Genome-Wide Identification and Characterization of Calcium-Dependent Protein Kinase (CDPK) and CDPK-Related Kinase (CRK) Gene Families in Medicago truncatula
Abstract
1. Introduction
2. Results
2.1. Identification of MtCRK Genes
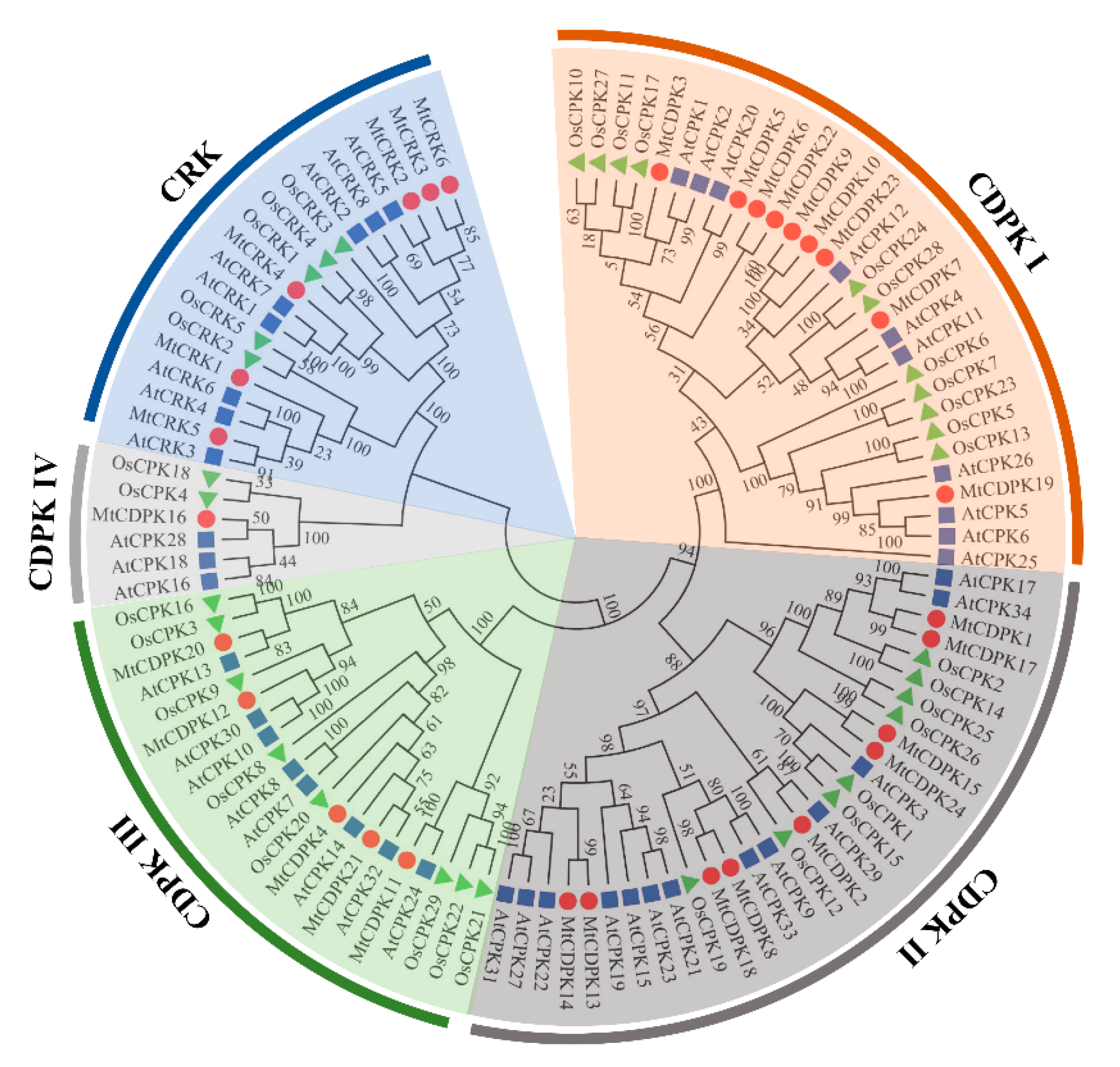
2.2. Phylogenetic and Chromosomal Distribution of MtCDPKs and MtCRKs
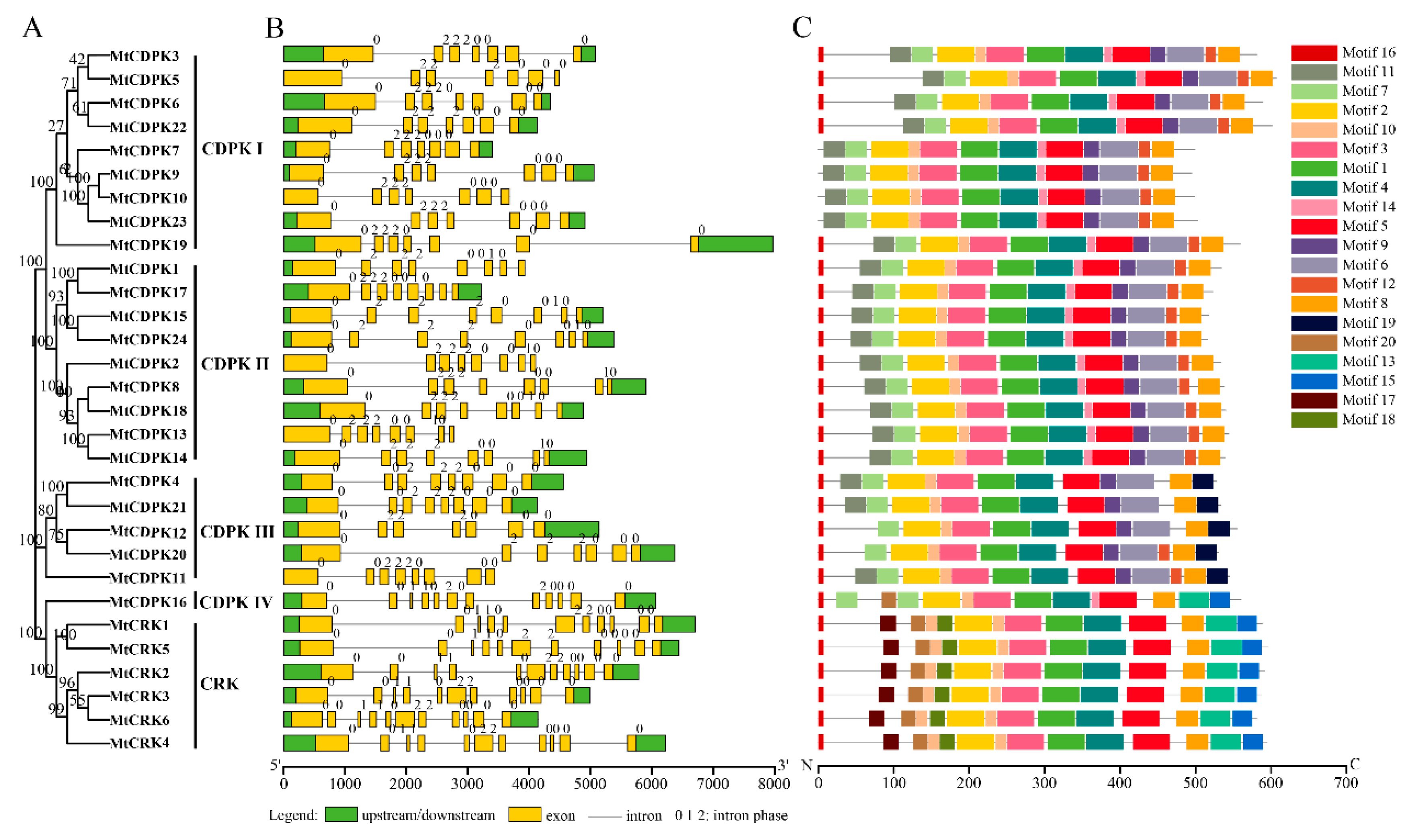
2.3. Analysis of Gene Structure and Conserved Motifs of MtCDPKs and MtCRKs
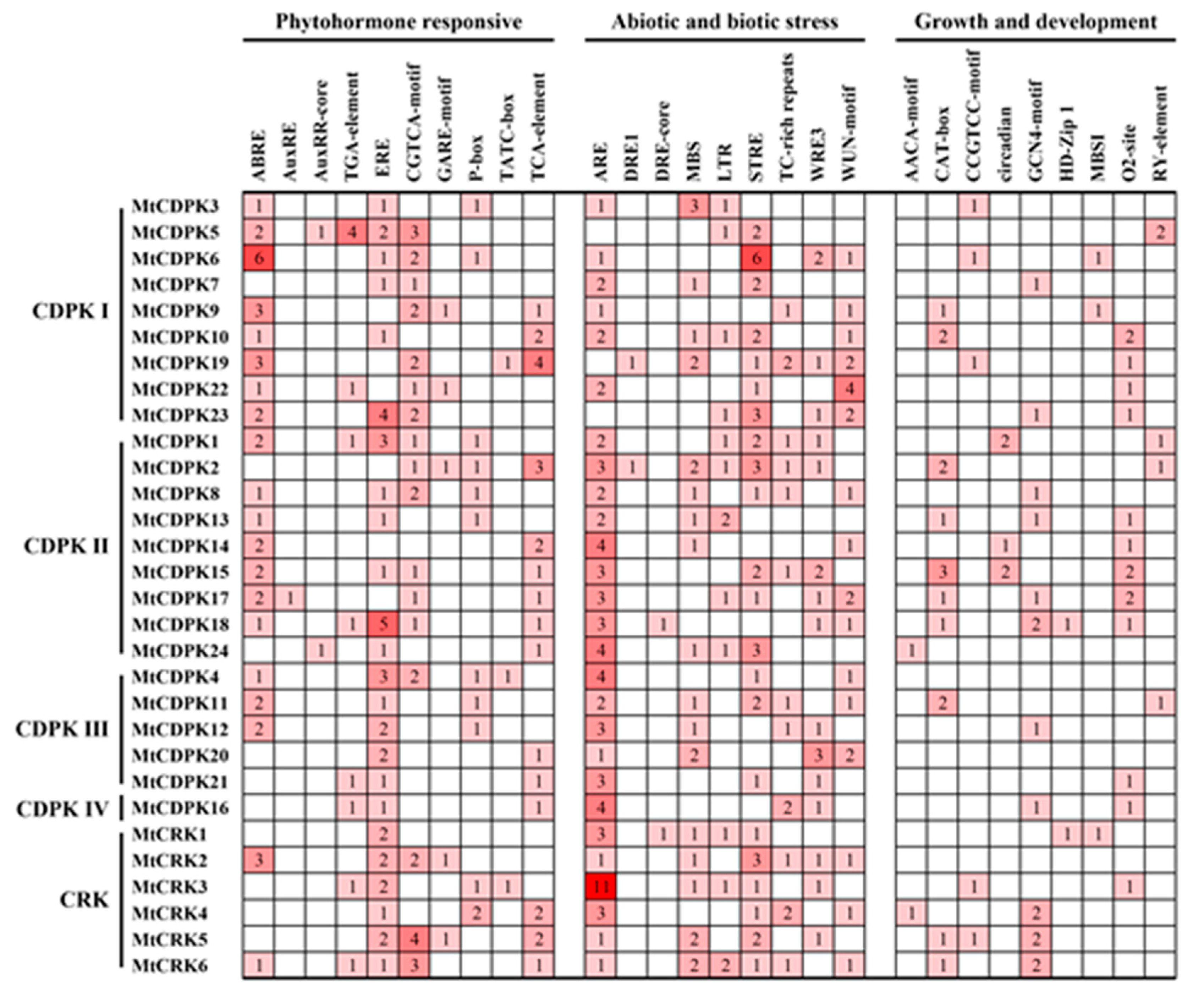
2.4. Cis-Acting Elements in the Promoter Region of MtCDPKs and MtCRKs
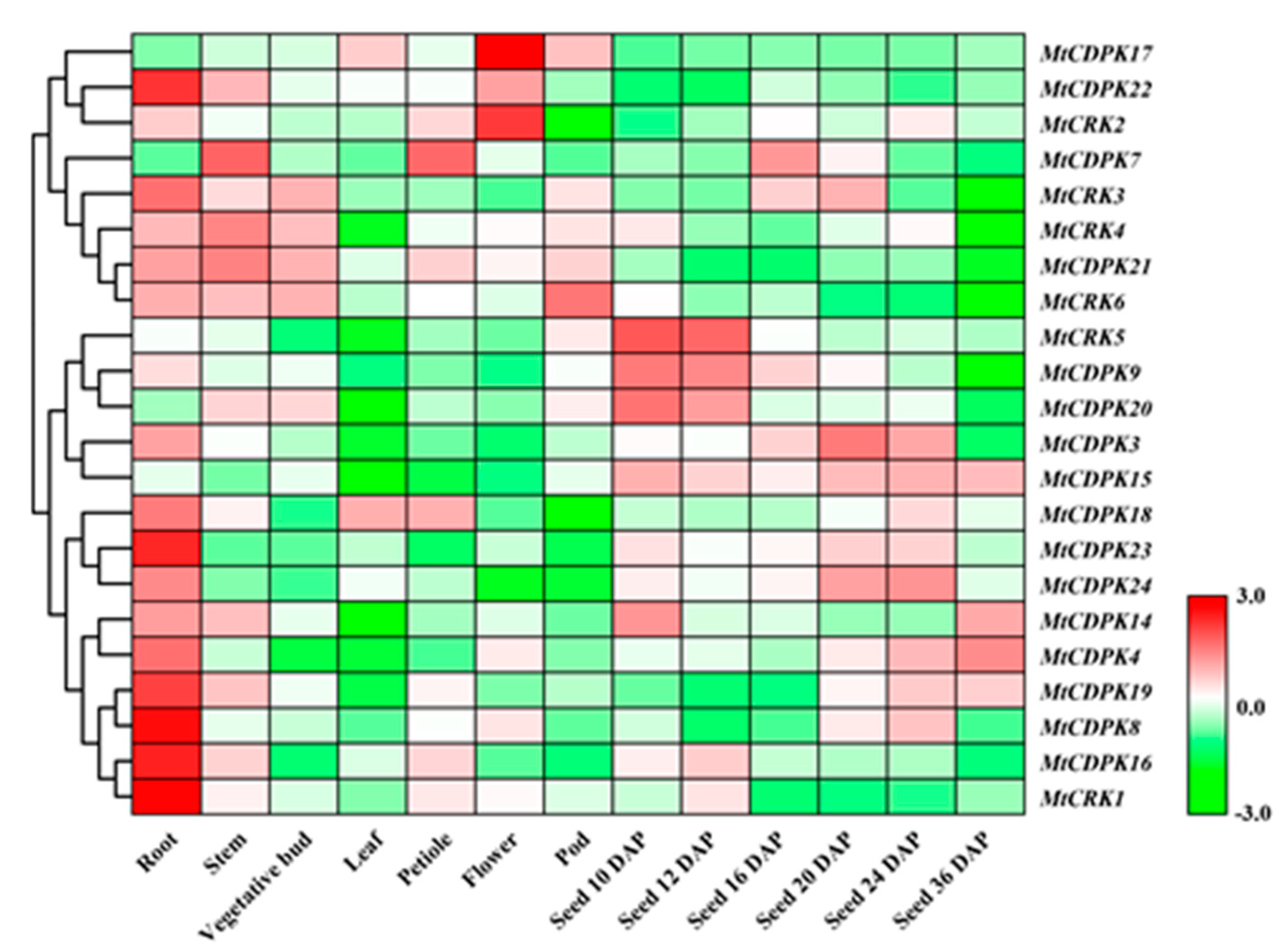
2.5. Spatial Expression Profiles of MtCDPK and MtCRK Genes
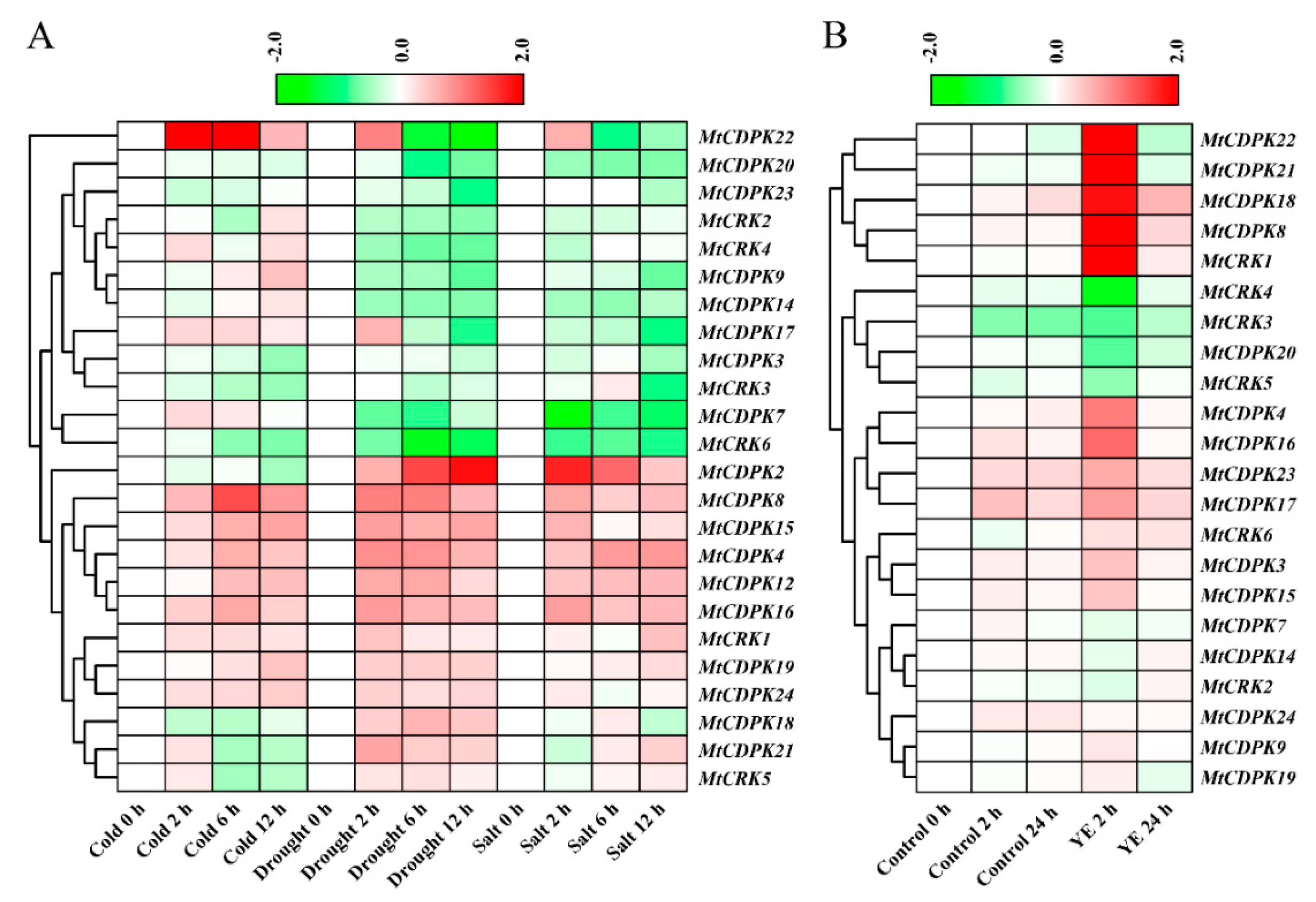
2.6. Expression Patterns of MtCDPK and MtCRK Genes in Response to Abiotic and Biotic Stresses
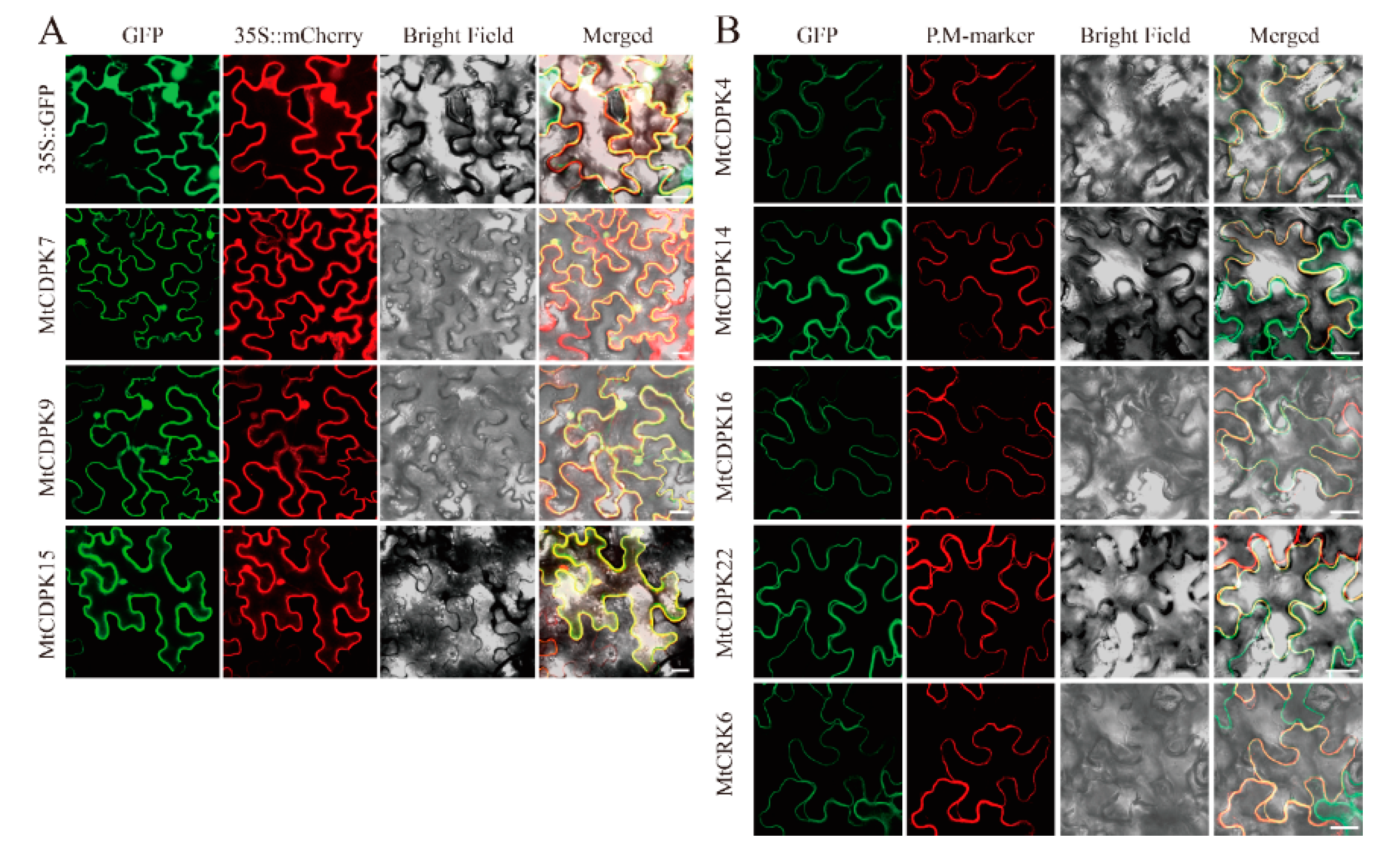
2.7. Subcellular Localization of Selected MtCDPKs and MtCRKs
3. Discussion
4. Materials and Methods
4.1. Identification of MtCRK Genes
4.2. Sequence Analysis of MtCDPKs and MtCRKs
4.3. Multiple Sequence Alignments and Phylogenetic Analysis
4.4. Gene Structure and Conserved Motifs Analysis
4.5. Chromosomal Location and Synteny Correlation Analysis
4.6. Calculation of Ka/Ks Ratios of Paralogous Gene Pairs of MtCDPKs and MtCRKs
4.7. Analysis of Cis-Acting Regulatory Elements in Promoter Regions
4.8. Analysis of Expression Profiles of MtCDPKs and MtCRKs
4.9. Analysis of Subcellular Localization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Trewavas, A.J.; Malho, R. Ca2+ signalling in plant cells: The big network! Curr. Opin. Plant Biol. 1998, 1, 428–433. [Google Scholar] [CrossRef]
- Dodd, A.N.; Kudla, J.; Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar] [CrossRef] [PubMed]
- Marcec, M.J.; Gilroy, S.; Poovaiah, B.W.; Tanaka, K. Mutual interplay of Ca(2+) and ROS signaling in plant immune response. Plant Sci. 2019, 283, 343–354. [Google Scholar] [CrossRef]
- Tian, W.; Wang, C.; Gao, Q.; Li, L.; Luan, S. Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat. Plants 2020, 6, 750–759. [Google Scholar] [CrossRef]
- Batistic, O.; Kudla, J. Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta 2012, 1820, 1283–1293. [Google Scholar] [CrossRef]
- Schulz, P.; Herde, M.; Romeis, T. Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol. 2013, 163, 523–530. [Google Scholar] [CrossRef]
- Harper, J.F.; Breton, G.; Harmon, A. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef]
- Valmonte, G.R.; Arthur, K.; Higgins, C.M.; MacDiarmid, R.M. Calcium-dependent protein kinases in plants: Evolution, expression and function. Plant Cell Physiol. 2014, 55, 551–569. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Harmon, A.C.; Gribskov, M.; Gubrium, E.; Harper, J.F. The CDPK superfamily of protein kinases. New Phytol. 2001, 151, 175–183. [Google Scholar] [CrossRef]
