Reactive Oxygen Species Accumulation Strongly Allied with Genetic Male Sterility Convertible to Cytoplasmic Male Sterility in Kenaf
Abstract
1. Introduction
2. Results
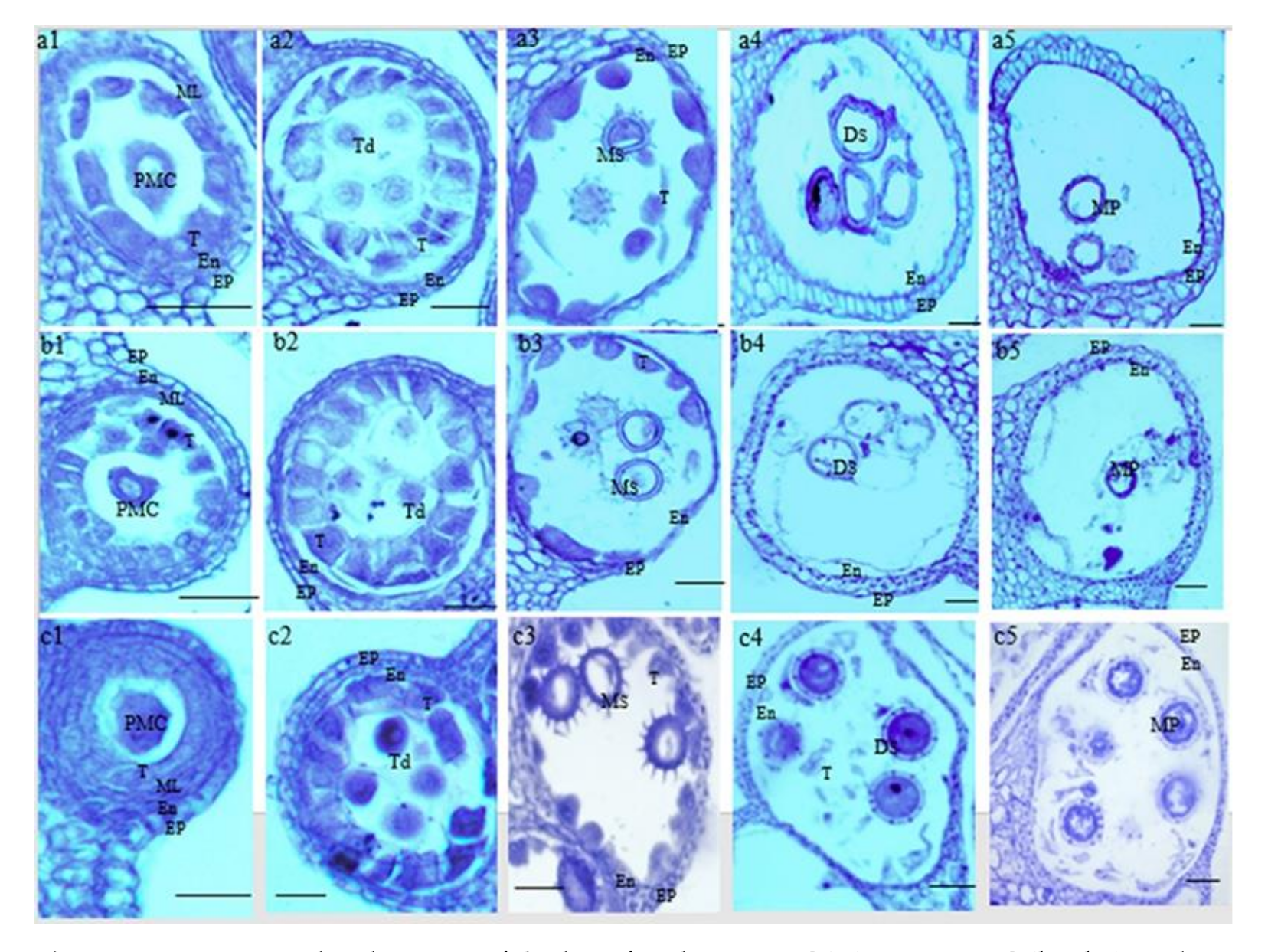
2.1. Phenotypic and Cytological Characterization of P9B, P9BS, and P9SA
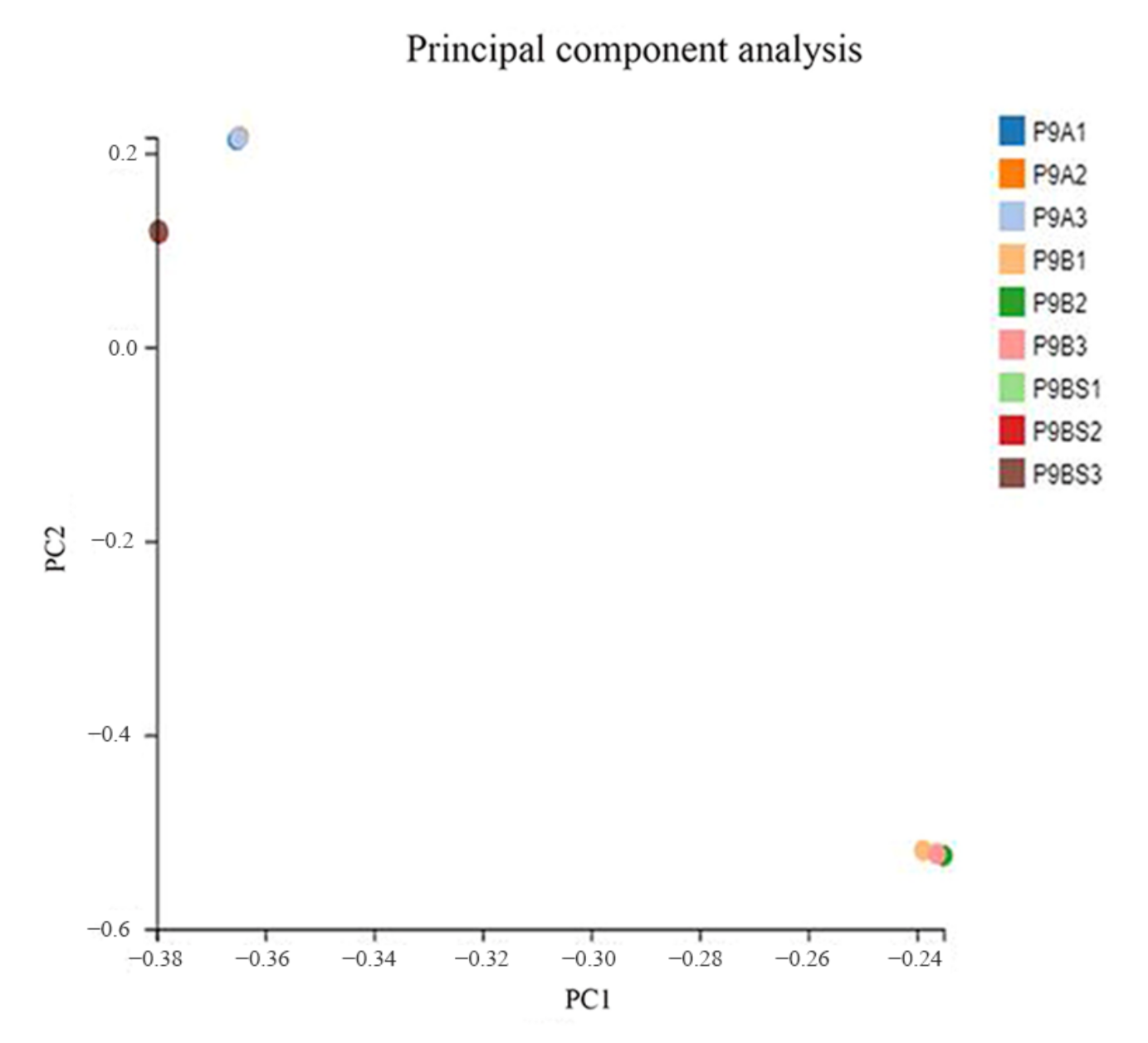
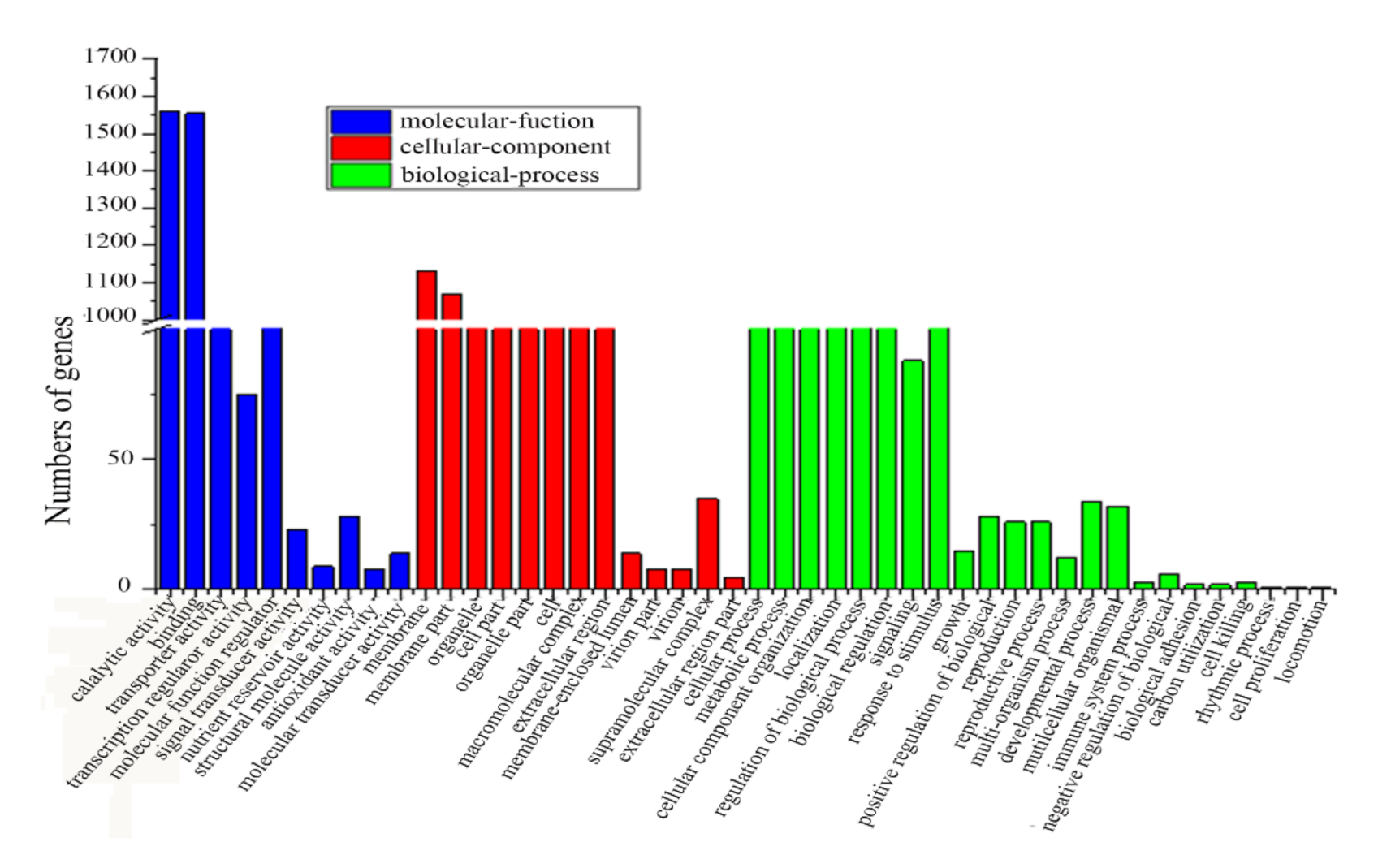
2.2. De Novo Assembly and Sequence Annotation
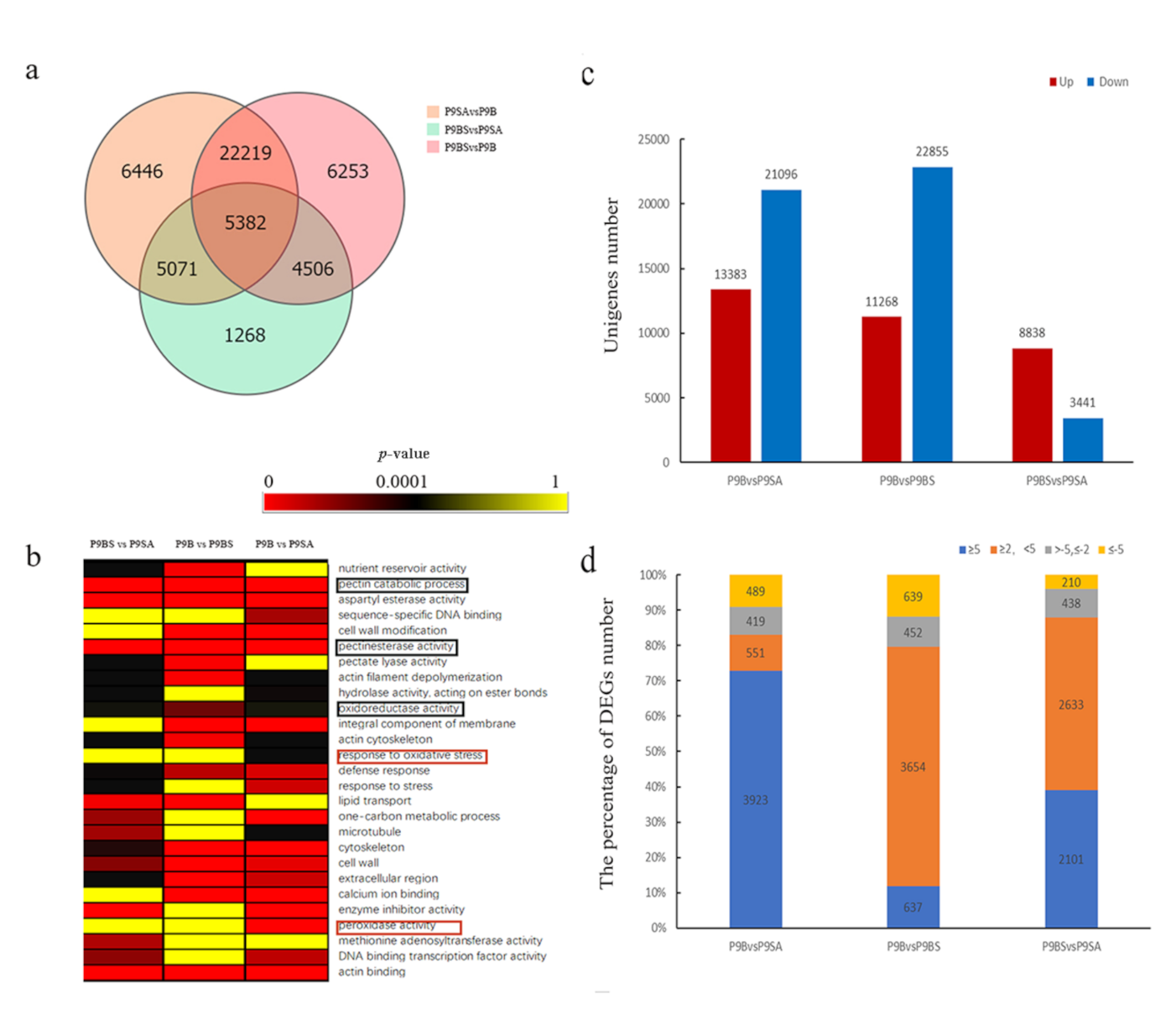
2.3. Identification of the DEGs among Different Kenaf Lines
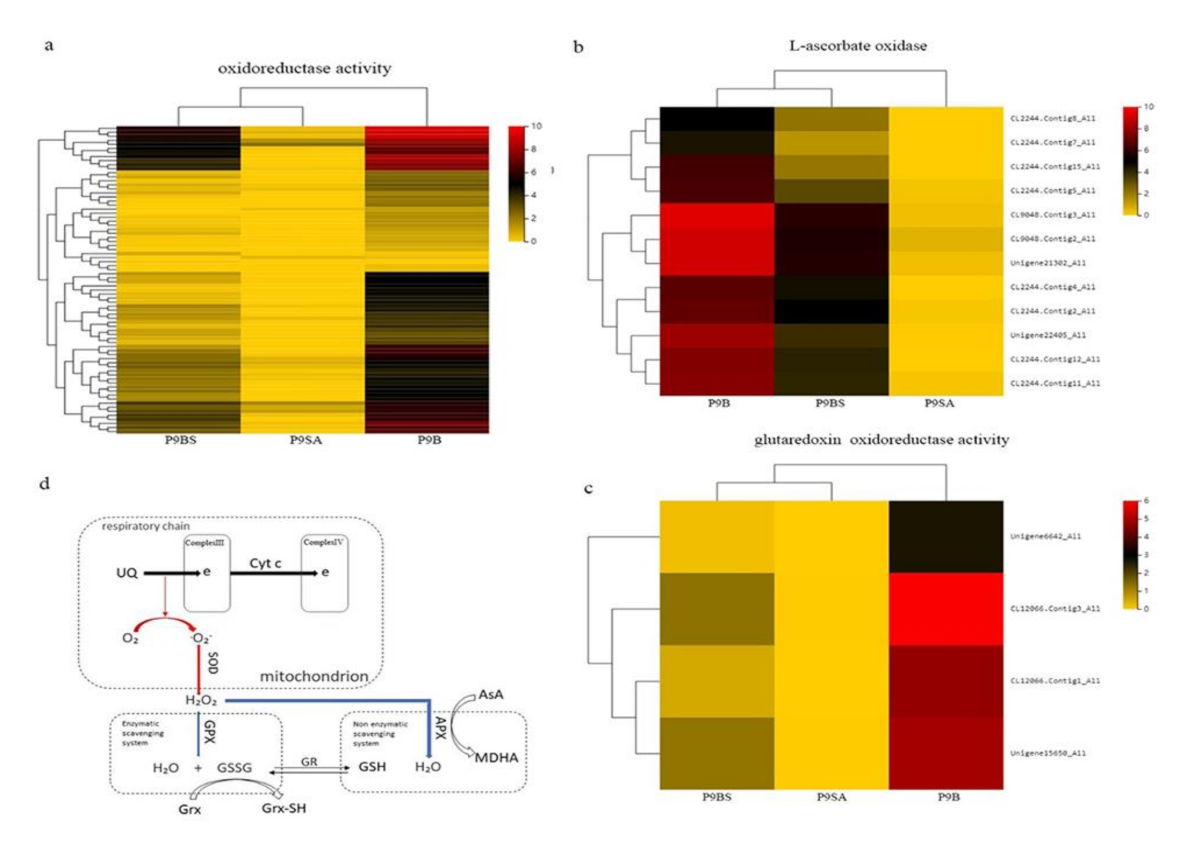
2.4. DEGs Related to ROS
2.5. DEGs Related to the Pollen Development Process
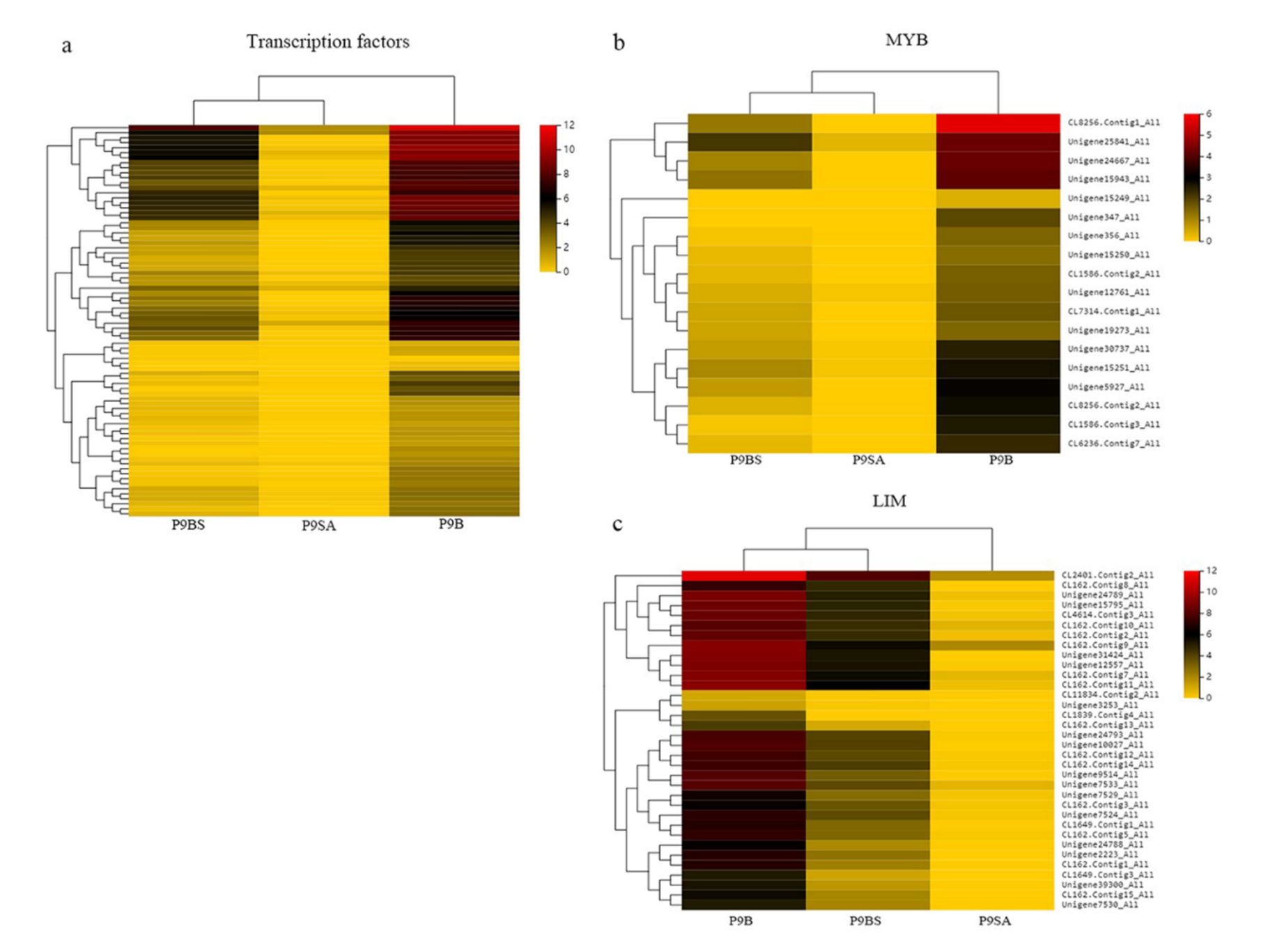
2.6. Transcription Factor Related to Male Sterility
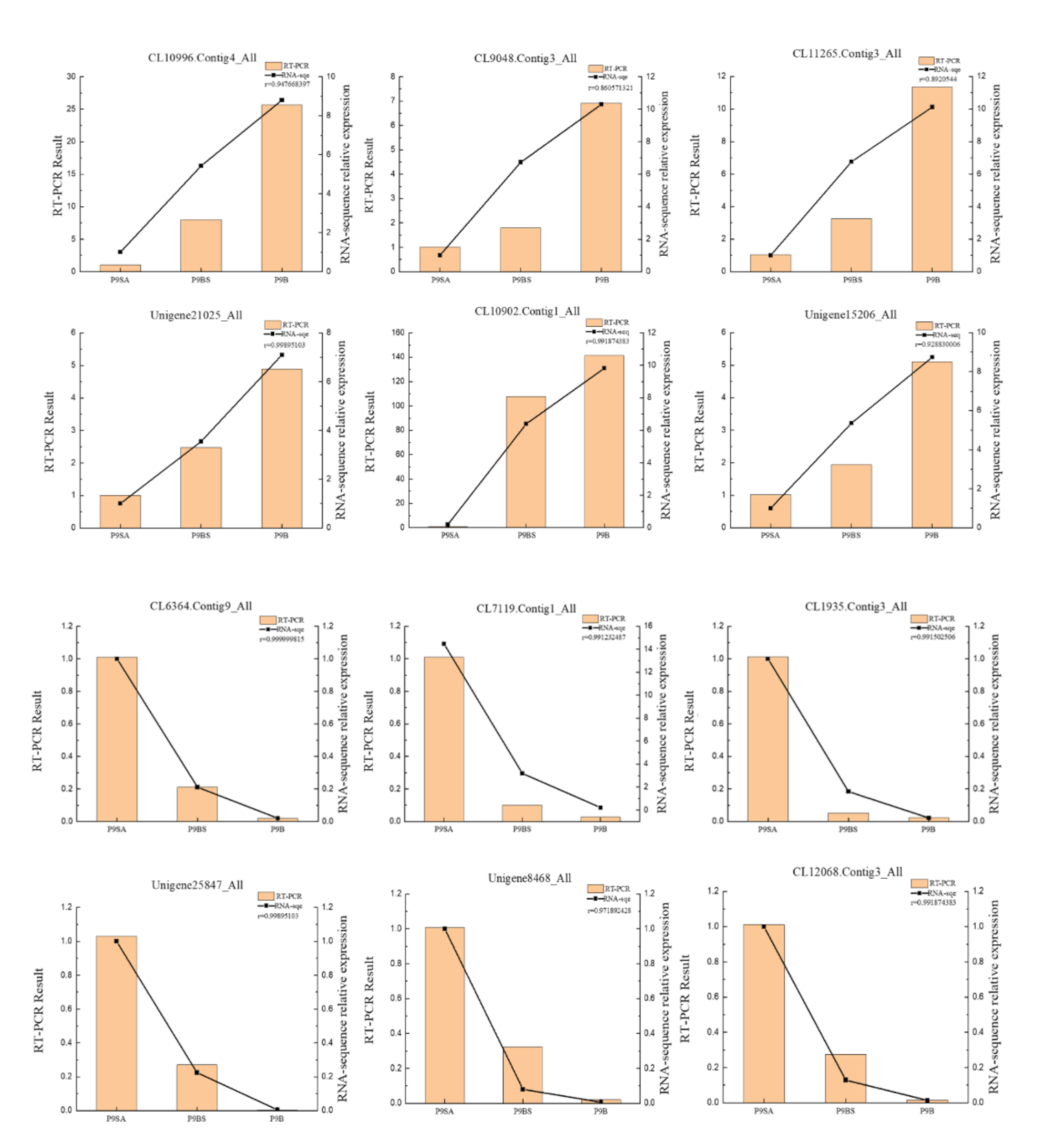
2.7. qRT-PCR Confirmation of DEG Data
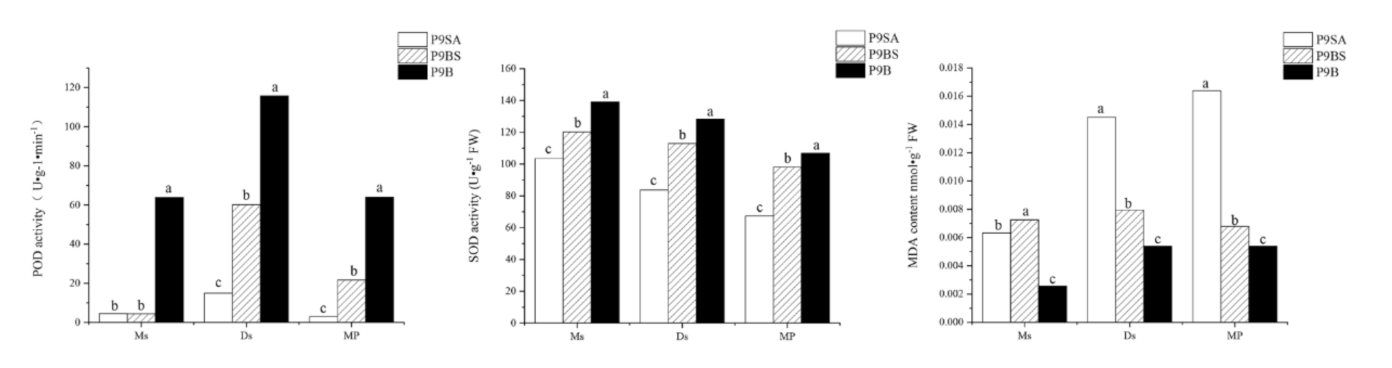
2.8. The Activity of Enzymes Involved in Reactive Oxygen Scavenging Pathways
3. Discussion
3.1. Importance of Materials for RNA-Seq Analysis
3.2. Production and Clearance of ROS in Anther Development
3.3. Burst of Reactive Oxygen Species Causes Premature PCD of Tapetum Cells and Abnormal Pollen Wall Development
4. Materials and Methods
4.1. Plant Materials
4.2. Morphological and Cytological Observations
4.3. Total RNA Extraction, cDNA Library Construction, and Deep Sequencing
4.4. RNA-Seq Data Processing Analysis
4.5. Quantitative Real-Time PCR for RNA-Seq Validation
4.6. Enzyme Activity and MDA Content Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CMS | Cytoplasmic male sterility |
| ROS | Reactive oxygen species |
| DEGs | Differentially expressed genes |
| POD | Peroxidase |
| SOD | Superoxide dismutase |
| MDA | Malondialdehyde |
| MS | Male sterility |
References
- Bel-Berger, P.; Hoven, T.V.; Ramaswamy, G.N.; Kimmel, L.; Bolyston, E. Cotton/kenaf fabrics: A viable natural fabric. J. Cotton Sci. 1999, 3, 60–70. [Google Scholar]
- Shahar, F.S.; Sultan, M.T.H.; Hua, L.S.; Jawaid, M.; Sivasankaran, P.N.A.P. A review on the orthotics and prosthetics and the potential of kenaf composites as alternative materials for ankle-foot orthosis. J. Mech. Behav. Biomed. 2019, 99, 169–185. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.O.; Othman, M.S.H.; Tahir, P.M.; Elhag, E.A.I. Feasibility of kenaf as bio-material for automotive components: The case of Malaysia. Int. J. Green Econ. 2016, 9, 273–290. [Google Scholar]
- Sheldon, A. Preliminary Evaluation of Kenaf as a Structural Material. Master’s Thesis, Clemson University, Clemson City, SC, USA, 2014. [Google Scholar]
