Aptamers Against the ?-Conglutin Allergen: Insights into the Behavior of the Shortest Multimeric(Intra)Molecular DNA G-Quadruplex
Abstract
:1. Introduction
2. Results
2.1. Structural Studies
2.1.1. Polyacrylamide Gel Electrophoresis (PAGE)
2.1.2. Liquid Chromatography Coupled with Electrospray Ionization Time-of-Flight Mass Spectrometry (HPLC-ESI-TOF-MS)
2.1.3. Thermal Melting Analysis
2.1.4. Circular Dichroism (CD)
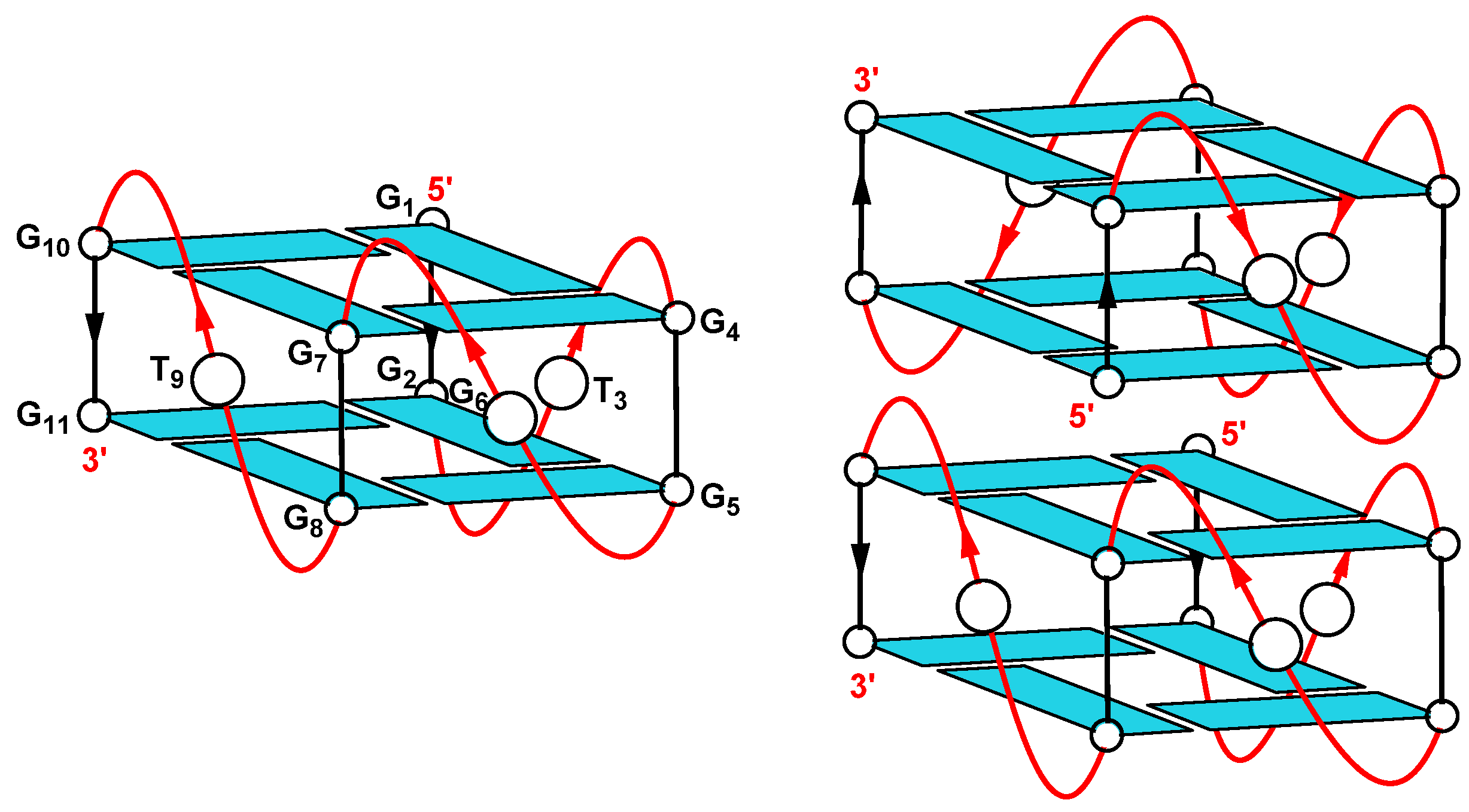
2.1.5. Nuclear Magnetic Resonance (NMR)
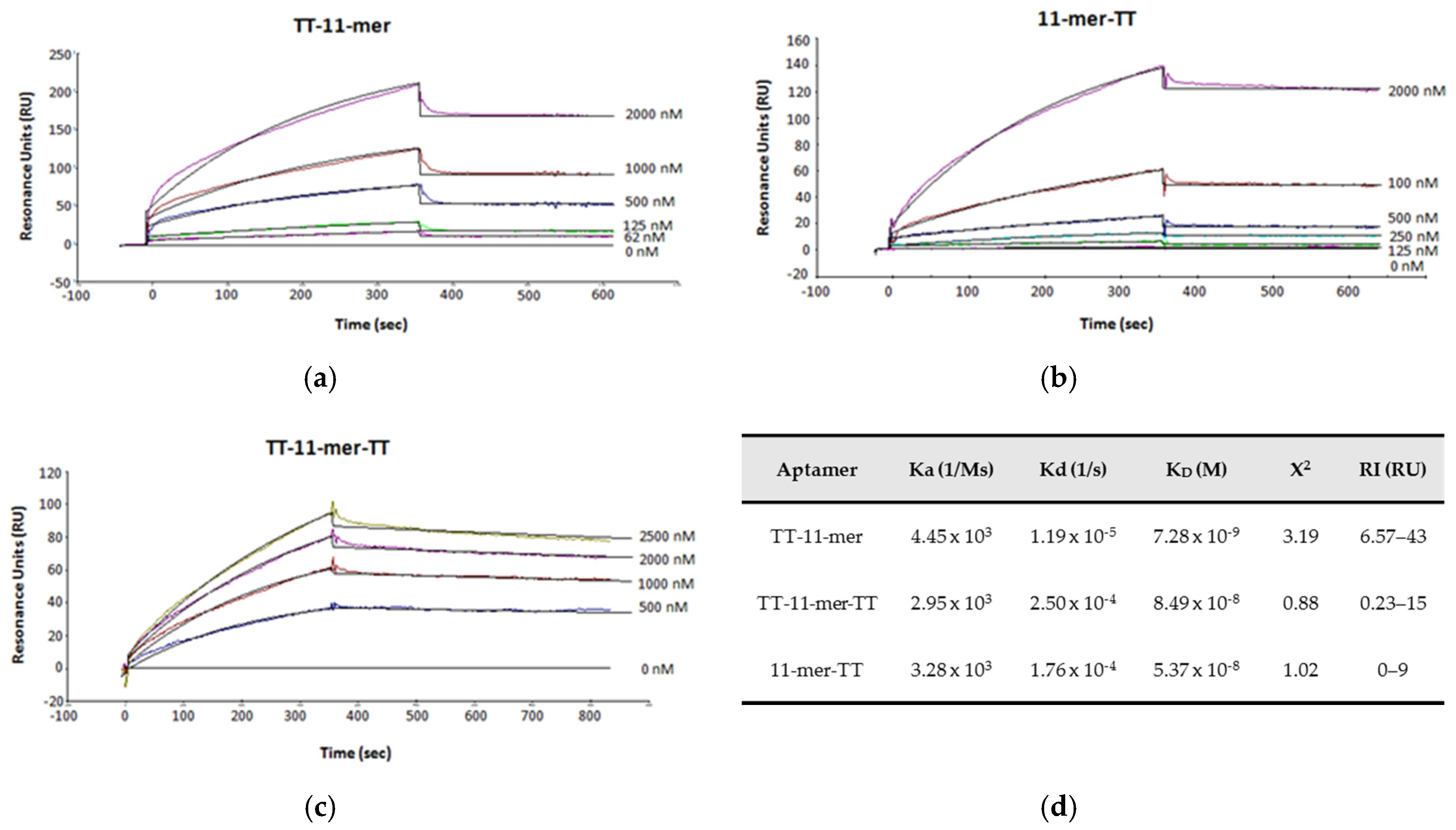
2.2. Binding Studies
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Gel Electrophoresis
4.3. Thermal Melting Analysis
4.4. Liquid Chromatography Coupled to Time-of-Flight Mass Spectrometry (HPLC-ESI-TOF-MS)
4.5. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF-MS)
4.6. Circular Dichroism (CD)
4.7. NMR
4.8. Surface Plasmon Resonance (SPR)
4.9. Biolayer Interferometry (BLI)
4.10. MicroScale Thermophoresis (MST)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BLI | Biolayer interferometry |
| CD | Circular dichroism |
| EDC | 3-(Ethyliminomethyleneamino)-N, N-dimethylpropan-1-amine |
| EDTA | Ethylenediaminetetraacetic acid |
| ESI-TOF-MS | Electrospray ionization time-of-flight mass spectrometry |
| FRET | Fluorescence resonance energy transfer |
| HPLC | High-performance liquid chromatography |
| KD | Affinity dissociation constant |
| MALDI | Matrix-assisted laser desorption/ionization |
| MST | Microscale thermophoresis |
| NHS | N-Hydroxysuccinimide |
| NMR | Nuclear magnetic resonance |
| PAGE | Polyacrylamide gel electrophoresis |
| SAX | High-sensitivity streptavidin-activated biosensors |
| SELEX | Systematic Evolution of Ligands by Exponential Enrichment |
| SPR | Surface plasmon resonance |
| SPRi | Surface plasmon resonance imaging |
| T15 | 15× thymine spacer |
| TBA | Thrombin binding aptamer |
| TBE | Tris-borate/EDTA |
| TEG | Triethylene glycol |
| UV | Ultraviolet |
References
- Skouridou, V.; Jauset-Rubio, M.; Ballester, P.; Bashammakh, A.S.; El-Shahawi, M.S.; Alyoubi, A.O.; O’Sullivan, C.K. Selection and characterization of DNA aptamers against the steroid testosterone. Microchim. Acta 2017, 184, 1631–1639. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Q.; Wei, X.; Zhang, J.; Mo, S. DNA aptamer for use in a fluorescent assay for the shrimp allergen tropomyosin. Microchim. Acta 2017, 184, 633–639. [Google Scholar] [CrossRef]