- Resh, M.D. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2006, 2, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Hamamoto, S.; Moriya, K.; Matsuura, A.; Sato, Y.; Muto, J.; Noguchi, H.; Yamauchi, S.; Tozawa, Y.; Ueda, M.; et al. N-myristoylation and S-acylation are common modifications of Ca2+-regulated Arabidopsis kinases and are required for activation of the SLAC1 anion channel. New Phytol. 2018, 218, 1504–1521. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.N.; Shen, L.K.; Zhang, W.Z.; Zhang, W.; Wang, Y.; Wu, W.H. Ca2+-dependent protein kinase11 and 24 modulate the activity of the inward rectifying K+ channels in Arabidopsis pollen tubes. Plant Cell 2013, 25, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhao, S.; Tian, H.; He, Y.; Xiong, W.; Guo, L.; Wu, Y. CPK3-phosphorylated RhoGDI1 is essential in the development of Arabidopsis seedlings and leaf epidermal cells. J. Exp. Bot. 2013, 64, 3327–3338. [Google Scholar] [CrossRef]
- Matschi, S.; Werner, S.; Schulze, W.X.; Legen, J.; Hilger, H.H.; Romeis, T. Function of calcium-dependent protein kinase CPK28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J. 2013, 73, 883–896. [Google Scholar] [CrossRef]
- Vijayakumar, P.; Datta, S.; Dolan, L. ROOT HAIR DEFECTIVE SIX-LIKE4 (RSL4) promotes root hair elongation by transcriptionally regulating the expression of genes required for cell growth. New Phytol. 2016, 212, 944–953. [Google Scholar] [CrossRef]
- Rigo, G.; Ayaydin, F.; Tietz, O.; Zsigmond, L.; Kovacs, H.; Pay, A.; Salchert, K.; Darula, Z.; Medzihradszky, K.F.; Szabados, L.; et al. Inactivation of plasma membrane-localized CDPK-RELATED KINASE5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis. Plant Cell 2013, 25, 1592–1608. [Google Scholar] [CrossRef]
- Baba, A.I.; Rigo, G.; Ayaydin, F.; Rehman, A.U.; Andrasi, N.; Zsigmond, L.; Valkai, I.; Urbancsok, J.; Vass, I.; Pasternak, T.; et al. Functional analysis of the Arabidopsis thaliana CDPK-related kinase family: AtCRK1 regulates responses to continuous light. Int. J. Mol. Sci. 2018, 19, 1282. [Google Scholar] [CrossRef]
- Kawamoto, N.; Endo, M.; Araki, T. Expression of a kinase-dead form of CPK33 involved in florigen complex formation causes delayed flowering. Plant Signal Behav. 2015, 10, e1086856. [Google Scholar] [CrossRef]
- Morello, L.; Frattini, M.; Giani, S.; Christou, P.; Breviario, D. Overexpression of the calcium-dependent protein kinase OsCDPK2 in transgenic rice is repressed by light in leaves and disrupts seed development. Transgenic Res. 2000, 9, 453–462. [Google Scholar] [CrossRef]
