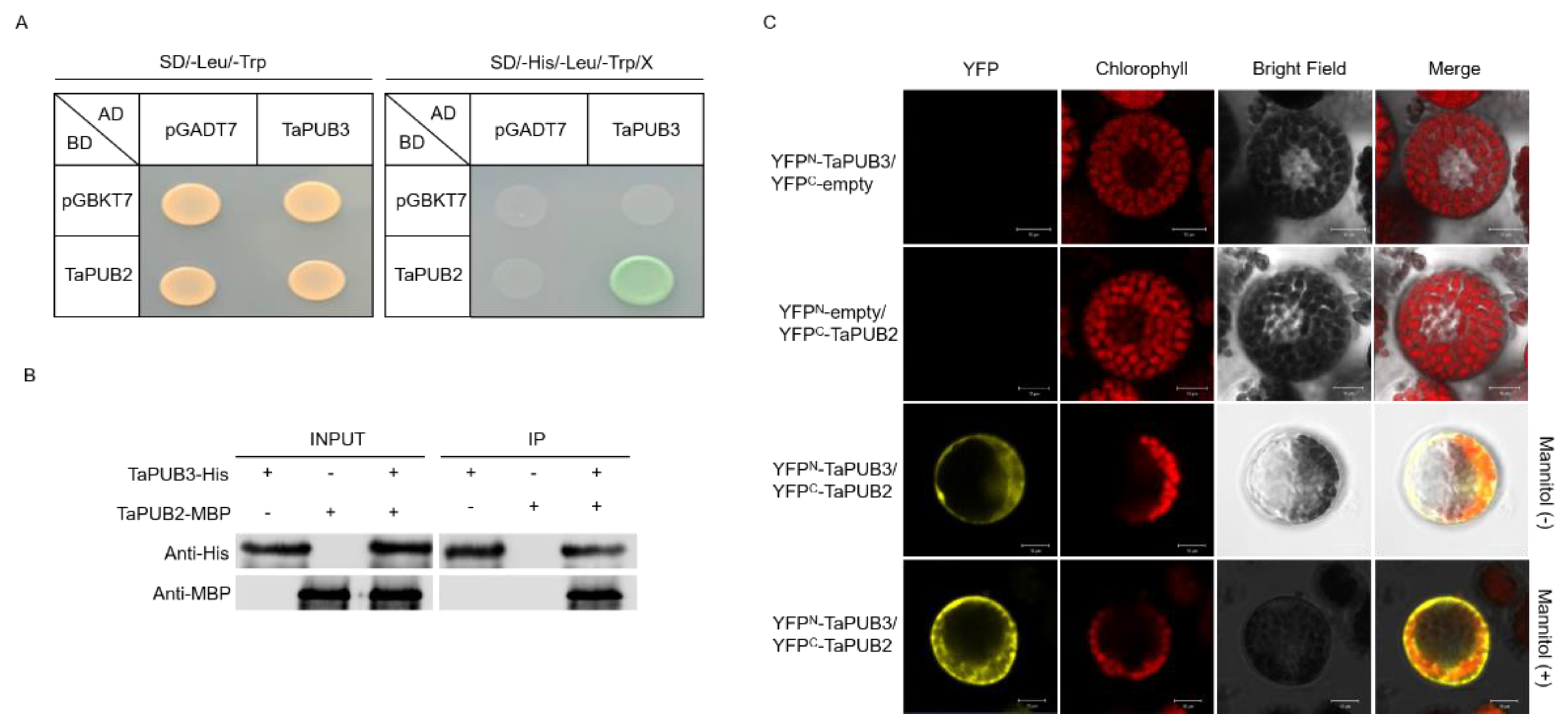
2.3. Subcellular Localization of TaPUB2 and TaPUB3 Proteins in Wheat Protoplast
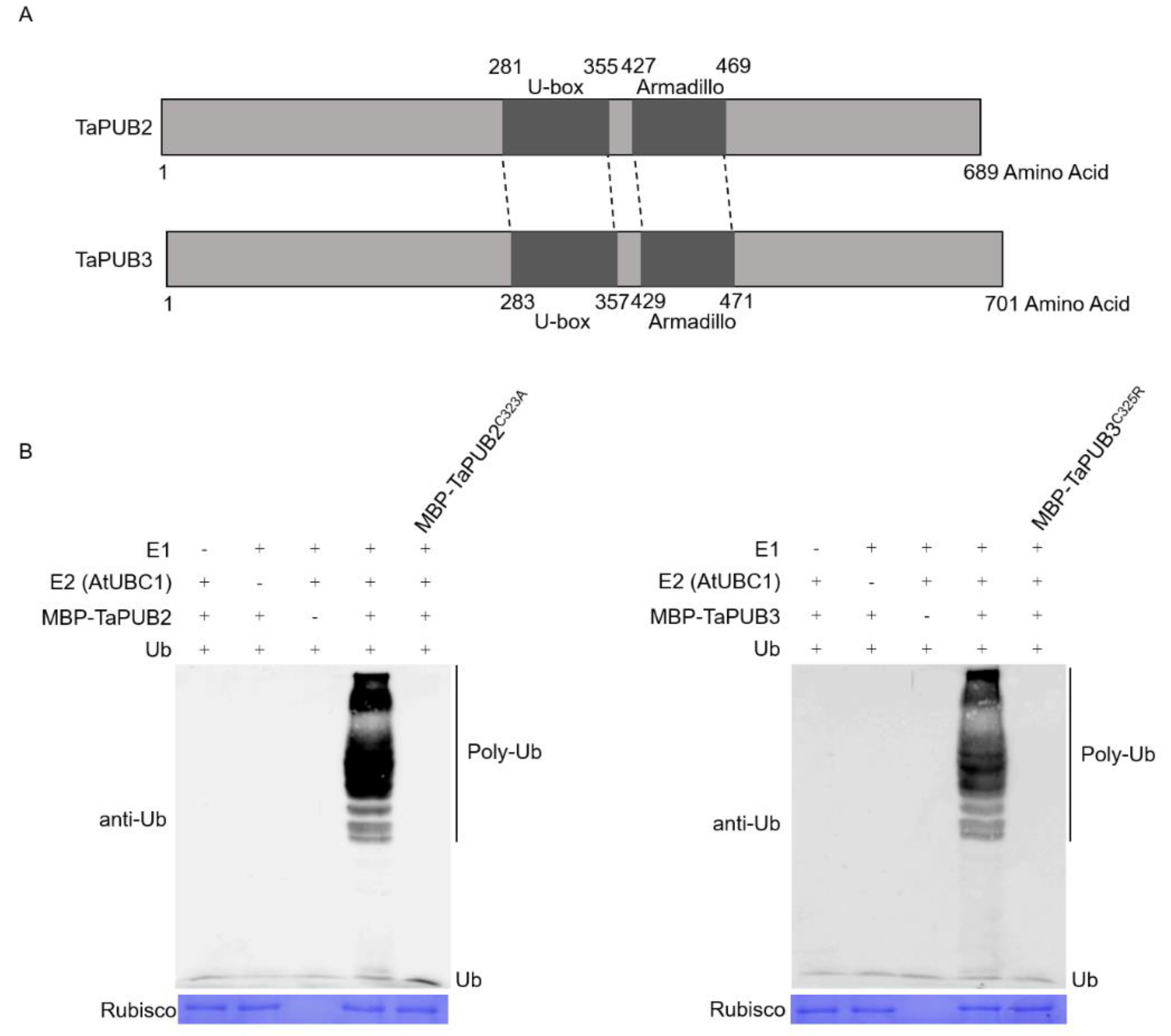
2.4. In Vitro Ubiquitination Assay of TaPUB2 and TaPUB3
2.5. TaPUB2 and TaPUB3 Form Heterodimers
2.6. Overexpression of TaPUB2 and TaPUB3 Confers Tolerance to Drought Stress in Arabidopsis Plants
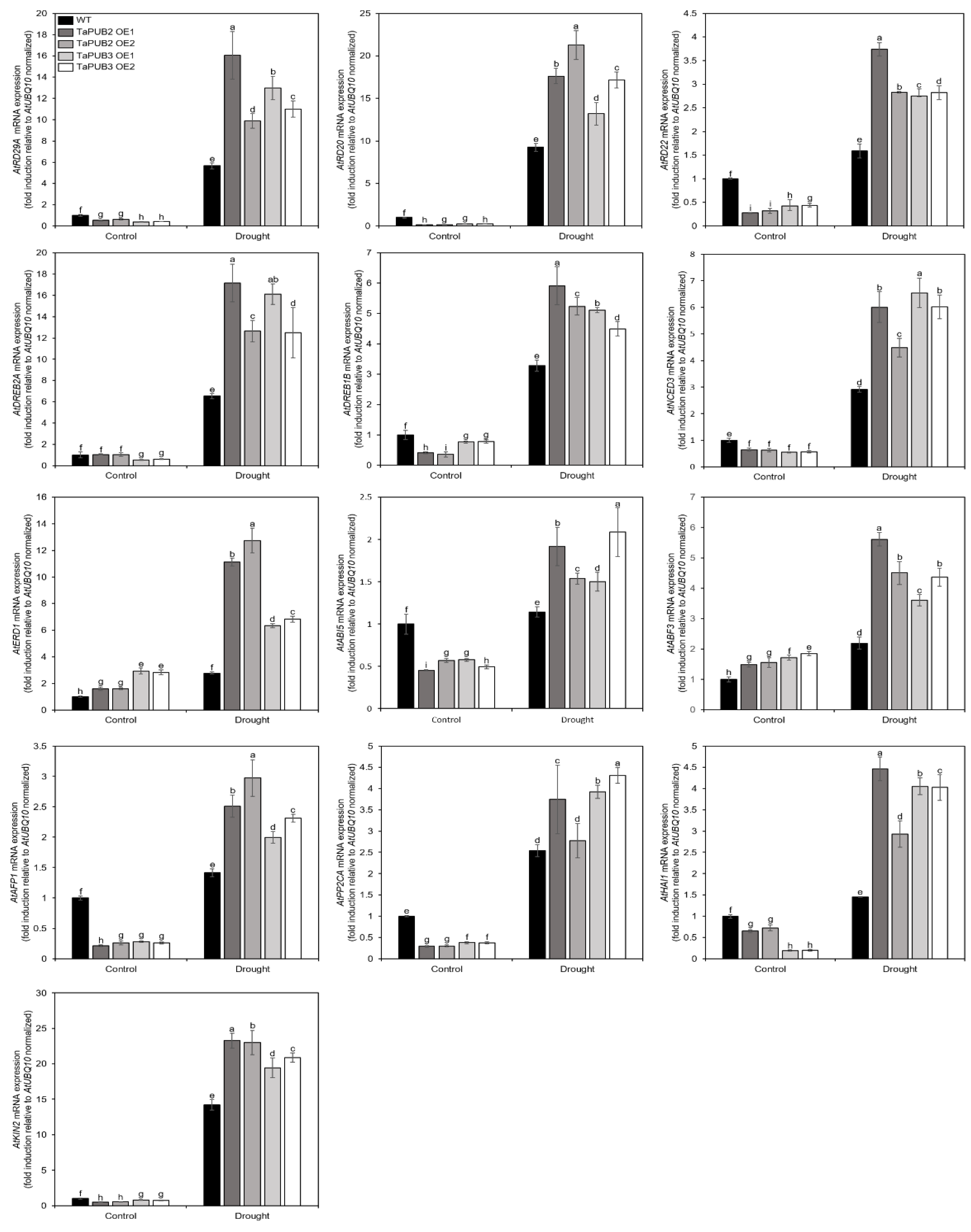
2.7. Transcriptional Analysis of Drought Stress Responsive Genes in TaPUB2 and TaPUB3-Overexpressing Plants
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Gene Cloning and Phylogeny Analysis
4.3. Gene Expression Studies
4.4. Subcellular Localization
4.5. Ubiquitination Assay
4.6. Yeast Two-Hybrid Assay
4.7. Pull-Down Assay
4.8. Generation of Transgenic Arabidopsis Plants
4.9. Dehydration and Drought Tolerance
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vij, S.; Tyagi, A.K. Emerging Trends in the Functional Genomics of the Abiotic Stress Response in Crop Plants. Plant Biotechnol. J. 2007, 5, 361–380. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.S.; Seo, Y.W. BdERF96 Interacts with BdASR1 to Specifically Respond to Drought and Oxidative Stress in Brachypodium Distachyon. J. Plant Biochem. Biotechnol. 2021, 30, 287–296. [Google Scholar] [CrossRef]
- Giraldo, P.; Benavente, E.; Manzano-Agugliaro, F.; Gimenez, E. Worldwide Research Trends on Wheat and Barley: A Bibliometric Comparative Analysis. Agronomy 2019, 9, 352. [Google Scholar] [CrossRef] [Green Version]
- Mwadzingeni, L.; Shimelis, H.; Dube, E.; Laing, M.D.; Tsilo, T.J. Breeding Wheat for Drought Tolerance: Progress and Technologies. J. Integr. Agric. 2016, 15, 935–943. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Cheng, X.; Liu, X.; Wu, H.; Bi, H.; Xu, H. The Wheat MYB Transcription Factor TaMYB31 Is Involved in Drought Stress Responses in Arabidopsis. Front. Plant Sci. 2018, 9, 1426. