Abstract
A library of novel imidazole-1,2,3-triazole hybrids were designed and synthesized based on the hybrid pharmacophore approach. Therefore, copper(I)catalyzed click reaction of thiopropargylated-imidazole 2 with several organoazides yielded two sets of imidazole-1,2,3-triazole hybrids carrying different un/functionalized alkyl/aryl side chains 4a–k and 6a–e. After full spectroscopic characterization using different spectral techniques (IR, 1H, 13C NMR) and elemental analyses, the resulted adducts were screened for their anticancer activity against four cancer cell lines (Caco-2, HCT-116, HeLa, and MCF-7) by the MTT assay and showed significant activity. In-silico molecular docking study was also investigated on one of the prominent cancer target receptors, i.e., glycogen synthase kinase-3β (GSK-3β), revealing a good binding interaction with our potent compound, 4k and was in agreement with the in vitro cytotoxic results. In addition, the ADMET profile was assessed for these novel derivatives to get an insight on their pharmacokinetic/dynamic attributes. Finally, this research design and synthesis offered click chemistry products with interesting biological motifs mainly 1,2,3 triazoles linked to phenyl imidazole as promising candidates for further investigation as anticancer drugs.
1. Introduction
Despite several anticancer drugs being available in the market hitherto, many impediments of current anticancer remedies have implored researchers to keep looking for new candidates [1,2,3]. However, heterocycles are well documented as fascinating skeletons when constructing molecules that will interact with the targets and disrupt the biological pathways associated with cancer progression. In addition, the relative ease in designing these ring systems with several substituents enables them to cover a wide range of chemical space and further making them as significant starting points for anticancer drug design and discovery [4,5].
Being a group of highly diversified structures, nitrogen rich heterocycles and their fused systems are widely incorporated into the structure of various pharmacologically active agents and synthetic drugs [6]. Among these bioactive heterocycles, imidazole derivatives [7], particularly substituted thioimidazoles are known in medicinal chemistry as anti-obesity [8], antitubercular [9], antidiabetic [10], anticancer [11,12,13], antimicrobial [14,15], and antioxidant agents [16].
1,2,3-triazoles have also marked their position as a significant pharmacophore from nitrogen rich heterocyclic compounds with spectacular therapeutic potential [17,18]. Rufinamide (anticonvulsant), TSAO (anti-HIV), cefatrizine (antibiotic), tazobactam (antibacterial), and CAI (anticancer) are some of the FDA approved drugs bearing 1,2,3-triazole moiety [19,20,21,22].
Lately, it has been observed that among the combinatorial libraries of derivatized heterocycles, the most active ones had a bi-heterocyclic structure, which provides better binding opportunities in comparison to mono-heterocycles [23,24,25]. Consequently, we envisioned the molecular coupling of thioimidazole derivative with the 1,2,3-triazole core through a flexible spacer. The resulting molecular hybrids could deliver better interactions with biological targets by means of their synergistic effects [26].
Physicochemical, pharmacokinetic, and pharmacodynamics features, over the past few decades, have emanated as one of the crucial phases in drug discovery. This “pharmaceutical profiling” strategy furnishes a variety of suitable “drug-like” traits that can be developed into a structure-property association. Jointly, this information is utilized to escort chemical synthesis and nominate the best candidates for further drug advancement. Drug discovery and research organizations have set up a capability for the structured in vitro computation of properties which are pivotal to the understanding of absorption, distribution, metabolism, excretion, and toxicity (ADMET) behavior in vivo. In silico ADMET forecasting is expected to minimize the probability of late-stage attrition of drug development procedure and to optimize screening and trials by gazing at the promising drug candidates only. Molecular docking is a computational simulation strategy of an aspirant ligand binding to a macromolecule (receptor) and foretells the favorable orientation of binding one compound to the second one to make a stable complex. In silico molecular docking is employed to forecast the binding affinity and activity of the small molecule to their target receptors by availing scoring functions. Hence, the docking approach plays an influential role in the rational design of drugs in the drug development process. The synthesized ligand 4k was appraised by the in silico docking analysis for procuring an additional comprehension of the binding pattern with the glycogen synthase kinase-3 (GSK-3). Glycogen synthase kinase-3 (GSK-3), a serine/threonine protein kinase, was initially described as a key enzyme involved in glycogen metabolism [27,28]. Recent reports have suggested that GSK-3β is a positive regulator of cancer cell proliferation and survival [29,30,31,32], thus, providing further support for GSK-3β as a therapeutic target in cancer.