- Durian, G.; Sedaghatmehr, M.; Matallana-Ramirez, L.P.; Schilling, S.M.; Schaepe, S.; Guerra, T.; Herde, M.; Witte, C.P.; Mueller-Roeber, B.; Schulze, W.X.; et al. Calcium-dependent protein kinase CPK1 controls cell death by in vivo phosphorylation of senescence master regulator ORE1. Plant Cell 2020, 32, 1610–1625. [Google Scholar] [CrossRef] [PubMed]
- Edel, K.H.; Kudla, J. Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 2016, 33, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.F.; Liu, D.Y.; Yang, B.; Liu, W.Z.; Mu, B.B.; Song, H.X.; Chen, B.Y.; Li, Y.; Ren, D.T.; Deng, H.Q.; et al. Arabidopsis CPK6 positively regulates ABA signaling and drought tolerance through phosphorylating ABA-responsive element-binding factors. J. Exp. Bot. 2020, 71, 188–203. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.J.; Li, X.D.; Ratnasekera, D.; Wang, C.; Liu, W.X.; Song, L.F.; Zhang, W.Z.; Wu, W.H. Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated Signaling and H2O2 Homeostasis in Stomatal Guard Cells under Drought Stress. Plant Cell 2015, 27, 1445–1460. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Hayashi, N.; Kobayashi, M.; Aoki, N.; Miyao, A.; Mitsuhara, I.; Ichikawa, H.; Komatsu, S.; Hirochika, H.; Kikuchi, S.; et al. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 2012, 69, 26–36. [Google Scholar] [CrossRef]
- Latz, A.; Mehlmer, N.; Zapf, S.; Mueller, T.D.; Wurzinger, B.; Pfister, B.; Csaszar, E.; Hedrich, R.; Teige, M.; Becker, D. Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol. Plant 2013, 6, 1274–1289. [Google Scholar] [CrossRef]
- Almadanim, M.C.; Alexandre, B.M.; Rosa, M.T.G.; Sapeta, H.; Leitao, A.E.; Ramalho, J.C.; Lam, T.T.; Negrao, S.; Abreu, I.A.; Oliveira, M.M. Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant Cell Environ. 2017, 40, 1197–1213. [Google Scholar] [CrossRef]
- Wang, S.; Tao, Y.; Zhou, Y.L.; Niu, J.; Shu, Y.J.; Yu, X.W.; Liu, S.S.; Chen, M.; Gu, W.H.; Ma, H. Translationally controlled tumor protein GmTCTP interacts with GmCDPKSK5 in response to high temperature and humidity stress during soybean seed development. Plant Growth Regul. 2017, 82, 187–200. [Google Scholar] [CrossRef]
- Tao, X.; Lu, Y. Loss of AtCRK1 gene function in Arabidopsis thaliana decreases tolerance to salt. J. Plant Biol. 2013, 56, 306–314. [Google Scholar] [CrossRef]
- Liu, H.T.; Gao, F.; Li, G.L.; Han, J.L.; Liu, D.L.; Sun, D.Y.; Zhou, R.G. The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. Plant J. 2008, 55, 760–773. [Google Scholar] [CrossRef]
- Coca, M.; Segundo, B. AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J. 2010, 63, 526–540. [Google Scholar] [CrossRef] [PubMed]
- Gravino, M.; Savatin, D.V.; Macone, A.; De Lorenzo, G. Ethylene production in Botrytis cinerea- and oligogalacturonide-induced immunity requires calcium-dependent protein kinases. Plant J. 2015, 84, 1073–1086. [Google Scholar] [CrossRef] [PubMed]