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Q.; Mao, X.; Li, A.; Jing, R. Wheat Transcription Factor TaAREB3 Participates in Drought and Freezing Tolerances in Arabidopsis. Int. J. Biol. Sci. 2016, 12, 257–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Yin, Y.; Liu, X.; Tong, S.; Xing, J.; Zhang, Y.; Pudake, R.N.; Izquierdo, E.M.; Peng, H.; Xin, M.; et al. The E3 Ligase TaSAP5 Alters Drought Stress Responses by Promoting the Degradation of DRIP Proteins. Plant Physiol. 2017, 175, 1878–1892. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Sun, X.; Yin, S.; Kong, X.; Zhou, S.; Xu, Y.; Luo, Y.; Wang, W. The Role of the F-Box Gene TaFBA1 from Wheat (Triticum aestivum L.) in Drought Tolerance. Plant Physiol. Biochem. 2014, 84, 213–223. [Google Scholar] [CrossRef]
- Friso, G.; Van Wijk, K.J. Posttranslational Protein Modifications in Plant Metabolism. Plant Physiol. 2015, 169, 1469–1487. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.C.; Chapagain, S.; Jang, C.S. A Negative Regulator in Response to Salinity in Rice: Oryza sativa Salt-, ABA- and Drought-Induced Ring Finger Protein 1 (OsSADR1). Plant Cell Physiol. 2018, 59, 575–589. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Hellmann, H. Plant E3 Ligases: Flexible Enzymes in a Sessile World. Mol. Plant 2013, 6, 1388–1404. [Google Scholar] [CrossRef] [Green Version]
- Ohi, M.D.; Vander Kooi, C.W.; Rosenberg, J.A.; Chazin, W.J.; Gould, K.L. Structural Insights into the U-Box, a Domain Associated with Multi-Ubiquitination. Nat. Struct. Biol. 2003, 10, 250–255. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, G.Q.; Kang, H.H.; Zhou, S.M.; Wang, W. TaPUB1, a Putative E3 Ligase Gene from Wheat, Enhances Salt Stress Tolerance in Transgenic Nicotiana Benthamiana. Plant Cell Physiol. 2017, 58, 1673–1688. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Lin, W.; Gao, X.; Wu, S.; Cheng, C.; Avila, J.; Heese, A.; Devarenne, T.P.; He, P.; Shan, L. Direct Ubiquitination of Pattern Recognition Receptor FLS2 Attenuates Plant Innate Immunity. Science 2011, 332, 1439–1442. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.R.; Qu, S.; Bordeos, A.; Yang, C.; Baraoidan, M.; Yan, H.; Xie, Q.; Nahm, B.H.; Leung, H.; Wang, G.L. Spotted leaf11, a Negative Regulator of Plant Cell Death and Defense, Encodes a U-Box/Armadillo Repeat Protein Endowed with E3 Ubiquitin Ligase Activity. Plant Cell 2004, 16, 2795–2808. [Google Scholar] [CrossRef] [Green Version]