- Mun, H.W.; Hoe, A.L.; Koo, L.D. Assessment of Pb uptake, translocation and immobilization in kenaf (Hibiscus cannabinus L.) for phytoremediation of sand tailings. J. Environ. Sci. 2008, 20, 1341–1347. [Google Scholar]
- Tao, L.; Min, Q.; Yang, Z.; Hui, L.; Ping, F.; Fen, T.; Tang, X. Effect of Cadmium Stress on the Growth, Antioxidative Enzymes and Lipid Peroxidation in Two Kenaf (Hibiscus cannabinus L.) Plant Seedlings. J. Integr. Agric. 2013, 12, 610–620. [Google Scholar]
- Li, H.; Li, D.; Chen, A.; Tang, H.; Li, J.; Huang, S. Progress and Prospect of Research on Kenaf Male Sterility. Crop. Res. 2015, 29, 206–209. [Google Scholar]
- Islam, M.S.; Studer, B.; Møller, I.M.; Asp, T. Genetics and biology of cytoplasmic male sterility and its applications in forage and turf grass breeding. Plant Breed. 2014, 133, 299–312. [Google Scholar] [CrossRef]
- Gils, M.; Marillonnet, S.; Werner, S.; Grützner, R.; Giritch, A.; Engler, C.; Schachschneider, R.; Klimyuk, V.; Gleba, Y. A novel hybrid seed system for plants. Plant Biotechnol. J. 2008, 6, 226–235. [Google Scholar] [CrossRef]
- Carlsson, J.; Glimelius, K. Cytoplasmic Male-Sterility and Nuclear Encoded Fertility Restoration. Plant Mitochondria 2011, 1, 469–491. [Google Scholar]
- Ma, X.; Xing, C.; Guo, L.; Gong, Y.; Wang, H.; Zhao, Y.; Wu, J. Analysis of Differentially Expressed Genes in Genic Male Sterility Cotton (Gossypium hirsutum L.) Using cDNA-AFLP. J. Genet. Genom. 2007, 36, 536–543. [Google Scholar] [CrossRef]
- Yang, X.; Ye, J.; Zhang, L.; Song, X. Blocked synthesis of sporopollenin and jasmonic acid leads to pollen wall defects and anther indehiscence in genic male sterile wheat line 4110S at high temperatures. Funct. Integr. Genom. 2020, 20, 383–396. [Google Scholar] [CrossRef]
- Huang, T.; Wang, Y.; Ma, B.; Ma, Y.; Li, S. Genetic Analysis and Mapping of Genes Involved in Fertility of Pingxiang Dominant Genic Male Sterile Rice. J. Genet. Genom. 2007, 34, 616–622. [Google Scholar] [CrossRef]
- Liu, B. Study on the classification and origin of dominant male Sterility genes in crops. Hereditas 1993, 6, 32–34. [Google Scholar]
- Zhang, D.; Wu, S.; An, X.; Xie, K.; Dong, Z.; Zhou, Y.; Xu, L.; Fang, W.; Liu, S.; Liu, S.; et al. Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol. J. 2018, 16, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.; Chen, Z.; Wang, N.; Xie, G.; Lu, J.; Yan, W.; Zhou, J.; Tang, X.; Deng, X.W. Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc. Natl. Acad. Sci. USA 2016, 113, 14145–14150. [Google Scholar] [CrossRef]
