Characterization and Expression Analysis of Insulin Growth Factor Binding Proteins (IGFBPs) in Pacific White Shrimp Litopenaeus vannamei
Abstract
1. Introduction
2. Results
2.1. Characteristics of IGFBP Sequences
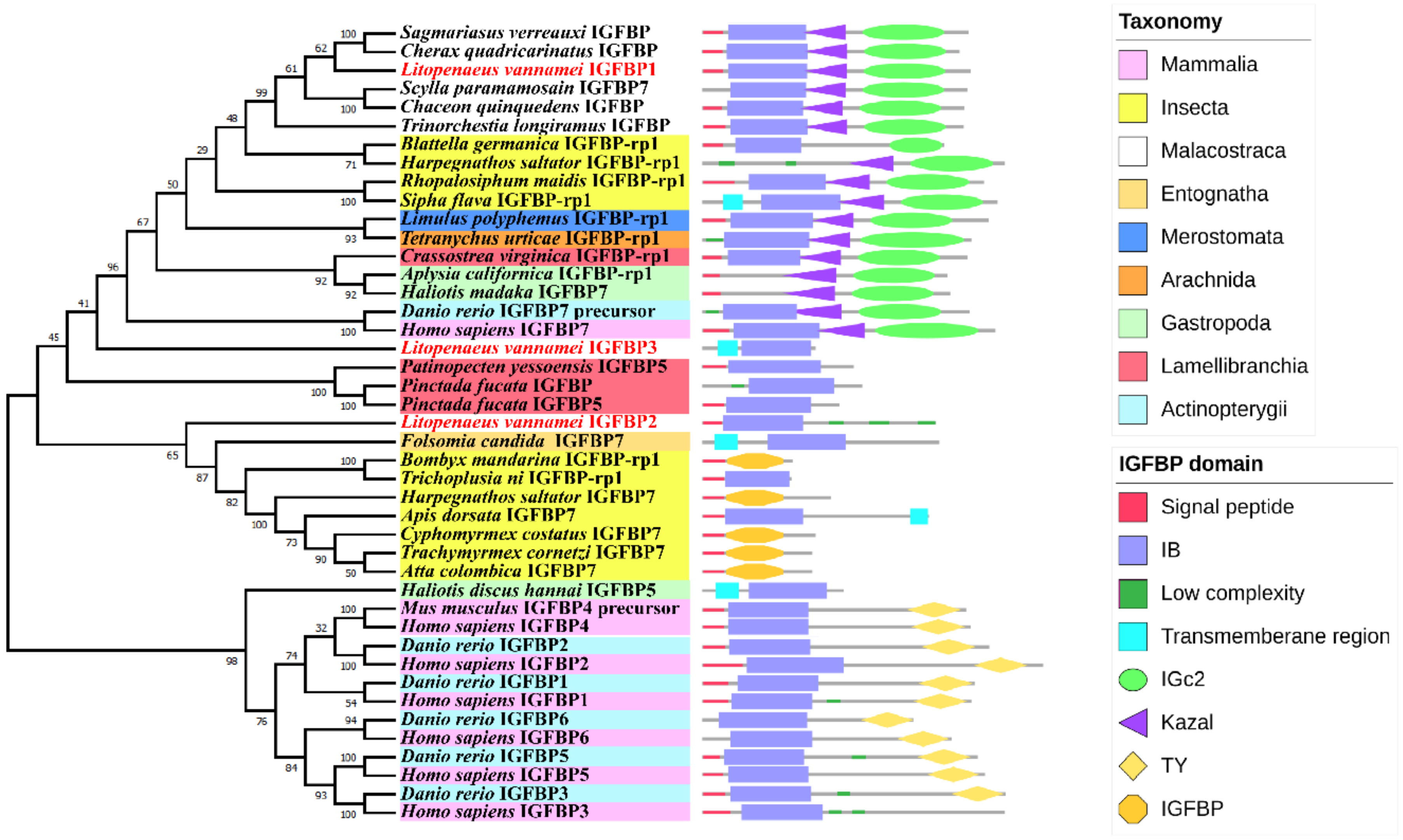
2.2. Phylogenetic Analysis of the IGFBPs
2.3. Multiple Sequence Alignment Analyses
2.4. Predicted 3D Structure of LvIGFBPs
2.5. Expression Profile of Three LvIGFBPs
2.6. LvIGFBP1 Gene RNA Interference
3. Discussion
3.1. Structure Characteristics of LvIGFBP Genes
3.2. Phylogeny Tree of LvIGFBPs
3.3. Possible Functions of LvIGFBPs
4. Materials and Methods
4.1. Experimental Animals and Ethical Statement
4.2. IGFBP Sequences Analyses
4.3. Phylogenetic Analysis
4.4. Gene Expression Profile
4.5. In Vivo RNAi Treatment
4.6. RNA Isolation and cDNA Synthesis
4.7. Real-Time Quantitative PCR (qRT-PCR)
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tatar, M.; Kopelman, A.; Epstein, D.; Tu, M.P.; Yin, C.M.; Garofalo, R.S. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 2001, 292, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, A.; White, M.F. Insulin-like signaling, nutrient homeostasis, and life span. Annu. Rev. Physiol. 2008, 70, 191–212. [Google Scholar] [CrossRef] [PubMed]
- Das, D.; Arur, S. Conserved insulin signaling in the regulation of oocyte growth, development, and maturation. Mol. Reprod. Dev. 2017, 84, 444–459. [Google Scholar] [CrossRef] [PubMed]
- Skorokhod, A.; Gamulin, V.; Gundacker, D.; Kavsan, V.; Muller, I.M.; Muller, W.E.G. Origin of insulin receptor-like tyrosine kinases in marine sponges. Biol. Bull. 1999, 197, 198–206. [Google Scholar] [CrossRef]
- Vitali, V.; Horn, F.; Catania, F. Insulin-like signaling within and beyond metazoans. Biol. Chem. 2018, 399, 851–857. [Google Scholar] [CrossRef]
- Duret, L.; Guex, N.; Peitsch, M.C.; Bairoch, A. New insulin-like proteins with atypical disulfide bond pattern characterized in Caenorhabditis elegans by comparative sequence analysis and homology modeling. Genome Res. 1998, 8, 348–353. [Google Scholar] [CrossRef]
- Zhang, H.; Shi, Y.; He, M. Molecular identification of an insulin growth factor binding protein (IGFBP) and its potential role in an insulin-like peptide system of the pearl oyster, Pinctada fucata. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017, 214, 27–35. [Google Scholar] [CrossRef]
- Denley, A.; Cosgrove, L.J.; Booker, G.W.; Wallace, J.C.; Forbes, B.E. Molecular interactions of the IGF system. Cytokine Growth Factor Rev. 2005, 16, 421–439. [Google Scholar] [CrossRef]
- Yamanaka, Y.; Wilson, E.M.; Rosenfeld, R.G.; Oh, Y. Inhibition of Insulin Receptor Activation by Insulin-like Growth Factor Binding Proteins. J. Biol. Chem. 1997, 272, 30729–30734. [Google Scholar] [CrossRef]
- Zeslawski, W.; Beisel, H.G.; Kamionka, M.; Kalus, W.; Engh, R.A.; Huber, R.; Lang, K.; Holak, T.A. The interaction of insulin-like growth factor-I with the N-terminal domain of IGFBP-5. EMBO J. 2001, 20, 3638–3644. [Google Scholar] [CrossRef]