- Dubiella, U.; Seybold, H.; Durian, G.; Komander, E.; Lassig, R.; Witte, C.P.; Schulze, W.X.; Romeis, T. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 2013, 110, 8744–8749. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.P.; Xu, Y.P.; Munyampundu, J.P.; Liu, T.Y.; Cai, X.Z. Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: Genome-wide identification and functional analyses in disease resistance. Mol. Genet Genomics 2016, 291, 661–676. [Google Scholar] [CrossRef]
- Ivashuta, S.; Liu, J.; Liu, J.; Lohar, D.P.; Haridas, S.; Bucciarelli, B.; VandenBosch, K.A.; Vance, C.P.; Harrison, M.J.; Gantt, J.S. RNA interference identifies a calcium-dependent protein kinase involved in Medicago truncatula root development. Plant Cell 2005, 17, 2911–2921. [Google Scholar] [CrossRef]
- Gargantini, P.R.; Gonzalez-Rizzo, S.; Chinchilla, D.; Raices, M.; Giammaria, V.; Ulloa, R.M.; Frugier, F.; Crespi, M.D. A CDPK isoform participates in the regulation of nodule number in Medicago truncatula. Plant J. 2006, 48, 843–856. [Google Scholar] [CrossRef]
- Yu, H.; Xiao, A.; Dong, R.; Fan, Y.; Zhang, X.; Liu, C.; Wang, C.; Zhu, H.; Duanmu, D.; Cao, Y.; et al. Suppression of innate immunity mediated by the CDPK-Rboh complex is required for rhizobial colonization in Medicago truncatula nodules. New Phytol. 2018, 220, 425–434. [Google Scholar] [CrossRef]
- Delormel, T.Y.; Boudsocq, M. Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 2019, 224, 585–604. [Google Scholar] [CrossRef]
- Vighi, I.L.; Crizel, R.L.; Perin, E.C.; Rombaldi, C.V.; Galli, V. Crosstalk During Fruit Ripening and Stress Response Among Abscisic Acid, Calcium-Dependent Protein Kinase and Phenylpropanoid. Crit. Rev. Plant Sci. 2019, 38, 99–116. [Google Scholar] [CrossRef]
- Gong, J.; Zhang, Y.; Li, F.; He, X.; Zhang, X.; Yi, Y. Bioinformation anlysis of Calcium-dependent protein kinase family of Medicago truncatula. Mol. Plant Breed. 2018, 9, 26–35. [Google Scholar]
- Asano, T.; Tanaka, N.; Yang, G.; Hayashi, N.; Komatsu, S. Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: Comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 2005, 46, 356–366. [Google Scholar] [CrossRef]
- Zuo, R.; Hu, R.; Chai, G.; Xu, M.; Qi, G.; Kong, Y.; Zhou, G. Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar (Populus trichocarpa). Mol. Biol. Rep. 2013, 40, 2645–2662. [Google Scholar] [CrossRef]
- Wu, P.; Wang, W.; Duan, W.; Li, Y.; Hou, X. Comprehensive analysis of the CDPK-SnRK superfamily genes in Chinese cabbage and its evolutionary implications in plants. Front. Plant Sci. 2017, 8, 162. [Google Scholar] [CrossRef] [PubMed]