- Song, S.; Wang, T.; Li, Y.; Hu, J.; Kan, R.; Qiu, M.; Deng, Y.; Liu, P.; Zhang, L.; Dong, H.; et al. A novel strategy for creating a new system of third-generation hybrid rice technology using a cytoplasmic sterility gene and a genic male-sterile gene. Plant Biotechnol. J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Liang, C.; Liu, H. Study on the physical variation of rice. Bull. Bot. 2000, 17, 232–241. [Google Scholar]
- Jing, N.; Zhixia, N. A Cytoplasmic Male Sterile Mutant was Obtained from the Anther Culture of the F2 of a Cross of Photoperiod-sensitive Genic Male Sterile Rice. Hybrid Rice 2001, 16, 40. [Google Scholar]
- Goldberg, B.R.B. A Novel Cell Ablation Strategy Blocks Tobacco Anther Dehiscence. Plant Cell 1997, 9, 1527–1545. [Google Scholar]
- Wu, H.M.; Cheun, A.Y. Programmed cell death in plant reproduction. Plant Mol. Biol. 2000, 44, 267–281. [Google Scholar] [CrossRef]
- Chen, P.-Y.; Wu, C.C.; Lin, C.-C.; Jane, W.-N.; Suen, D.-F. 3D imaging of tapetal mitochondria suggests the importance of mitochondrial fission in pollen growth. Plant Physiol. 2019, 180, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.; Yoon, H.; Chung, S.Y.Y. Male-sterility of thermosensitive genic male-sterile rice is associated with premature programmed cell death of the tapetum. Planta 2003, 217, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.A.; Zhu, J.; Gao, J.A.; Wang, C.; Li, H.; Li, H.; Zhang, H.I.; Zhang, S.; Wang, D.I.; Wang, Q.I. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 2010, 52, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Higginson, T.; Song, F.L.; Parish, R.W. AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 2010, 35, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Luo, L.; Xu, J.; Xin, P.; Guo, H.; Wu, J.; Bai, L.; Wang, G.; Chu, J.; Zuo, J. Malate transported from chloroplast to mitochondrion triggers production of ROS and PCD in Arabidopsis thaliana. Cell Res. 2018, 28, 448–461. [Google Scholar] [CrossRef]
- Yan, M.Y.; Xie, D.L.; Cao, J.J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Zhou, J.; Foyer, C.H.; Yu, J.Q. Brassinosteroid-mediated reactive oxygen species are essential for tapetum degradation and pollen fertility in tomato. Plant J. 2020, 102, 931–947. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tang, H.; Chen, Y.; Chen, A.; Huang, S.; Li, D. Cytomorphological Observation on Cytoplasm Nuclear Male Sterile line of Kenaf. Plant Fiber Sci. China 2015, 37, 229–232. [Google Scholar]