- Castellanos, M.; Jimenez-Vega, F.; Vargas-Albores, F. Single IB domain (SIBD) protein from Litopenaeus vannamei, a novel member for the IGFBP family. Comp. Biochem. Physiol. Part D Genom. Proteom. 2008, 3, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Kalus, W.; Zweckstetter, M.; Renner, C.; Sanchez, Y.; Georgescu, J.; Grol, M.; Demuth, D.; Schumacher, R.; Dony, C.; Lang, K.; et al. Structure of the IGF-binding domain of the insulinlike growth factor-binding protein-5 (IGFBP-5): Implications for IGF and IGF-I receptor interactions. EMBO J. 1998, 17, 6558–6572. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.N.; Delgado, C.H.; Owens, G.A.; Steller, M.A. Insulin-like growth factor-II participates in the biphasic effect of a gonadotropin-releasing hormone agonist on ovarian cancer cell growth. Fertil. Steril. 1997, 67, 870–876. [Google Scholar] [CrossRef]
- Wang, J.F.; Hampton, B.; Mehlman, T.; Burgess, W.H.; Rechler, M.M. Isolation of a biologically active fragment from the carboxy terminus of the fetal rat binding protein for insulin-like growth factors. Biochem. Biophys. Res. Commun. 1988, 157, 718–726. [Google Scholar] [CrossRef]
- Spencer, E.M.; Chan, K. A 3-dimensional model for the insulin-like growth factor binding proteins (IGFBPs) supporting evidence using the structural determinants of the IGF binding site on IGFBP-3. Prog. Growth Factor Res. 1995, 6, 209–214. [Google Scholar] [CrossRef]
- Ferry, R.J., Jr.; Katz, L.E.; Grimberg, A.; Cohen, P.; Weinzimer, S.A. Cellular actions of insulin-like growth factor binding proteins. Horm. Metab. Res. 1999, 31, 192–202. [Google Scholar] [CrossRef]
- Bach, L.A. What Happened to the IGF Binding Proteins? Endocrinology 2018, 159, 570–578. [Google Scholar] [CrossRef]
- Chandler, J.C.; Aizen, J.; Elizur, A.; Hollander-Cohen, L.; Battaglene, S.C.; Ventura, T. Discovery of a novel insulin-like peptide and insulin binding proteins in the Eastern rock lobster Sagmariasus verreauxi. Gen. Comp. Endocrinol. 2015, 215, 76–87. [Google Scholar] [CrossRef]
- Rosen, O.; Weil, S.; Manor, R.; Roth, Z.; Khalaila, I.; Sagi, A. A crayfish insulin-like-binding protein: Another piece in the androgenic gland insulin-like hormone puzzle is revealed. J. Biol. Chem. 2013, 288, 22289–22298. [Google Scholar] [CrossRef]
- Huang, X.; Ye, H.; Feng, B.; Huang, H. Insights into insulin-like peptide system in invertebrates from studies on IGF binding domain-containing proteins in the female mud crab, Scylla paramamosain. Mol. Cell Endocrinol. 2015, 416, 36–45. [Google Scholar] [CrossRef]