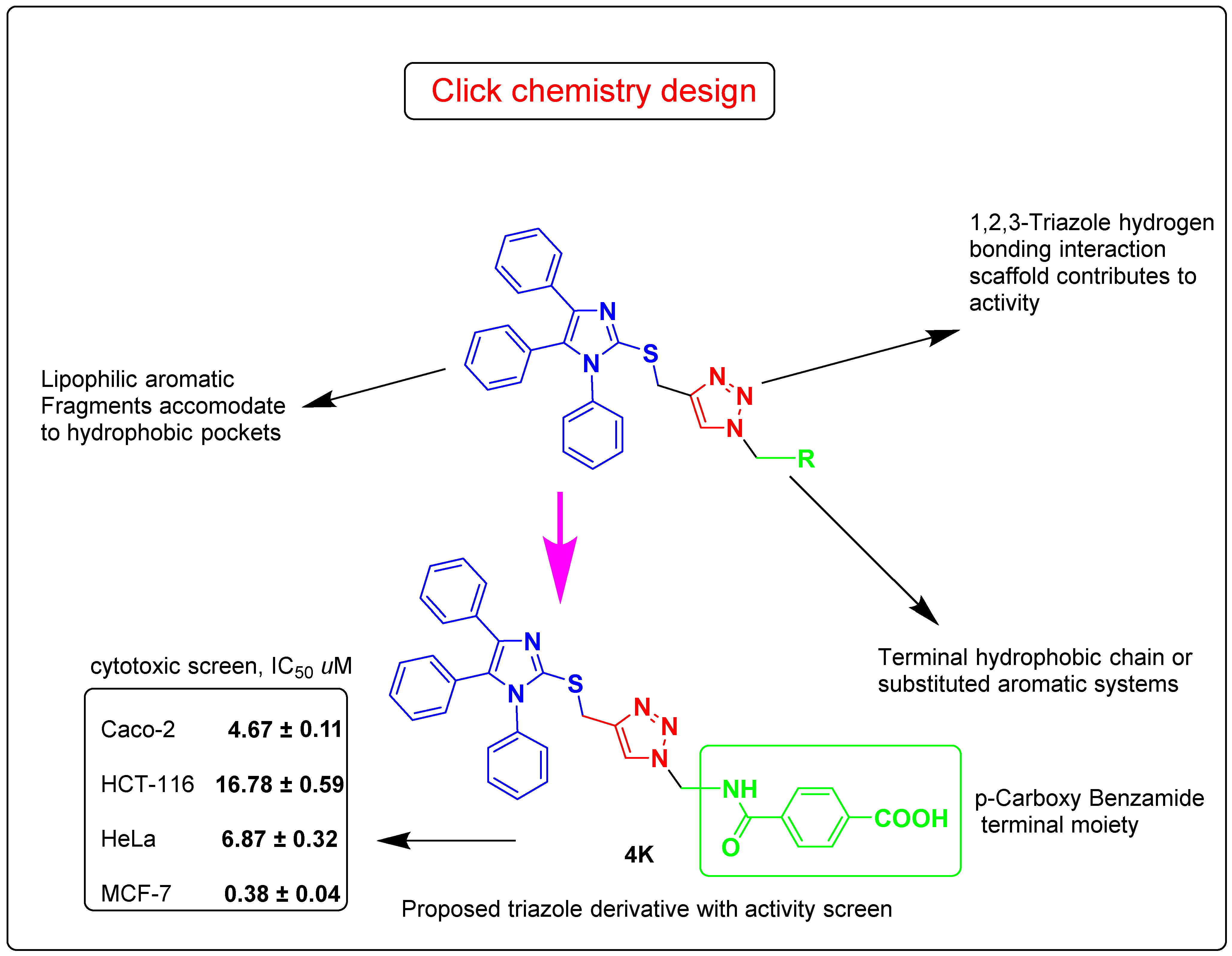
Considering the facts mentioned above, and pursuing our ongoing interest in the design of bioactive 1,2,3-triazoles molecular conjugates [33,34,35,36,37,38,39,40,41,42,43], we describe herein the design and synthesis of focused 1,4-disubstituted 1,2,3-triazoles, linked to the imidazole ring via regioselective copper catalyzed-1,3-dipolar cycloaddition reaction of S-propargylimidazole and some selected un/functionalized alkyl and aryl azides. The newly synthesized adducts were subjected to in vitro anticancer evaluation utilizing an MTT colorimetric assay against four different human cancer cell lines (Caco-2, HCT-116, HeLa, and MCF-7). The physicochemical and ADMET properties were also forecasted. Further mechanistic computational analysis was done by molecular docking screening of the synthesized click product 4k using GSK-3β as the target receptor. A schematic diagram for the study work is given in Figure 1.
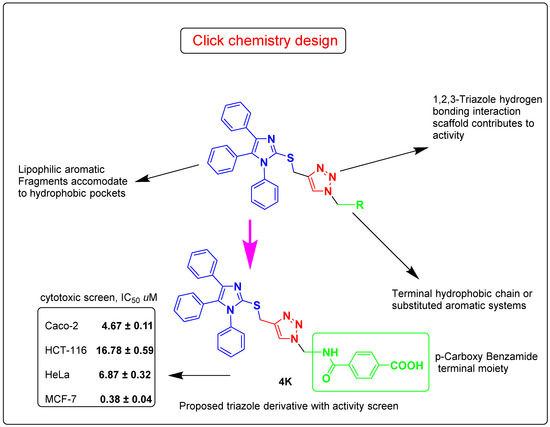
Figure 1.
Schematic diagram for the study work.