- Rival, P.; de Billy, F.; Bono, J.J.; Gough, C.; Rosenberg, C.; Bensmihen, S. Epidermal and cortical roles of NFP and DMI3 in coordinating early steps of nodulation in Medicago truncatula. Development 2012, 139, 3383–3391. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Che, Z.; Zeng, X.; Zhou, X.; Sitoe, H.M.; Wang, H.; Yu, D. Genome-wide analysis of calcium-dependent protein kinases and their expression patterns in response to herbivore and wounding stresses in soybean. Funct. Integr. Genomics 2016, 16, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.F.; Wei, C.H.; Yang, X.Z.; Chen, H.J.; Yang, Y.C.; Mo, Y.L.; Li, H.; Zhang, Y.; Ma, J.X.; Yang, J.Q.; et al. Genome-wide identification and expression analysis of calcium-dependent protein kinase and its related kinase gene families in melon (Cucumis melo L.). PLoS ONE 2017, 12, e0176352. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Zhang, R.; Yang, X.; Zhu, C.; Li, H.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X. Comparative analysis of calcium-dependent protein kinase in Cucurbitaceae and expression studies in watermelon. Int. J. Mol. Sci. 2019, 20, 2527. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Freeling, M.; Tang, H.; Wang, X. Insights from the comparison of plant genome sequences. Annu. Rev. Plant Biol. 2010, 61, 349–372. [Google Scholar] [CrossRef]
- Young, N.D.; Debelle, F.; Oldroyd, G.E.D.; Geurts, R.; Cannon, S.B.; Udvardi, M.K.; Benedito, V.A.; Mayer, K.F.X.; Gouzy, J.; Schoof, H.; et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 2011, 480, 520–524. [Google Scholar] [CrossRef]
- Hu, Z.; Lv, X.; Xia, X.; Zhou, J.; Shi, K.; Yu, J.; Zhou, Y. Genome-wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front. Plant Sci. 2016, 7, 469. [Google Scholar] [CrossRef]
- Myers, C.; Romanowsky, S.M.; Barron, Y.D.; Garg, S.; Azuse, C.L.; Curran, A.; Davis, R.M.; Hatton, J.; Harmon, A.C.; Harper, J.F. Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J. 2009, 59, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, J.; Matschi, S.; Shorinola, O.; Rovenich, H.; Matei, A.; Segonzac, C.; Malinovsky, F.G.; Rathjen, J.P.; MacLean, D.; Romeis, T.; et al. The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 2014, 16, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Li, C.L.; Wang, M.; Wu, X.M.; Chen, D.H.; Lv, H.J.; Shen, J.L.; Qiao, Z.; Zhang, W. THI1, a thiamine thiazole synthase, interacts with Ca2+-dependent protein kinase CPK33 and modulates the S-Type anion channels and stomatal closure in Arabidopsis. Plant Physiol. 2016, 170, 1090–1104. [Google Scholar] [CrossRef]
- Huang, K.; Peng, L.; Liu, Y.; Yao, R.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. Arabidopsis calcium-dependent protein kinase AtCPK1 plays a positive role in salt/drought-stress response. Biochem. Biophys. Res. Commun. 2018, 498, 92–98. [Google Scholar] [CrossRef]