- Zhao, Y.; Chen, P.; Liao, X.; Zhou, B.; Liao, J. A Comparative Analysis of the atp8 Gene Between a Cytoplasmic Male Sterile Line and Its Maintainer and Further Development of a Molecular Marker Specific to Male Sterile Cytoplasm in Kenaf (Hibiscus cannabinus L.). Mol. Breed. 2013, 32, 969–976. [Google Scholar] [CrossRef]
- Bujin, Z.; Peng, C.; Aziz, K.; Yanhong, Z.; Lihong, C.; Dongmei, L.; Xiaofang, L.; Xiangjun, K.; Ruiyang, Z. Candidate Reference Genes Selection and Application for RT-qPCR Analysis in Kenaf with Cytoplasmic Male Sterility Background. Front. Plant Sci. 2017, 8, 1520. [Google Scholar]
- Zhou, B.; Liu, Y.; Chen, Z.; Liu, D.; Wang, Y.; Zheng, J.; Liao, X.; Zhou, R. Comparative Transcriptome Analysis Reveals the Cause for Accumulation of Reactive Oxygen Species During Pollen Abortion in Cytoplasmic Male-Sterile Kenaf Line 722HA. Int. J. Mol. Sci. 2019, 20, 5515. [Google Scholar] [CrossRef]
- Guan, L. Two structurally similar maize cytosolic superoxide dismutase genes, Sod4 and Sod4A, respond differentially to abscisic acid and high osmoticum. Plant Physiol. 1998, 117, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Liang-De, L.I.; Ding-Feng, W.; Guang-Yuan, W.U. Progresses on Producing Sites of Reactive Oxygen Species in Mitochondria. Life Sci. Res. 2015, 6, 530–535. [Google Scholar]
- Siedow, J.N.; Umbach, A.L. Plant Mitochondrial Electron Transfer and Molecular Biology. Plant Cell 1995, 7, 821. [Google Scholar] [CrossRef] [PubMed]
- Noster, J.; Persicke, M.; Chao, T.C.; Krone, L.; Heppner, B.; Hensel, M.; Hansmeier, N. Impact of ROS-Induced Damage of TCA Cycle Enzymes on Metabolism and Virulence of Salmonella enterica serovar Typhimurium. Front Microbiol. 2019, 10, 762. [Google Scholar] [CrossRef] [PubMed]
- Yesodi, V.; Hauschner, H.; Tabib, Y.; Firon, N. An intact F1ATPase alpha-subunit gene and a pseudogene with differing genomic organization are detected in both male-fertile and CMS petunia mitochondria. Curr. Genet. 1997, 32, 348–357. [Google Scholar] [CrossRef]
- Li, S.; Wan, C.; Hu, C.; Gao, F.; Huang, Q.; Wang, K.; Wang, T.; Zhu, Y. Mitochondrial mutation impairs cytoplasmic male sterility rice in response to HO stress. Plant Sci. 2012, 195, 143–150. [Google Scholar] [CrossRef]
- Suzuki, N.; Miller, G.; Morales, J.; Shulaev, V.; Torres, M.A.; Mittler, R. Respiratory burst oxidases: The engines of ROS signaling. Curr. Opin. Plant Biol. 2011, 14, 691–69914. [Google Scholar] [CrossRef]
- Geng, X.; Ye, J.; Yang, X.; Li, S.; Zhang, L.; Song, X. Identification of Proteins Involved in Carbohydrate Metabolism and Energy Metabolism Pathways and Their Regulation of Cytoplasmic Male Sterility in Wheat. Int. J. Mol. Sci. 2018, 19, 324. [Google Scholar] [CrossRef]