2. Results and Discussion
2.1. Chemistry
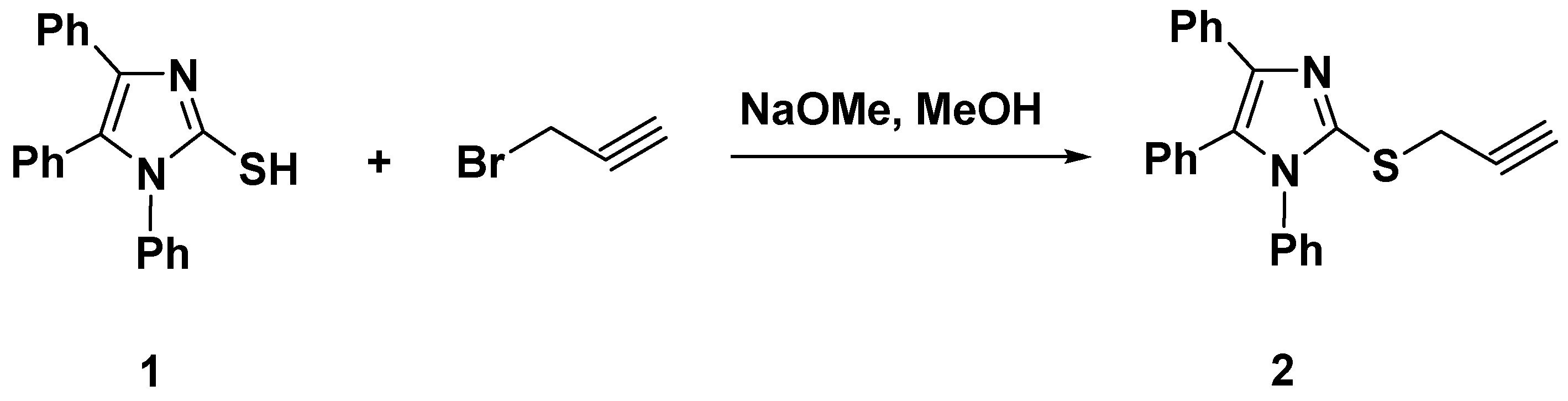
Our strategy for designing two sets of novel bi-heterocyclic based on imidazole-1,2,3-triazole combined systems was driven by employing a substituted imidazole, inserting thiomethylene bridge as a flexible linker between imidazole and triazole moieties, along with derivatizing triazole at the N-1 position with flexible (Scheme 2) and rigid (Scheme 3) groups to enhance the solubility and/or bioavailability of the resulting products, which is incredibly essential for crossing the cell membrane and reaching the target. Initially, 1,4,5-triphenylimidazole-2-thione (1) upon reaction with propargyl bromide in refluxing the methanolic solution of sodium methoxide for 2 h, yielded the desired thioimidazole-based alkyne 2 in 90% yield, Scheme 1.
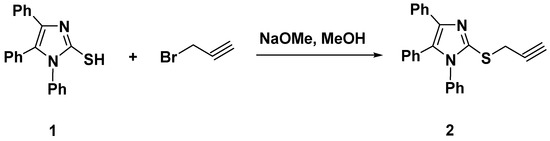
Scheme 1.
Synthesis of the thioimidazole-based alkyne 2.
The formation of the thiopropargylated imidazole 2 has been approved based on its spectral analysis. The sharp band observed at 3300 cm−1 in the IR spectrum indicated the presence of the acetylenic hydrogen (≡C–H), while the (C≡C) group appeared also as a sharp band at 2140 cm−1. Additionally, its 