- Quang, N.N.; Miodek, A.; Cibiel, A.; Ducongé, F. Selection of aptamers against whole living cells: From cell-SELEX to identification of biomarkers. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2017; Volume 1575, pp. 253–272. [Google Scholar] [CrossRef]
- Zou, X.; Wu, J.; Gu, J.; Shen, L.; Mao, L. Application of aptamers in virus detection and antiviral therapy. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamay, G.S.; Ivanchenko, T.I.; Zamay, T.N.; Grigorieva, V.L.; Glazyrin, Y.E.; Kolovskaya, O.S.; Garanzha, I.V.; Barinov, A.A.; Krat, A.V.; Mironov, G.G.; et al. DNA Aptamers for the Characterization of Histological Structure of Lung Adenocarcinoma. Mol. Ther. Nucleic Acids 2017, 6, 150–162. [Google Scholar] [CrossRef] [Green Version]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Moccia, F.; Riccardi, C.; Musumeci, D.; Leone, S.; Oliva, R.; Petraccone, L.; Montesarchio, D. Insights into the G-rich VEGF-binding aptamer V7t1: When two G-quadruplexes are better than one! Nucleic Acids Res. 2019, 47, 8318–8331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G-quadruplex-forming aptamers—Characteristics, applications, and perspectives. Molecules 2019, 24, 3781. [Google Scholar] [CrossRef] [Green Version]
- Neidle, S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009, 19, 239–250. [Google Scholar] [CrossRef]
- Wong, H.M.; Payet, L.; Huppert, J.L. Function and targeting of G-quadruplexes. Curr. Opin. Mol. Ther. 2009, 11, 146–155. [Google Scholar] [PubMed]
- Zhao, X.; Liu, B.; Yan, J.; Yuan, Y.; An, L.; Guan, Y. Structure variations of TBA G-quadruplex induced by 2′-O-methyl nucleotide in K+ and Ca2+ environments. Acta Biochim. Biophys. Sin. (Shanghai) 2014, 46, 837–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Sato, H.; Sannohe, Y.; Shinohara, K.I.; Sugiyama, H. Stable lariat formation based on a G-quadruplex scaffold. J. Am. Chem. Soc. 2008, 130, 16470–16471. [Google Scholar] [CrossRef]