- Du, X.M.; Yin, W.X.; Zhao, Y.X.; Zhang, H. The production and scavenging of reactive oxygen species in plants. Sheng Wu Gong Cheng Xue Bao 2001, 17, 121–125. [Google Scholar]
- Bian, Y.W.; Lv, D.W.; Cheng, Z.W.; Gu, A.Q.; Cao, H.; Yan, Y.M. Integrative proteome analysis of Brachypodium distachyon roots and leaves reveals a synergetic responsive network under H2O2 stress. J. Proteom. 2015, 128, 388–402. [Google Scholar] [CrossRef]
- Mock; Hans-Peter; Dietz; Karl-Josef, Redox proteomics for the assessment of redox-related posttranslational regulation in plants. Biochim. Biophys. Acta 2016, 1864, 967–973. [CrossRef] [PubMed]
- Navrot, N.; Rouhier, N.; Gelhaye, E.; Jacquot, J.P. ROS generation and antioxidant systems in plant mitochondria. Physiol. Plant. 2006, 129, 185–195. [Google Scholar] [CrossRef]
- Rodríguez-Manzaneque, M.T.; Ros, J.; Cabiscol, E.; Sorribas, A.; Herrero, E. Grx5 Glutaredoxin Plays a Central Role in Protection against Protein Oxidative Damage in Saccharomyces cerevisiae. Mol. Cell Biol. 1999, 19, 8180–8190. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.J.; Kim, D.Y.; Lee, T.G.; Jeon, W.B.; Seo, Y.W. Functional characterization of pectin methylesterase inhibitor (PMEI) in wheat. Genes. Genet. Syst. 2010, 85, 97–106. [Google Scholar] [CrossRef]
- Rocchi, V.; Janni, M.; Bellincampi, D.; Giardina, T.; D Ovidio, R. Intron retention regulates the expression of pectin methyl esterase inhibitor (Pmei) genes during wheat growth and development. Plant Biol. 2012, 14, 365–373. [Google Scholar] [CrossRef]
- Röckel, N.; Wolf, S.; Kost, B.; Rausch, T.; Greiner, S. Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J. 2008, 53, 133–143. [Google Scholar] [CrossRef]
- Safadi, F.; Reddy, V.S.; Reddy, A.S.N. A Pollen-specific Novel Calmodulin-binding Protein with Tetratricopeptide Repeats. J. Biol. Chem. 2000, 275, 35457–35470. [Google Scholar] [CrossRef]
- Zhefeng, L.; Chin-Wen, H.; Don, G. AtTRP1 encodes a novel TPR protein that interacts with the ethylene receptor ERS1 and modulates development in Arabidopsis. J. Exp. Bot. 2009, 60, 3697–3714. [Google Scholar]
- Liang, X.; Zhou, J.M. Receptor-Like Cytoplasmic Kinases: Central Players in Plant Receptor Kinase–Mediated Signaling. Annu. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef]
- Morrison, D.K.; Davis, R.J. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 2003, 19, 91–118. [Google Scholar] [CrossRef]