- Wiborg, J.; O’Shea, C.; Skriver, K. Biochemical Function of Typical and Variant Arabidopsis thaliana U-Box E3 Ubiquitin-Protein Ligases. Biochem. J. 2008, 413, 447–457. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.R.; Park, C.H.; Venu, R.C.; Gough, J.; Wang, G.L. Classification, Expression Pattern, and E3 Ligase Activity Assay of Rice U-Box-Containing Proteins. Mol. Plant 2008, 1, 800–815. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.Y.; Lee, Y.J.; Hong, M.J.; Kim, J.H.; Seo, Y.W. Genome Wide Analysis of u-box e3 Ubiquitin Ligases in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2021, 22, 2699. [Google Scholar] [CrossRef]
- Cho, S.K.; Chung, H.S.; Ryu, M.Y.; Park, M.J.; Lee, M.M.; Bahk, Y.Y.; Kim, J.; Pai, H.S.; Kim, W.T. Heterologous Expression and Molecular and Cellular Characterization of CaPUB1 Encoding a Hot Pepper U-Box E3 Ubiquitin Ligase Homolog. Plant Physiol. 2006, 142, 1664–1682. [Google Scholar] [CrossRef] [Green Version]
- González-Lamothe, R.; Tsitsigiannis, D.I.; Ludwig, A.A.; Panicot, M.; Shirasu, K.; Jones, J.D.G. The U-Box Protein CMPG1 Is Required for Efficient Activation of Defense Mechanisms Triggered by Multiple Resistance Genes in Tobacco and Tomato. Plant Cell 2006, 18, 1067–1083. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.W.; González-Lamothe, R.; Ewan, R.A.; Rowland, O.; Yoshioka, H.; Shenton, M.; Ye, H.; O’Donnell, E.; Jones, J.D.G.; Sadanandom, A. The E3 Ubiquitin Ligase Activity of Arabidopsis PLANT U-BOX17 and Its Functional Tobacco Homolog ACRE276 Are Required for Cell Death and Defense. Plant Cell 2006, 18, 1084–1098. [Google Scholar] [CrossRef] [Green Version]