- Morris, M.J.; Basu, S. An unusually stable G-quadruplex within the 5′-UTR of the MT3 matrix metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry 2009, 48, 5313–5319. [Google Scholar] [CrossRef]
- Xiao, C.-D.; Shibata, T.; Yamamoto, Y.; Xu, Y. An intramolecular antiparallel G-quadruplex formed by human telomere RNA. Chem. Commun. 2018, 54, 3944–3946. [Google Scholar] [CrossRef] [PubMed]
- Salazar, M.; Fedoroff, O.Y.; Miller, J.M.; Ribeiro, N.S.; Reid, B.R. The DNA Strand in DNA·RNA Hybrid Duplexes is neither B-form nor A-form in Solution. Biochemistry 1993, 32, 4207–4215. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.N.; Lee, M.P.H.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Yu, H.; Gu, X.; Nakano, S.I.; Miyoshi, D.; Sugimoto, N. Beads-on-a-string structure of long telomeric dnas under molecular crowding conditions. J. Am. Chem. Soc. 2012, 134, 20060–20069. [Google Scholar] [CrossRef]
- Yuan, W.-F.; Wan, L.Y.; Peng, H.; Zhong, Y.-M.; Cai, W.-L.; Zhang, Y.-Q.; Ai, W.-B.; Wu, J.-F. The influencing factors and functions of DNA G-quadruplexes. Cell Biochem. Funct. 2020, 524–532. [Google Scholar] [CrossRef]
- Gatto, B.; Palumbo, M.; Sissi, C. Nucleic Acid Aptamers Based on the G-Quadruplex Structure: Therapeutic and Diagnostic Potential. Curr. Med. Chem. 2009, 16, 1248–1265. [Google Scholar] [CrossRef]
- Jia, M.; Sha, J.; Li, Z.; Wang, W.; Zhang, H. High affinity truncated aptamers for ultra-sensitive colorimetric detection of bisphenol A with label-free aptasensor. Food Chem. 2020, 317. [Google Scholar] [CrossRef] [PubMed]
- Mairal Lerga, T.; Jauset-Rubio, M.; Skouridou, V.; Bashammakh, A.S.; El-Shahawi, M.S.; Alyoubi, A.O.; O’Sullivan, C.K. High Affinity Aptamer for the Detection of the Biogenic Amine Histamine. Anal. Chem. 2019, 91, 7104–7111. [Google Scholar] [CrossRef] [PubMed]
- Nadal, P.; Pinto, A.; Svobodova, M.; Canela, N.; O’Sullivan, C.K. DNA aptamers against the lup an 1 food allergen. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Nadal, P.; Svobodova, M.; Mairal, T.; O’Sullivan, C.K. Probing high-affinity 11-mer DNA aptamer against Lup an 1 (β-conglutin). Anal. Bioanal. Chem. 2013, 405, 9343–9349. [Google Scholar] [CrossRef]