- Okuda Satohiro, T.H.S.K. Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 2009, 458, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Zimei, S.; Yuanfang, T.; Runyan, L.; Hongbin, W.; Lan, Z.; Min, Y. Genome-wide Analysis of Lectin-like Receptor Kinases(LecRLKs) Family in Soybean. Mol. Plant Breed. 2019, 17, 699. [Google Scholar]
- Qiu, W.M.; Zhu, A.D.; Wang, Y.; Chai, L.J.; Ge, X.X.; Deng, X.X.; Guo, W.W. Comparative transcript profiling of gene expression between seedless Ponkan mandarin and its seedy wild type during floral organ development by suppression subtractive hybridization and cDNA microarray. BMC Genom. 2012, 13, 397. [Google Scholar] [CrossRef]
- Gao, R.; Liang, X.Q.; Cheedipudi, S.; Cordero, J.; Jiang, X.; Zhang, Q.Q.; Ren, Y.G. Pioneering function of Isl1 in the epigenetic control of cardiomyocyte cell fate. Cell Res. 2019, 29, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2010, 50. [Google Scholar] [CrossRef]
- Chen, P.; Ran, S.; Li, R.; Huang, Z.; Qian, J.; Yu, M.; Zhou, R. Transcriptome de novo assembly and differentially expressed genes related to cytoplasmic male sterility in kenaf (Hibiscus cannabinus L.). Mol. Breed. 2014, 34, 1879–1891. [Google Scholar] [CrossRef]
- Zheng, J.; Kong, X.; Li, B.; Khan, A.; Li, Z.; Liu, Y.; Kang, H.; Ullah, D.F.; Zhou, R. Comparative Transcriptome Analysis between a Novel Allohexaploid Cotton Progeny CMS Line LD6A and Its Maintainer Line LD6B. Int. J. Mol. Sci. 2019, 20, 6127. [Google Scholar] [CrossRef]
- Kovtun, Y.; Chiu, W.; Tena, G. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 2000, 97, 2940–2945. [Google Scholar] [CrossRef]
- Fluhr, R.G.A.R. UV-B-Induced PR-1 Accumulation Is Mediated by Active Oxygen Species. Plant Cell 1995, 7, 203–212. [Google Scholar]
- Giampieri, F.; Gasparrini, M.; Forbes-Hernández, T.Y.; Manna, P.P.; Zhang, J.; Reboredo-Rodríguez, P.; Cianciosi, D.; Quiles, J.L.; Fernández-Piñar, C.T.; Orantes-Bermejo, F.J. Beeswax by-Products Efficiently Counteract the Oxidative Damage Induced by an Oxidant Agent in Human Dermal Fibroblasts. Int. J. Mol. Sci. 2018, 19, 2842. [Google Scholar] [CrossRef]
- Yong, Z.M. Identification of a Rice cDNA Encoding the Acyl CoA binding Protein (ACBP). Acta Photophysiol. Sin. 1999, 4, 327–331. [Google Scholar]
- Wang, K.; Gao, F.; Ji, Y.X.; Liu, Y.; Dan, Z.W.; Yang, P.F.; Zhu, Y.G.; Li, S.Q. ORFH79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in Honglian cytoplasmic male sterile rice. New Phytol. 2013, 198, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Dussert, S.; Davey, M.W.; Laffargue, A.; Doulbeau, S.; Swennen, R.; Etienne, H. Oxidative stress, phospholipid loss and lipid hydrolysis during drying and storage of intermediate seeds. Physiol. Plant. 2010, 127, 192–204. [Google Scholar] [CrossRef]