1H NMR revealed the disappearance of the SH proton of its starting material 1 and the appearance of a new significant singlet in the aliphatic region at δH 3.25 and 3.99 ppm assigning the sp-CH and SCH2 protons, respectively, guaranteed the incorporation of a propargyl side chain. The aromatic protons of the phenyl rings have been recorded at their usual chemical shifts (δH 7.18–7.46 ppm). Moreover, the 13C NMR spectrum exhibited the diagnostic propargyl carbon signals (SCH2 and C≡C) at δC 21.54, 74.81, and 80.47 ppm, respectively. The signals observed at δC 126.72–141.39 ppm were assigned to the aromatic and C=N carbons.
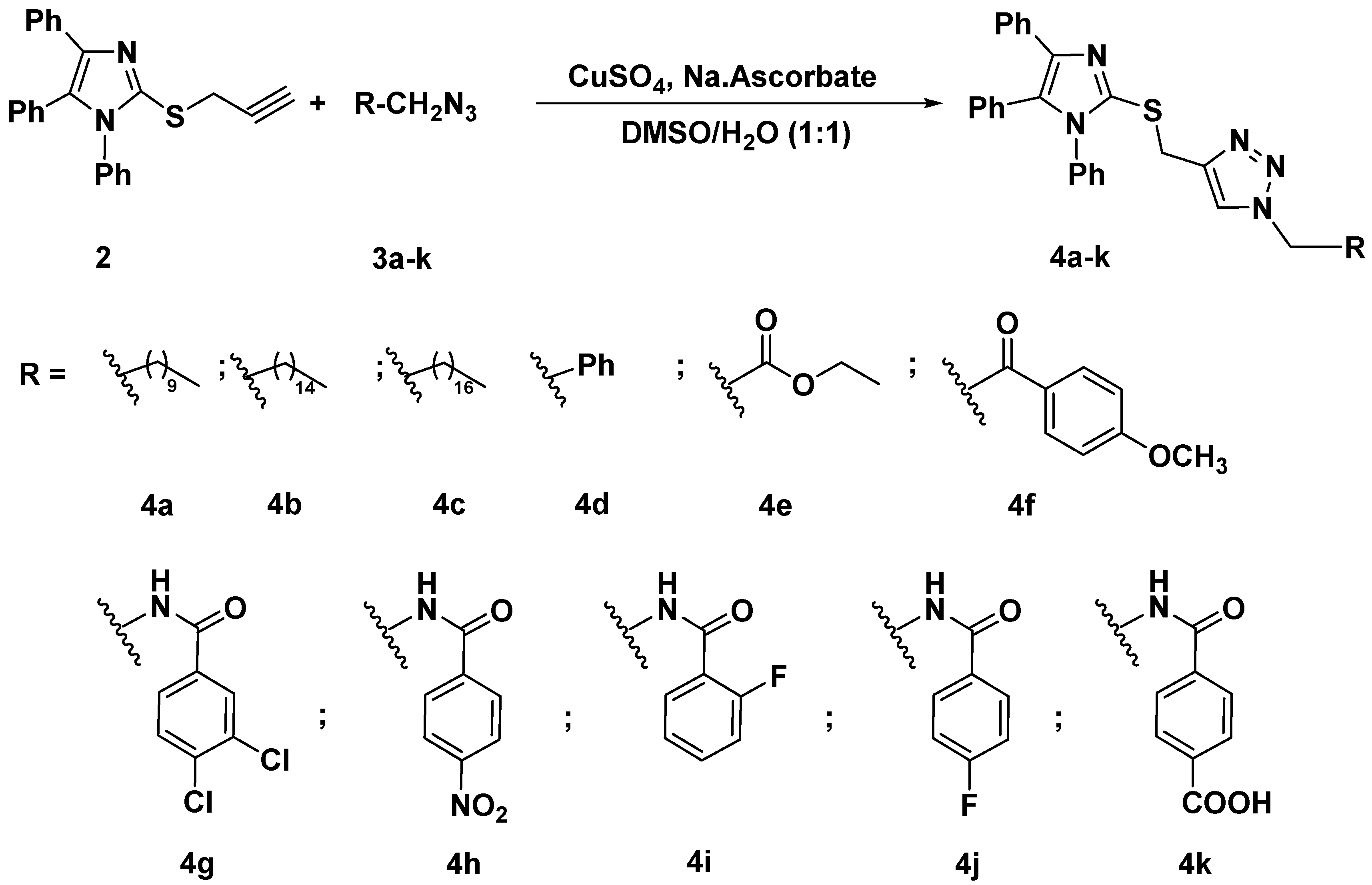
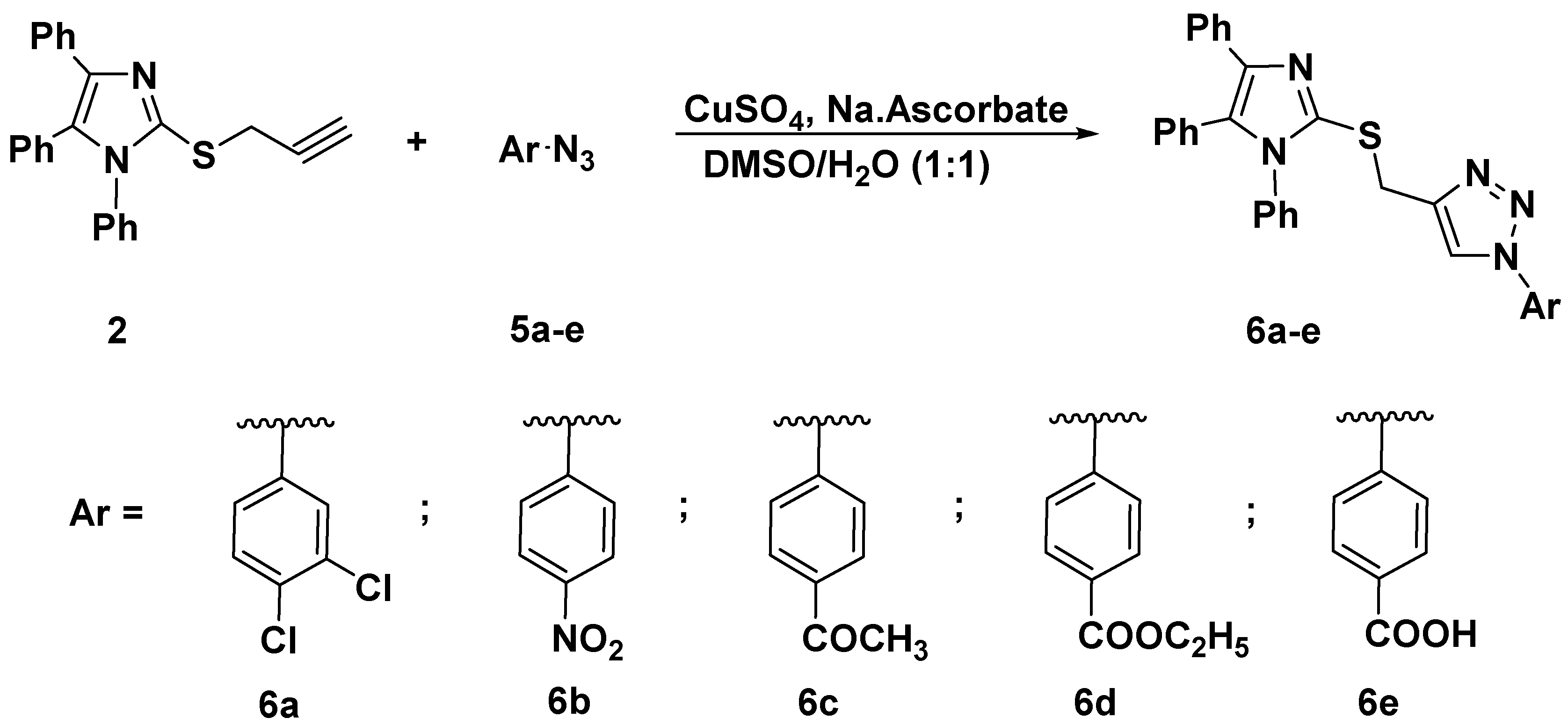
The synthesized alkyne derivative 2 was then coupled with a rich variety of organic azides 3a–k and 5a–e via the 1,3-dipolar cycloaddition reaction catalyzed by copper sulphate and sodium ascorbate in a mixture of DMSO:H2O (1:1) at room temperature to afford the targeted imidazole-1,2,3-triazole hybrids carrying un-/functionalized alkyl and/or phenyl acetamide side chain 4a–k (86–93%) and 6a–e (85–86%) and possessing rigid functionalized aromatic units; as described in Scheme 2 and Scheme 3, respectively.