- Li, F.; Bai, H.; Xiong, Y.; Fu, H.; Jiang, S.; Jiang, F.; Jin, S.; Sun, S.; Qiao, H.; Zhang, W. Molecular characterization of insulin-like androgenic gland hormone-binding protein gene from the oriental river prawn Macrobrachium nipponense and investigation of its transcriptional relationship with the insulin-like androgenic gland hormone gene. Gen. Comp. Endocrinol. 2015, 216, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.; Wang, L.; Song, L.; Zhao, J.; Qiu, L.; Xing, K. A putative endocrine factor SIBD (single insulin binding domain protein) involved in immune response of Chinese mitten crab Eriocheir sinensis. Fish Shellfish Immunol. 2010, 28, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, Z.; Zhang, L.; Wang, S.; Zou, Z.; Wang, G.; Wang, Y. Insulin-like growth factor binding protein 7, a member of insulin-like growth factor signal pathway, involved in immune response of small abalone Haliotis diversicolor. Fish Shellfish Immunol. 2012, 33, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Li, X.; Yu, Q.; Ning, X.; Dou, J.; Zou, J.; Zhang, L.; Wang, S.; Hu, X.; Bao, Z. A scallop IGF binding protein gene: Molecular characterization and association of variants with growth traits. PLoS ONE 2014, 9, e89039. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zhang, X.; Yu, Y.; Huang, H.; Li, F.; Xiang, J. Comparative transcriptomic characterization of the early development in Pacific white shrimp Litopenaeus vannamei. PLoS ONE 2014, 9, e106201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yuan, J.; Sun, Y.; Li, S.; Gao, Y.; Yu, Y.; Liu, C.; Wang, Q.; Lv, X.; Zhang, X.; et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, X.; Wei, J.; Sun, X.; Yuan, J.; Li, F.; Xiang, J. Whole Transcriptome Analysis Provides Insights into Molecular Mechanisms for Molting in Litopenaeus vannamei. PLoS ONE 2015, 10, e0144350. [Google Scholar] [CrossRef]
- Headey, S.J.; Keizer, D.W.; Yao, S.; Brasier, G.; Kantharidis, P.; Bach, L.A.; Norton, R.S. C-terminal domain of insulin-like growth factor (IGF) binding protein-6: Structure and interaction with IGF-II. Mol. Endocrinol. 2004, 18, 2740–2750. [Google Scholar] [CrossRef][Green Version]
- Bach, L.A. IGF-binding proteins. J. Mol. Endocrinol. 2018, 61, T11–T28. [Google Scholar] [CrossRef]
- Chernausek, S.D.; Smith, C.E.; Duffin, K.L.; Busby, W.H.; Wright, G.; Clemmons, D.R. Proteolytic cleavage of insulin-like growth factor binding protein 4 (IGFBP-4). J. Biol. Chem. 1995, 270, 11377–11382. [Google Scholar] [CrossRef]
- Wetterau, L.A.; Moore, M.G.; Lee, K.-W.; Shim, M.L.; Cohen, P. Novel Aspects of the Insulin-like Growth Factor Binding Proteins. Mol. Genet. Metab. 1999, 68, 161–181. [Google Scholar] [CrossRef] [PubMed]
- Bach, L.A.; Headey, S.J.; Norton, R.S. IGF-binding proteins--the pieces are falling into place. Trends Endocrinol. Metab. 2005, 16, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Sitar, T.; Popowicz, G.M.; Siwanowicz, I.; Huber, R.; Holak, T.A. Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins. Proc. Natl. Acad. Sci. USA 2006, 103, 13028–13033. [Google Scholar] [CrossRef] [PubMed]