- Morant, M.; Jorgensen, K.; Schaller, H.; Pinot, F.; Moller, B.L.; Werck-Reichhart, D.; Bak, S. CYP703 Is an Ancient Cytochrome P450 in Land Plants Catalyzing in-Chain Hydroxylation of Lauric Acid to Provide Building Blocks for Sporopollenin Synthesis in Pollen. Plant. Cell 2007, 19, 1473–1487. [Google Scholar] [CrossRef]
- Phan, H.A.; Li, S.F.; Parish, R.W. MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant Mol. Biol. 2012, 78, 171–183. [Google Scholar] [CrossRef]
- Paxson-Sowders, D.M.; Dodrill, C.H.; Owen, H.A.; Makaroff, C.A. DEX1, a Novel Plant Protein, Is Required for Exine Pattern Formation during Pollen Development in Arabidopsis. Plant Physiol. 2001, 127, 1739–1749. [Google Scholar] [CrossRef]
- Berghe, T.V.; Linkermann, A.; Jouan-Lanhouet, S.; Walczak, H.; Vandenabeele, P. Regulated necrosis: The expanding network of non-apoptotic cell death pathways. Nat. Rev. Mol. Cell Biol. 2014, 15, 135–147. [Google Scholar] [CrossRef]
- Luo, D.; Xu, H.; Liu, Z.; Guo, J.; Liu, Y.G. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat. Genet. 2013, 45, 573. [Google Scholar] [CrossRef]
- Hu, L.; Liang, W.; Yin, C.; Cui, X.; Zong, J.; Wang, X.; Hu, J.; Zhang, D. Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 2011, 23, 515–533. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, Z.; Cao, J. Pollen wall development: The associated enzymes and metabolic pathways. Plant Biol. 2012, 15, 249–263. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, B.; Chen, Z.; Zhang, D.; Zhang, H.; Wang, H.; Zhang, Y.; Cai, D.; Liu, J.; Xiao, S. A PECTIN METHYLESTERASE gene at the maize Ga1 locus confers male function in unilateral cross-incompatibility. Nat. Commun. 2018, 9, 3678. [Google Scholar] [CrossRef] [PubMed]
- Pina, C.; Pinto, F.; Feijó, J.A.; Becker, J.D. Gene Family Analysis of the Arabidopsis Pollen Transcriptome Reveals Biological Implications for Cell Growth, Division Control, and Gene Expression Regulation. Plant Physiol. 2005, 138, 744–756. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.; Miao, J.; Liu, B.J.; Yang, Y.Y. The expression of pectin methylesterase in onion flower buds is associated with the dominant male-fertility restoration allele. Plant Breed. 2012, 131, 211–216. [Google Scholar] [CrossRef]
- Shuping, W.; Gaisheng, Z.; Yingxin, Z.; Qilu, S.; Zheng, C.; Junsheng, W.; Jialin, G.; Na, N.; Junwei, W.; Shoucai, M. Comparative studies of mitochondrial proteomics reveal an intimate protein network of male sterility in wheat (Triticum aestivum L.). J. Exp. Bot 2015, 66, 6191–6203. [Google Scholar]