- Mairal, T.; Nadal, P.; Svobodova, M.; O’Sullivan, C.K. FRET-based dimeric aptamer probe for selective and sensitive Lup an 1 allergen detection. Biosens. Bioelectron. 2014, 54, 207–210. [Google Scholar] [CrossRef]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roe, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA 1993, 90, 3745–3749. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Langley, D.R.; Rangan, A.; Hurley, L.H. Selective interactions of cationic porphyrins with G-quadruplex structures. J. Am. Chem. Soc. 2001, 123, 8902–8913. [Google Scholar] [CrossRef]
- Do, N.Q.; Lim, K.W.; Teo, M.H.; Heddi, B.; Phan, A.T. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity. Nucleic Acids Res. 2011, 39, 9448–9457. [Google Scholar] [CrossRef]
- Zhou, J.; Yuan, G. Effect of pH and cations on the formation and structure of human telomeric G-quadruplex DNA. Acta Chim. Sin. 2007, 65, 1728–1732. [Google Scholar]
- Mukundan, V.T.; Do, N.Q.; Phan, A.T. HIV-1 integrase inhibitor T30177 forms a stacked dimeric G-quadruplex structure containing bulges. Nucleic Acids Res. 2011, 39, 8984–8991. [Google Scholar] [CrossRef]
- Marchand, A.; Gabelica, V. Folding and misfolding pathways of G-quadruplex DNA. Nucleic Acids Res. 2016, 44, 10999–11012. [Google Scholar] [CrossRef] [PubMed]
- Kypr, J.; Kejnovská, I.; Renčiuk, D.; Vorlíčková, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karsisiotis, A.I.; Hessari, N.M.A.; Novellino, E.; Spada, G.P.; Randazzo, A.; Webba Da Silva, M. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem. Int. Ed. 2011, 50, 10645–10648. [Google Scholar] [CrossRef]
- Nici, F.; Oliviero, G.; Falanga, A.P.; D’Errico, S.; Marzano, M.; Musumeci, D.; Montesarchio, D.; Noppen, S.; Pannecouque, C.; Piccialli, G.; et al. Anti-HIV activity of new higher order G-quadruplex aptamers obtained from tetra-end-linked oligonucleotides. Org. Biomol. Chem. 2018, 16, 2349–2355. [Google Scholar] [CrossRef] [PubMed]
- Novoseltseva, A.A.; Ivanov, N.M.; Novikov, R.A.; Tkachev, Y.V.; Bunin, D.A.; Gambaryan, A.S.; Tashlitsky, V.N.; Arutyunyan, A.M.; Kopylov, A.M.; Zavyalova, E.G. Structural and functional aspects of G-quadruplex aptamers which bind a broad range of influenza a viruses. Biomolecules 2020, 10, 119. [Google Scholar] [CrossRef] [Green Version]