- Hwa, V.; Oh, Y.; Rosenfeld, R.G. The Insulin-Like Growth Factor-Binding Protein (IGFBP) Superfamily. Endocr. Rev. 1999, 20, 761–787. [Google Scholar] [CrossRef] [PubMed]
- Fowlkes, J.L.; Thrailkill, K.M.; GeorgeNascimento, C.; Rosenberg, C.K.; Serra, D.M. Heparin-binding, highly basic regions within the thyroglobulin type-1 repeat of insulin-like growth factor (IGF)-binding proteins (IGFBPs) -3, -5, and -6 inhibit IGFBP-4 degradation. Endocrinology 1997, 138, 2280–2285. [Google Scholar] [CrossRef]
- Klauwer, D.; Blum, W.F.; Hanitsch, S.; Rascher, W.; Lee, P.D.; Kiess, W. IGF-I, IGF-11, free IGF-I and IGFBP-1, -2 and -3 levels in venous cord blood- relationship to birthweight, length and gestational age in healthy newborns. Acta Paediatr. 1997, 86, 826–833. [Google Scholar]
- Zapf, J. Physiological role of the insulin-like growth factor binding proteins. Eur. J. Endocrinol. 1995, 132, 645–654. [Google Scholar] [CrossRef]
- Sun, M.; Li, S.; Zhang, X.; Xiang, J.; Li, F. Isolation and transcriptome analysis of three subpopulations of shrimp hemocytes reveals the underlying mechanism of their immune functions. Dev. Comp. Immunol. 2020, 108, 103689. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]





| Gene Name | Deduced Amino Acids | Molecular Weight (kDa) | Isoelectric Point (pI) | Signal Peptide | Accession Number | Location on Genome |
|---|---|---|---|---|---|---|
| LvIGFBP1 | 258 | 27.03 | 4.54 | 1–20 | XP_027212706.1 | LVANscaffold_1676:698634–704445(−) |
| LvIGFBP2 | 225 | 24.35 | 10.10 | 1–19 | XP_027219293.1 | LVANscaffold_1140:490253–502498(−) |
| LvIGFBP3 | 109 | 12.00 | 8.86 | - | XP_027214055.1 | LVANscaffold_1781:46638–49525(−) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, Y.; Zhang, X.; Yuan, J.; Zhang, X.; Xiang, J.; Li, F. Characterization and Expression Analysis of Insulin Growth Factor Binding Proteins (IGFBPs) in Pacific White Shrimp Litopenaeus vannamei. Int. J. Mol. Sci. 2021, 22, 1056. https://doi.org/10.3390/ijms22031056
Pang Y, Zhang X, Yuan J, Zhang X, Xiang J, Li F. Characterization and Expression Analysis of Insulin Growth Factor Binding Proteins (IGFBPs) in Pacific White Shrimp Litopenaeus vannamei. International Journal of Molecular Sciences. 2021; 22(3):1056. https://doi.org/10.3390/ijms22031056
Chicago/Turabian StylePang, Ying, Xiaojun Zhang, Jianbo Yuan, Xiaoxi Zhang, Jianhai Xiang, and Fuhua Li. 2021. "Characterization and Expression Analysis of Insulin Growth Factor Binding Proteins (IGFBPs) in Pacific White Shrimp Litopenaeus vannamei" International Journal of Molecular Sciences 22, no. 3: 1056. https://doi.org/10.3390/ijms22031056
APA StylePang, Y., Zhang, X., Yuan, J., Zhang, X., Xiang, J., & Li, F. (2021). Characterization and Expression Analysis of Insulin Growth Factor Binding Proteins (IGFBPs) in Pacific White Shrimp Litopenaeus vannamei. International Journal of Molecular Sciences, 22(3), 1056. https://doi.org/10.3390/ijms22031056