- Hemsley, P.A. The importance of lipid modified proteins in plants. New Phytol. 2015, 205, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhong, S.; Guo, X.; Hao, L.; Wei, X.; Huang, Q.; Hou, Y.; Shi, J.; Wang, C.; Gu, H.; et al. Membrane-bound RLCKs LIP1 and LIP2 are essential male factors controlling male-female attraction in Arabidopsis. Curr. Biol. 2013, 23, 993–998. [Google Scholar] [CrossRef] [PubMed]
- Hurst, C.H.; Hemsley, P.A. Current perspective on protein S-acylation in plants: More than just a fatty anchor? J. Exp. Bot. 2015, 66, 1599–1606. [Google Scholar] [CrossRef]
- Simeunovic, A.; Mair, A.; Wurzinger, B.; Teige, M. Know where your clients are: Subcellular localization and targets of calcium-dependent protein kinases. J. Exp. Bot. 2016, 67, 3855–3872. [Google Scholar] [CrossRef]
- Zheng, L.; Liu, P.; Liu, Q.; Wang, T.; Dong, J. Dynamic protein s-acylation in plants. Int. J. Mol. Sci. 2019, 20, 560. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, M.; Ke, K.; Lu, Y.T. Cellular localization and biochemical characterization of a novel calcium-dependent protein kinase from tobacco. Cell Res. 2005, 15, 604–612. [Google Scholar] [CrossRef]
- Galbiati, F.; Guzzi, F.; Magee, A.I.; Milligan, G.; Parenti, M. N-terminal fatty acylation of the alpha-subunit of the G-protein G(I)1-only the myristoylated protein is a substrate for palmitoylation. Biochem. J. 1994, 303, 697–700. [Google Scholar] [CrossRef] [PubMed]
- Resh, M.D. Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. BBA-Mol. Cell Res. 1999, 1451, 1–16. [Google Scholar] [CrossRef]
- Hemsley, P.A.; Weimar, T.; Lilley, K.S.; Dupree, P.; Grierson, C.S. A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytol. 2013, 197, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Aicart-Ramos, C.; Valero, R.A.; Rodriguez-Crespo, I. Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta 2011, 1808, 2981–2994. [Google Scholar] [CrossRef]
- Duan, M.; Zhang, R.X.; Zhu, F.G.; Zhang, Z.Q.; Gou, L.M.; Wen, J.Q.; Dong, J.L.; Wang, T. A lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase I expression during drought stress. Plant Cell 2017, 29, 1748–1772. [Google Scholar] [CrossRef]
- Liu, K.H.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Sun Chung, H.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2020, 49, D458–D460. [Google Scholar] [CrossRef]
- Xie, Y.; Zheng, Y.; Li, H.; Luo, X.; He, Z.; Cao, S.; Shi, Y.; Zhao, Q.; Xue, Y.; Zuo, Z.; et al. GPS-Lipid: A robust tool for the prediction of multiple lipid modification sites. Sci. Rep. 2016, 6, 28249. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Benedito, V.A.; Torres-Jerez, I.; Murray, J.D.; Andriankaja, A.; Allen, S.; Kakar, K.; Wandrey, M.; Verdier, J.; Zuber, H.; Ott, T.; et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008, 55, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Mo, X.; Yang, H.; Yue, L.; Song, J.; Mo, B. The U-box family genes in Medicago truncatula: Key elements in response to salt, cold, and drought stresses. PLoS ONE 2017, 12, e0182402. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene ID | Chromosome Location | No. of aa | MW (kDa) | pI | GRAVY | No. of EF Hands | N-Acylation Prediction (No.) |