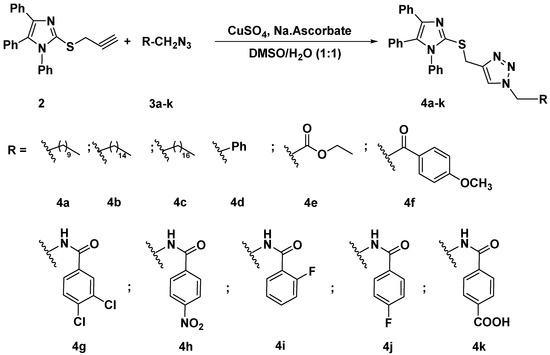
Scheme 2.
Click synthesis of imidazole-1,2,3-triazole hybrids carrying un/functionalized alkyl and/or phenyl acetamide side chain 4a–k.
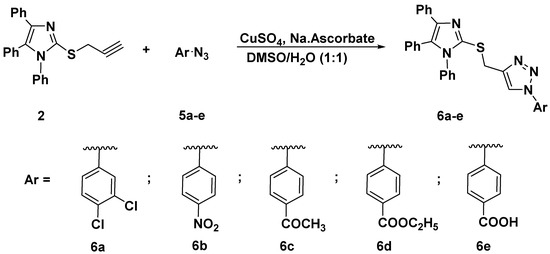
Scheme 3.
Click synthesis of imidazole-1,2,3-triazole hybrids carrying un/functionalized aromatic unit 6a–e.
The spectroscopic data including IR, 1H NMR, 13C NMR, and elemental analyses of all newly designed hybrids 4a–k and 6a–e approved the success of the click reaction. By taking the adduct 1,2,3-triazole 4e as a model, its IR spectrum disclosed the absence of absorption bands related to the C≡C and ≡C-H groups and the presence of characteristic bands at 1725, 2889, and 2967 cm−1 related to the C=O group and aliphatic hydrogens, respectively. Additionally, the 1H NMR spectrum exhibited a diagnostic singlet at δH 8.04 ppm attributed to the CH-1,2,3-triazole ring. The spectrum also showed characteristic triplet and quartet at δH 1.20 and 4.14-4.19 ppm belonging to the ester functionality. While the SCH2 and NCH2 protons resonated as two singlets at δH 4.50 and 5.39 ppm, respectively. Furthermore, the 13C NMR analysis supported the proposed structure 4e by the absence of the alkyne carbons of its precursor alkyne 2 and the presence of new signals at δC 14.43 and 61.34 ppm attributed to the ester side chain carbons. The presence of the ester functionality has been also confirmed by the appearance of a downfield signal at δC 167.74 ppm assigned to the ester carbonyl carbon.
2.2. Biological Screening
2.2.1. In Vitro Anticancer Study
The synthesized imidazole-1,2,3-triazole hybrids were screened for their anticancer activity against three different types of cancers namely colon, cervical, and breast cancers using doxorubicin as the reference drug. Cancer cell lines included Caco2 and HCT116 (human colon carcinoma), HeLa (human cervical carcinoma), and MCF-7 (human breast adenocarcinoma). The results were reported as half maximal inhibitory concentration (IC50) caused by the tested candidates (Table 1).

Table 1.
Anticancer screening of the synthesized compounds expressed as IC50 (μM) using the MTT assay. IC50 values are expressed as the mean ±SD of three independent experiments.
Initial screening revealed that most of the synthesized triazoles showed very good to good activity, i.e., their IC50 lies in the range of 4.67–20.69 µM (Caco-2), 4.80–30.41 µM (HeLa), and 0.38–27.29 µM (MCF-7), except 2, 4d, 4e, 6c, and 6d. Interestingly, a preliminary structure–activity relationship analysis urged the low activity of the acetylated 4e, 6c, and 6d derivatives due to the presence of acetyl functionality in their structures in the form of ketone or ester composition. The S-propargylimidazole 2 (high IC50) have not displayed any significant anticancer activity against all the used cell lines, confirming the benefit of the molecular hybridization. Among the newly synthesized click adducts, the triazole 4k was found to be one of the most potent derivatives against MCF-7 cell lines with IC50 of 0.38 µM and demonstrated similar potency to the standard drug, doxorubicin. This could be presumably due to the presence of a lipophilic long alkyl side chain appended to the 1,2,3-triazole ring in compound 4k. Moreover, the imidazole triazole conjugates, 4k and 6e bearing aromatic carboxylic group in their structures, displayed better potency amongst the synthesized compounds against the Caco-2 cell line with IC50 of 4.67 ± 0.11 µM and 5.22 ± 0.20 µM, respectively, significantly similar to the standard doxorubicin (IC50 = 5.17 ± 0.25 µM). The replacement of aromatic carboxylic group by the fluorinated phenyl group decreased the anticancer potency against the Caco-2 cell line, compound 4i (IC50 = 6.31 ± 0.17 µM) and compound 4g (IC50 = 8.45 ± 0.18 µM). Whereas, triazoles 4g and 4h having electron withdrawing groups (Cl, NO2) showed a moderate anticancer activity ranging from 12.31 ± 0.22 µM to 15.78 ± 0.31 µM. These results suggested that, the designed triazoles are promising candidates for the future anticancer molecule discovery and research.