- Kochba, J.; Lavee, S.; Spiegel-Roy, P. Differences in peroxidase activity and isoenzymes in embryogenic ane non-embryogenic ’Shamouti’ orange ovular callus lines. Plant Cell Physiol. 1977, 18, 463–467. [Google Scholar] [CrossRef]



| Categories | P9SA | P9BS | P9B |
|---|---|---|---|
| Total raw reads (Gb) | 20.40 | 20.57 | 20.41 |
| Clean Reads Ratio (%) | 96.24 | ||
| Total clean reads (Gb) | 19.59 | 19.86 | 19.61 |
| Total transcripts | 588,381 | ||
| Average length (transcripts) | 1281 | ||
| N50 (transcripts) | 1862 | ||
| Total unigenes | 125,778 | ||
| Total length (unigenes) | 217,394,220 | ||
| Average length (unigenes) | 1728 | ||
| N50 (unigenes) | 2325 | ||
| GC (%) | 42.13 | ||
| Generation | Fertile | Sterility | Totally | Actual Separation Ratio | Expected Separation Ratio |
|---|---|---|---|---|---|
| P1 (P9B) | 120 | 0 | |||
| P2 (P9BS) | 0 | 5 | |||
| F1 (P2 × P1) | 68 | 70 | 138 | 1:1.03 | 1:1 |
| F2 (F1S × F1F) | 48 | 50 | 98 | 1:1.04 | 1:1 |
| BC1 (F2S × P1) | 50 | 60 | 110 | 1:1.2 | |
| BC2 (BC1S × P1) | 48 | 60 | 108 | 1:1.25 | |
| BC7 (BC6 × P1) | 0 | 96 | 96 | ||
| BC8 (BC7 × P1) | 0 | 132 | 132 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Zhou, B.; Khan, A.; Zheng, J.; Dawar, F.U.; Akhtar, K.; Zhou, R. Reactive Oxygen Species Accumulation Strongly Allied with Genetic Male Sterility Convertible to Cytoplasmic Male Sterility in Kenaf. Int. J. Mol. Sci. 2021, 22, 1107. https://doi.org/10.3390/ijms22031107
Liu Y, Zhou B, Khan A, Zheng J, Dawar FU, Akhtar K, Zhou R. Reactive Oxygen Species Accumulation Strongly Allied with Genetic Male Sterility Convertible to Cytoplasmic Male Sterility in Kenaf. International Journal of Molecular Sciences. 2021; 22(3):1107. https://doi.org/10.3390/ijms22031107
Chicago/Turabian StyleLiu, Yiding, Bujin Zhou, Aziz Khan, Jie Zheng, Farman Ullah Dawar, Kashif Akhtar, and Ruiyang Zhou. 2021. "Reactive Oxygen Species Accumulation Strongly Allied with Genetic Male Sterility Convertible to Cytoplasmic Male Sterility in Kenaf" International Journal of Molecular Sciences 22, no. 3: 1107. https://doi.org/10.3390/ijms22031107
APA StyleLiu, Y., Zhou, B., Khan, A., Zheng, J., Dawar, F. U., Akhtar, K., & Zhou, R. (2021). Reactive Oxygen Species Accumulation Strongly Allied with Genetic Male Sterility Convertible to Cytoplasmic Male Sterility in Kenaf. International Journal of Molecular Sciences, 22(3), 1107. https://doi.org/10.3390/ijms22031107