- Stone, S.L.; Anderson, E.M.; Mullen, R.T.; Goring, D.R. ARC1 Is an E3 Ubiquitin Ligase and Promotes the Ubiquitination of Proteins During the Rejection of Self-Incompatible Brassica Pollen. Plant Cell 2003, 15, 885–898. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Sherman-Broyles, S.; Nasrallah, M.E.E.; Nasrallah, J.B. A Cryptic Modifier Causing Transient Self-Incompatibility in Arabidopsis thaliana. Curr. Biol. 2007, 17, 734–740. [Google Scholar] [CrossRef]
- Amador, V.; Monte, E.; García-Martínez, J.L.; Prat, S. Gibberellins Signal Nuclear Import of PHOR1, a Photoperiod-Responsive Protein with Homology to Drosophila armadillo. Cell 2001, 106, 343–354. [Google Scholar] [CrossRef]
- Kim, M.; Cho, H.S.; Kim, D.M.; Lee, J.H.; Pai, H.S. CHRK1, a Chitinase-Related Receptor-Like Kinase, Interacts with NtPUB4, an Armadillo Repeat Protein, in Tobacco. Biochim. Biophys. Acta 2003, 1651, 50–59. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, M.; Zhao, Z.; Ren, Y.; Li, Q.; Wang, W. Wheat TaPUB1 Modulates Plant Drought Stress Resistance by Improving Antioxidant Capability. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Byun, M.Y.; Cui, L.H.; Oh, T.K.; Jung, Y.J.; Lee, A.; Park, K.Y.; Kang, B.G.; Kim, W.T. Homologous U-Box E3 Ubiquitin Ligases OsPUB2 and OsPUB3 Are Involved in the Positive Regulation of Low Temperature Stress Response in Rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 16. [Google Scholar] [CrossRef]
- Hashizume, R.; Fukuda, M.; Maeda, I.; Nishikawa, H.; Oyake, D.; Yabuki, Y.; Ogata, H.; Ohta, T. The RING Heterodimer BRCA1-BARD1 Is a Ubiquitin Ligase Inactivated by a Breast Cancer-Derived Mutation. J. Biol. Chem. 2001, 276, 14537–14540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chun, H.J.; Lim, L.H.; Cheong, M.S.; Baek, D.; Park, M.S.; Cho, H.M.; Lee, S.H.; Jin, B.J.; Cha, Y.J.; Lee, Y.B.; et al. Arabidopsis ccoaomt1 Plays a Role in Drought Stress Response via Ros- and Aba-Dependent Manners. Plants 2021, 10, 831. [Google Scholar] [CrossRef]
- Chung, E.; Cho, C.W.; So, H.A.; Kang, J.S.; Chung, Y.S.; Lee, J.H. Overexpression of VrUBC1, a Mung Bean E2 Ubiquitin-Conjugating Enzyme, Enhances Osmotic Stress Tolerance in Arabidopsis. PLoS ONE 2013, 8, e66056. [Google Scholar] [CrossRef] [Green Version]