2.2.2. In Silico ADMET Analysis
In silico predictions of the physicochemical descriptors, drug likeness or ADMET (adsorption, distribution, metabolism, excretion, and toxicity) properties, have increased the possibility of detecting new lead compounds in a much shorter time span as compared to the conventional/traditional procedures. In silico studies were conducted to confirm the precision of in vitro biological outcomes. Various physicochemical traits, i.e., the presence of a particular class of atoms or bonds, lipophilicity, molar refractivity, and solubility in water and TPSA (topological polar surface area) were computed. These physicochemical properties are given in Table 2. The forecasted physicochemical characteristics are in agreement with the exercised criteria and are presumed to possess a good bioavailability score as all the compounds have TPSA ≤ 140 Å2 except compound 4h.

Table 2.
Physicochemical properties of the selected compounds.
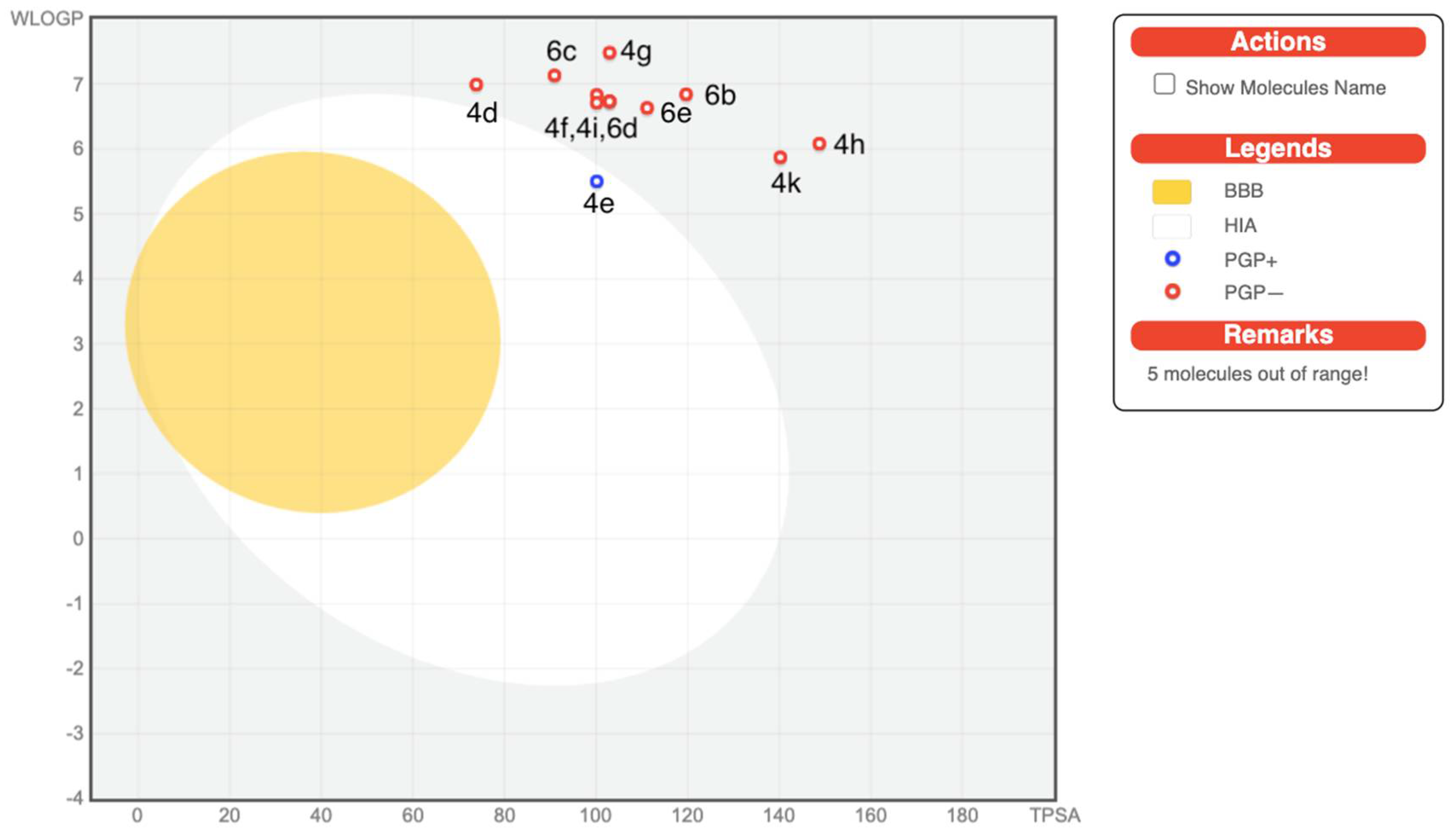
On the other hand, ADME predictions are documented in Table 3, revealing that all the compounds have low gastrointestinal absorptions (GI) except compound 4e. All the compounds displayed no blood brain barrier (BBB) permeation. Few of the tested compounds are P-gp (p-glycoprotein) inhibitors, i.e., 2, 4a–4c, and 4e. The BOILED-Egg diagram which is a pooled built-in graphical classification model for the prediction of BBB permeations, passive human gastrointestinal absorption (HIA), and P-gp substrates, is shown in Figure 2. In terms of metabolism, all the synthesized compounds were tested for inhibition of the Cytochrome P450 isomers, i.e., CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4. Most of the compounds are non-inhibitors of CYP1A2 and CYP2D6 with the exception of 4b, as it is a CYP1A2 inhibitor and 4a which also inhibits both CYP1A2 and CYP2D6. Most of the tested molecules are CYP3A4 inhibitors. The tested compounds emerged to be non-inhibitors of CYP2D6. The skin permeability coefficient (log Kp; with Kp in cm/s) values display low permeability through the skin for most of the compounds except, compound 2, 4a–4c which displayed good skin permeability.