|---|---|---|---|---|---|---|---|---|
| MtCDPK1 | Medtr0028s0170.1 | scaffold0028:72630..76559 | 534 | 59.85 | 5.65 | −0.542 | 4 | N-Myr (1)–Palm (2) |
| MtCDPK2 | Medtr1g026190.1 | chr1:8404402..8408507 | 533 | 60.47 | 6.21 | −0.591 | 4 | N-Myr (1)–Palm (1) |
| MtCDPK3 | Medtr1g041150.1 | chr1:18398807..18403890 | 581 | 65.08 | 5.37 | −0.376 | 4 | N-Myr (1)–Palm (1) |
| MtCDPK4 | Medtr1g052530.1 | chr1:21386135..21390691 | 528 | 60.13 | 6.16 | −0.501 | 4 | N-Myr (1)–Palm (2) |
| MtCDPK5 | Medtr1g054865.1 | chr1:24041840..24046332 | 607 | 66.45 | 5.51 | −0.334 | 4 | N-Myr (1)–Palm (1) |
| MtCDPK6 | Medtr1g055255.1 | chr1:24417898..24422244 | 589 | 66.07 | 6.52 | −0.478 | 4 | N-Myr (2)–Palm (1) |
| MtCDPK7 | Medtr1g096490.1 | chr1:43462266..43465665 | 499 | 56.18 | 5.21 | −0.380 | 4 | N-Palm (1) |
| MtCDPK8 | Medtr3g051770.1 | chr3:20500557..20506458 | 538 | 60.60 | 5.77 | −0.460 | 4 | N-Myr (1)–Palm (1) |
| MtCDPK9 | Medtr3g098070.1 | chr3:44756354..44761409 | 495 | 55.92 | 5.17 | −0.342 | 4 | - (0) |
| MtCDPK10 | Medtr3g098090.1 | chr3:44766535..44770211 | 498 | 56.37 | 5.36 | −0.395 | 4 | - (0) |
| MtCDPK11 | Medtr4g066660.1 | chr4:25214155..25217596 | 545 | 62.02 | 6.06 | −0.549 | 4 | N-Myr (1)–Palm (1) |
| MtCDPK12 | Medtr4g107490.1 | chr4:44509014..44514153 | 555 | 63.02 | 6.37 | −0.470 | 4 | N-Myr (1)–Palm (2) |
| MtCDPK13 | Medtr4g132040.1 | chr4:55151186..55153959 | 543 | 60.88 | 5.83 | −0.415 | 4 | N-Myr (1)–Palm (1) |
| MtCDPK14 | Medtr4g132070.1 | chr4:55167475..55172407 | 539 | 60.56 | 6.16 | −0.463 | 4 | N-Myr (1)–Palm (1) |
| MtCDPK15 | Medtr5g009830.1 | chr5:2478209..2483414 | 517 | 58.73 | 6.44 | −0.534 | 4 | N-Myr (1) |
| MtCDPK16 | Medtr5g022030.1 | chr5:8628890..8634949 | 560 | 63.50 | 9.10 | −0.615 | 4 | N-Myr (1)–Palm (1) |
| MtCDPK17 | Medtr5g089320.1 | chr5:38807982..38811203 | 523 | 58.50 | 5.43 | −0.535 | 4 | N-Myr (1)–Palm (2) |
| MtCDPK18 | Medtr5g092810.1 | chr5:40530948..40535829 | 540 | 61.32 | 7.67 | −0.615 | 4 | N-Myr (2)–Palm (1) |
| MtCDPK19 | Medtr5g099240.1 | chr5:43501966..43509933 | 559 | 62.93 | 5.68 | −0.443 | 4 | N-Myr (1)–Palm (1) |
| MtCDPK20 | Medtr7g068710.1 | chr7:25219104..25225477 | 530 | 59.97 | 6.03 | −0.457 | 4 | N-Myr (1)–Palm (2) |
| MtCDPK21 | Medtr7g091890.1 | chr7:36380965..36385094 | 533 | 60.59 | 6.16 | −0.554 | 4 | N-Myr (1)–Palm (2) |