- Sato, H.; Takasaki, H.; Takahashi, F.; Suzuki, T.; Iuchi, S.; Mitsuda, N.; Ohme-Takagi, M.; Ikeda, M.; Seo, M.; Yamaguchi-Shinozaki, K.; et al. Arabidopsis thaliana NGATHA1 Transcription Factor Induces ABA Biosynthesis by Activating NCED3 Gene During Dehydration Stress. Proc. Natl. Acad. Sci. USA 2018, 115, E11178–E11187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hrmova, M.; Hussain, S.S. Plant transcription factors involved in drought and associated stresses. Int. J. Mol. Sci. 2021, 22, 5662. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dong, C.; Sun, D.; Hu, Y.; Xie, J. Genome-wide identification and analysis of U-box E3 ubiquitin–protein ligase gene family in banana. Int. J. Mol. Sci. 2018, 19, 3874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, H.; Zhu, J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [Green Version]
- Adler, G.; Konrad, Z.; Zamir, L.; Mishra, A.K.; Raveh, D.; Bar-Zvi, D. The Arabidopsis paralogs, PUB46 and PUB48, encoding U-box E3 ubiquitin ligases, are essential for plant response to drought stress. BMC Plant Biol. 2017, 17, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Cruz De Carvalho, M.H. Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signal. Behav. 2008, 3, 156–165. [Google Scholar] [CrossRef] [Green Version]
- Noctor, G.; Foyer, C.H. Ascorbate and Glutathione: Keeping Active Oxygen under Control. Annu. Rev. Plant Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef]
- Chen, C.; Cui, X.; Zhang, P.; Wang, Z.; Zhang, J. Expression of the pyrroline-5-carboxylate reductase (P5CR) gene from the wild grapevine Vitis yeshanensis promotes drought resistance in transgenic Arabidopsis. Plant Physiol. Biochem. 2021, 168, 188–201. [Google Scholar] [CrossRef]
- Tu, M.; Wang, X.; Zhu, Y.; Wang, D.; Zhang, X.; Cui, Y.; Li, Y.; Gao, M.; Li, Z.; Wang, Y.; et al. VlbZIP30 of Grapevine Functions in Dehydration Tolerance via the Abscisic Acid Core Signaling Pathway. Hortic. Res. 2018, 5, 49. [Google Scholar] [CrossRef]