- Pavc, D.; Wang, B.; Spindler, L.; Drevenšek-Olenik, I.; Plavec, J.; Šket, P. GC ends control topology of DNA G-quadruplexes and their cation-dependent assembly. Nucleic Acids Res. 2020, 48, 2749–2761. [Google Scholar] [CrossRef]
- Kogut, M.; Kleist, C.; Czub, J. Why do G-quadruplexes dimerize through the 5′-ends? Driving forces for G4 DNA dimerization examined in atomic detail. PLoS Comput. Biol. 2019, 15. [Google Scholar] [CrossRef]
- Jauset Rubio, M.; Svobodová, M.; Mairal, T.; O’Sullivan, C.K. Surface plasmon resonance imaging (SPRi) for analysis of DNA aptamer: β-conglutin interactions. Methods 2016, 97, 20–26. [Google Scholar] [CrossRef]
- Nadal, P.; Canela, N.; Katakis, I.; O’Sullivan, C.K. Extraction, isolation, and characterization of globulin proteins from lupinus albus. J. Agric. Food Chem. 2011, 59, 2752–2758. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’ Sullivan, C.K.; Mairal, T.; Jauset-Rubio, M.; Svobodova, M.; Skouridou, V.; Esposito, V.; Virgilio, A.; Galeone, A. Aptamers Against the ?-Conglutin Allergen: Insights into the Behavior of the Shortest Multimeric(Intra)Molecular DNA G-Quadruplex. Int. J. Mol. Sci. 2021, 22, 1150. https://doi.org/10.3390/ijms22031150
O’ Sullivan CK, Mairal T, Jauset-Rubio M, Svobodova M, Skouridou V, Esposito V, Virgilio A, Galeone A. Aptamers Against the ?-Conglutin Allergen: Insights into the Behavior of the Shortest Multimeric(Intra)Molecular DNA G-Quadruplex. International Journal of Molecular Sciences. 2021; 22(3):1150. https://doi.org/10.3390/ijms22031150
Chicago/Turabian StyleO’ Sullivan, Ciara K., Teresa Mairal, Miriam Jauset-Rubio, Marketa Svobodova, Vasso Skouridou, Veronica Esposito, Antonella Virgilio, and Aldo Galeone. 2021. "Aptamers Against the ?-Conglutin Allergen: Insights into the Behavior of the Shortest Multimeric(Intra)Molecular DNA G-Quadruplex" International Journal of Molecular Sciences 22, no. 3: 1150. https://doi.org/10.3390/ijms22031150
APA StyleO’ Sullivan, C. K., Mairal, T., Jauset-Rubio, M., Svobodova, M., Skouridou, V., Esposito, V., Virgilio, A., & Galeone, A. (2021). Aptamers Against the ?-Conglutin Allergen: Insights into the Behavior of the Shortest Multimeric(Intra)Molecular DNA G-Quadruplex. International Journal of Molecular Sciences, 22(3), 1150. https://doi.org/10.3390/ijms22031150