| MtCDPK22 | Medtr7g106710.1 | chr7:43452426..43456560 | 602 | 68.15 | 5.34 | −0.451 | 4 | N-Myr (1)–Palm (1) |
| MtCDPK23 | Medtr8g095440.1 | chr8:39939663..39944572 | 503 | 56.82 | 5.20 | −0.344 | 4 | - (0) |
| MtCDPK24 | Medtr8g099095.1 | chr8:41618323..41623707 | 516 | 58.06 | 5.93 | −0.410 | 4 | N-Myr (1)–Palm (1) |
| MtCRK1 | Medtr3g092230.1 | chr3:42122948..42129657 | 588 | 65.61 | 8.74 | −0.318 | 0 | N-Myr (1)–Palm (2) |
| MtCRK2 | Medtr4g086660.1 | chr4:33966651..33972436 | 592 | 66.57 | 9.07 | −0.398 | 0 | N-Myr (2)–Palm (1) |
| MtCRK3 | Medtr5g017830.1 | chr5:6589564..6594550 | 587 | 65.86 | 8.49 | −0.309 | 2 | N-Myr (1)–Palm (1) |
| MtCRK4 | Medtr7g089480.1 | chr7:35036401..35042624 | 595 | 66.80 | 8.84 | −0.305 | 0 | N-Myr (1)–Palm (1) |
| MtCRK5 | Medtr7g118020.1 | chr7:48973096..48979529 | 596 | 66.62 | 8.92 | −0.415 | 3 | N-Myr (1)–Palm (1) |
| MtCRK6 | Medtr8g079790.1 | chr8:34200112..34204254 | 581 | 65.15 | 9.00 | −0.309 | 1 | N-Myr (1)–Palm (1) |
| Gene 1 | Gene 2 | Ka | Ks | Ka/Ks | Purifying Selection | Duplicate Type |
|---|---|---|---|---|---|---|
| MtCDPK6 | MtCDPK22 | 0.243 | 0.791 | 0.307 | Yes | Segmental |
| MtCDPK8 | MtCDPK18 | 0.112 | 0.659 | 0.171 | Yes | Segmental |
| MtCDPK9 | MtCDPK23 | 0.100 | 0.720 | 0.139 | Yes | Segmental |
| MtCDPK15 | MtCDPK24 | 0.098 | 0.768 | 0.128 | Yes | Segmental |
| MtCRK2 | MtCRK3 | 0.168 | 1.936 | 0.087 | Yes | Segmental |
| MtCRK3 | MtCRK6 | 0.136 | 0.708 | 0.192 | Yes | Segmental |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, P.; Liu, Y.; Kong, W.; Ji, J.; Cai, T.; Guo, Z. Genome-Wide Identification and Characterization of Calcium-Dependent Protein Kinase (CDPK) and CDPK-Related Kinase (CRK) Gene Families in Medicago truncatula. Int. J. Mol. Sci. 2021, 22, 1044. https://doi.org/10.3390/ijms22031044
Zhao P, Liu Y, Kong W, Ji J, Cai T, Guo Z. Genome-Wide Identification and Characterization of Calcium-Dependent Protein Kinase (CDPK) and CDPK-Related Kinase (CRK) Gene Families in Medicago truncatula. International Journal of Molecular Sciences. 2021; 22(3):1044. https://doi.org/10.3390/ijms22031044
Chicago/Turabian StyleZhao, Pengcheng, Yajie Liu, Weiyi Kong, Jiayi Ji, Tianyu Cai, and Zhenfei Guo. 2021. "Genome-Wide Identification and Characterization of Calcium-Dependent Protein Kinase (CDPK) and CDPK-Related Kinase (CRK) Gene Families in Medicago truncatula" International Journal of Molecular Sciences 22, no. 3: 1044. https://doi.org/10.3390/ijms22031044
APA StyleZhao, P., Liu, Y., Kong, W., Ji, J., Cai, T., & Guo, Z. (2021). Genome-Wide Identification and Characterization of Calcium-Dependent Protein Kinase (CDPK) and CDPK-Related Kinase (CRK) Gene Families in Medicago truncatula. International Journal of Molecular Sciences, 22(3), 1044. https://doi.org/10.3390/ijms22031044