- Xu, J.; Chua, N.H. Dehydration Stress Activates Arabidopsis MPK6 to Signal DCP1 Phosphorylation. EMBO J. 2012, 31, 1975–1984. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Pandey, A.; Pandey, G.K. Role of Plant U-BOX (PUB) Protein in Stress and Role of Plant U-BOX (PUB) Protein in Stress and Development. Plant Stress 2013, 7, 1–9. [Google Scholar]
- Navarro, L.; Zipfel, C.; Rowland, O.; Keller, I.; Robatzek, S.; Boller, T.; Jones, J.D.G. The Transcriptional Innate Immune Response to flg22. Interplay and Overlap with Avr Gene-Dependent Defense Responses and Bacterial Pathogenesis. Plant Physiol. 2004, 135, 1113–1128. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Li, B.; Wang, J.; Chang, X.; Mao, X.; Jing, R. TaPUB15, a U-Box E3 Ubiquitin Ligase Gene from Wheat, Enhances Salt Tolerance in Rice. Food Energy Secur. 2021, 10, e250. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [Green Version]
- Zheng, D.; Song, F. Expression Patterns and Functional Analysis of E3 Ubiquitin Ligase Genes in Rice. ResearchSquare 2021. [Google Scholar] [CrossRef]
- Hatakeyama, S.; Nakayama, K.I.I. U-Box Proteins as a New Family of Ubiquitin Ligases. Biochem. Biophys. Res. Commun. 2003, 302, 635–645. [Google Scholar] [CrossRef]
- Trujillo, M. News from the PUB: Plant U-Box type E3 Ubiquitin Ligases. J. Exp. Bot. 2018, 69, 371–384. [Google Scholar] [CrossRef] [Green Version]
- Koegl, M.; Hoppe, T.; Schlenker, S.; Ulrich, H.D.; Mayer, T.U.; Jentsch, S. A Novel Ubiquitination Factor, E4, Is Involved in Multiubiquitin Chain Assembly. Cell 1999, 96, 635–644. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Dai, Y.; Cui, S.; Ma, L. Histone H2B Monoubiquitination in the Chromatin of Flowering Locus C Regulates Flowering Time in Arabidopsis. Plant Cell 2008, 20, 2586–2602. [Google Scholar] [CrossRef] [Green Version]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A Multicolored Set of In Vivo Organelle Markers for Co-Localization Studies in Arabidopsis and Other Plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, M.S.; Prasad, D.; Jung, W.J.; Seo, Y.W. Molecular Characterization of the Wheat Putative Proline-Rich Protein TaELF7 and Its Involvement in the Negative Regulation of Arabidopsis Flowering. J. Plant Physiol. 2021, 262, 153439. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Mega, X. MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kim, J.H.; Khan, I.U.; Kim, M.S.; Seo, Y.W. Functional characterization of wheat histone H2B monoubiquitination enzyme TaHUB2 in response to vernalization in Keumkang (Triticum aestivum L.). J. Plant Interact. 2021, 16, 93–103. [Google Scholar] [CrossRef]
- Lim, S.D.; Cho, H.Y.; Park, Y.C.; Ham, D.J.; Lee, J.K.; Jang, C.S. The Rice Ring Finger E3 Ligase, OsHCI1, Drives Nuclear Export of Multiple Substrate Proteins and Its Heterogeneous Overexpression Enhances Acquired Thermotolerance. J. Exp. Bot. 2013, 64, 2899–2914. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral Dip: A Simplified Method for Agrobacterium-Mediated Transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.H.; Kim, M.S.; Kim, D.Y.; Amoah, J.N.; Seo, Y.W. Molecular Characterization of U-box E3 Ubiquitin Ligases (TaPUB2 and TaPUB3) Involved in the Positive Regulation of Drought Stress Response in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 13658. https://doi.org/10.3390/ijms222413658
Kim JH, Kim MS, Kim DY, Amoah JN, Seo YW. Molecular Characterization of U-box E3 Ubiquitin Ligases (TaPUB2 and TaPUB3) Involved in the Positive Regulation of Drought Stress Response in Arabidopsis. International Journal of Molecular Sciences. 2021; 22(24):13658. https://doi.org/10.3390/ijms222413658
Chicago/Turabian StyleKim, Jae Ho, Moon Seok Kim, Dae Yeon Kim, Joseph Noble Amoah, and Yong Weon Seo. 2021. "Molecular Characterization of U-box E3 Ubiquitin Ligases (TaPUB2 and TaPUB3) Involved in the Positive Regulation of Drought Stress Response in Arabidopsis" International Journal of Molecular Sciences 22, no. 24: 13658. https://doi.org/10.3390/ijms222413658