Table 3.
Pharmacokinetic/adsorption, distribution, metabolism, excretion, and toxicity (ADME) properties of the selected compounds.
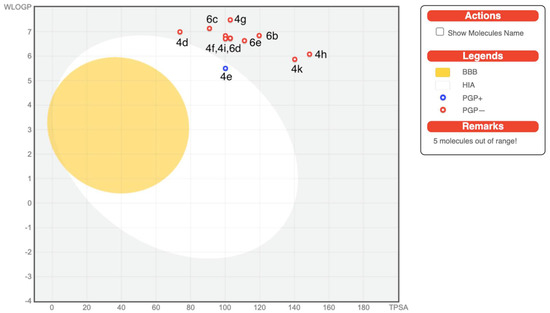
Figure 2.
BOILED-Egg diagram of the selected compounds.
Five different rule-based sieves, with diverse ranges of characteristics inside of which the drug candidate is defined as drug-like were appraised, i.e., Lipinski [44], Ghose [45], Veber [46], Egan [47], and Muegge [48] rules. Drug-like predictions with a number of violations to the above-mentioned rules, their bioavailability scores, and toxicity profiles are documented in Table 4. All the tested compounds violated the Ghose and Muegge rules. The Lipinski and Egan rules are violated by most of the compounds except compound 4e. Most of the compounds follow the Veber rule with few exceptions. Our results indicated that the tested compounds were in good agreement in terms of the bioavailability score ranging from 0.17–0.55. Compounds 4e, 6a, 6d, and 6e are predicted to be non-toxic in nature by AMES, i.e., they are not mutagenic. All the synthesized compounds are forecasted as non-carcinogens. Doxorubicin was predicted as AMES toxic, mutagenic, and non-carcinogenic in nature.

Table 4.
Drug-likeness and toxicity predictions of the selected compounds.
2.2.3. Molecular Docking Simulations
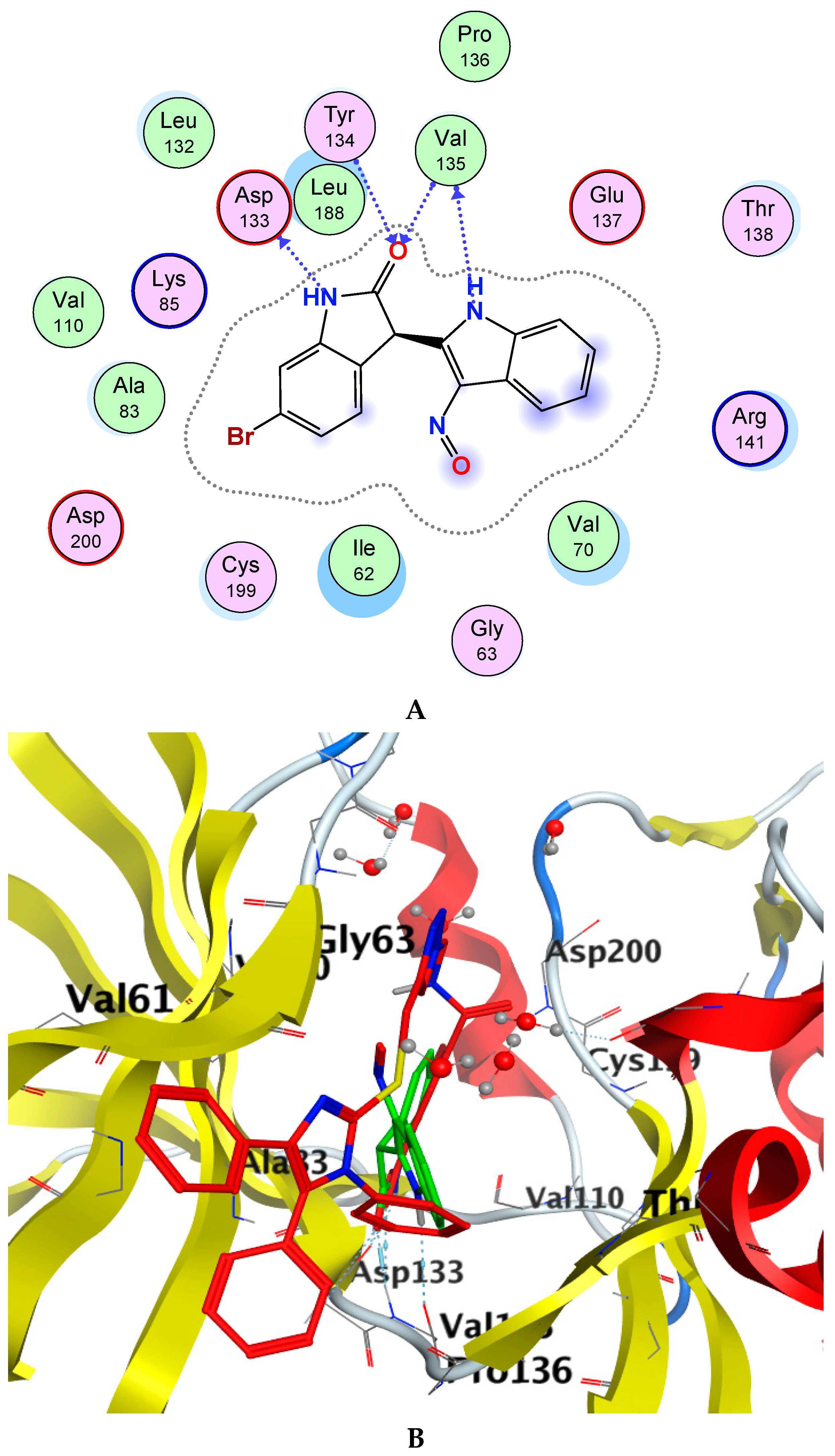
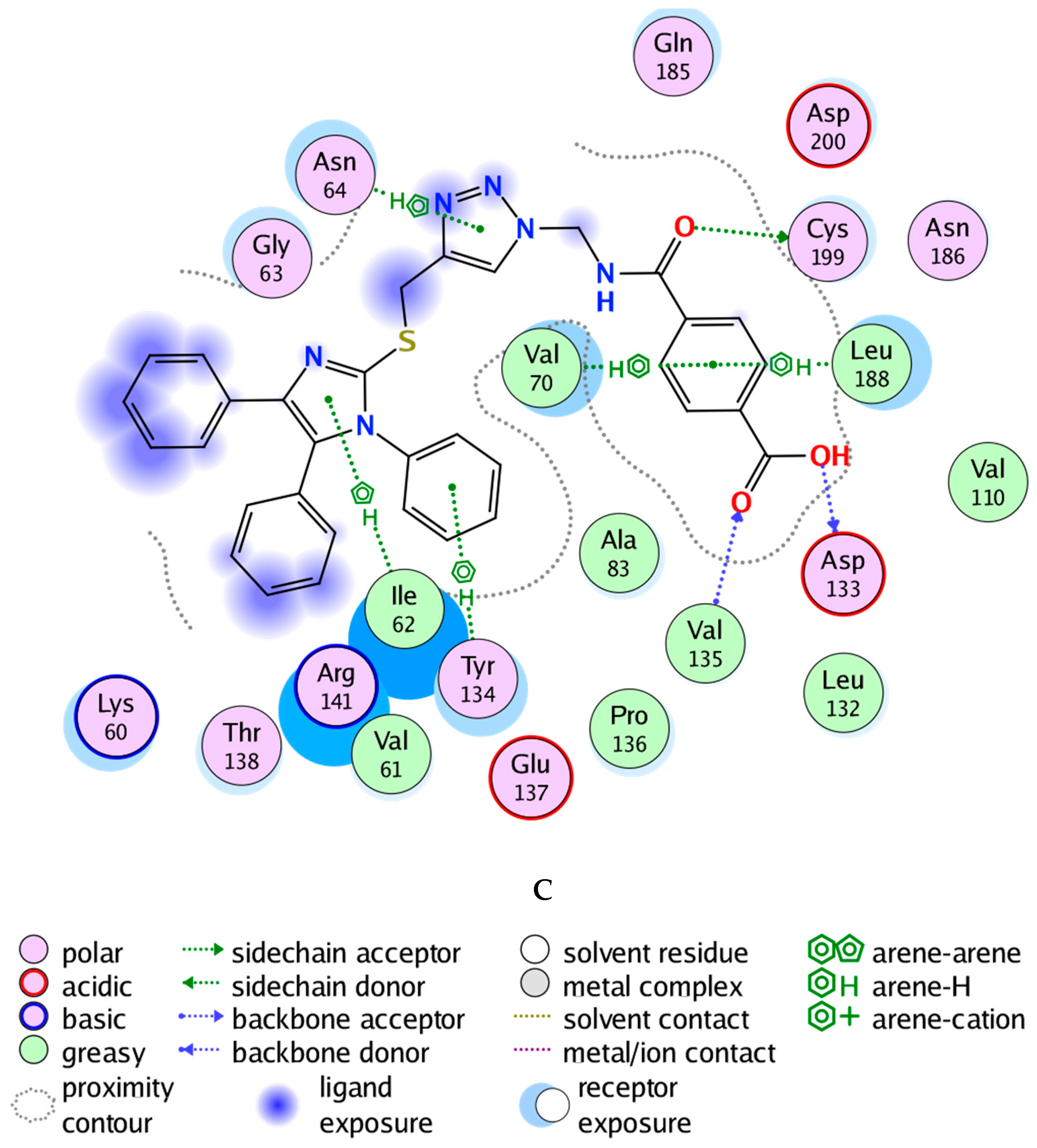
Glycogen synthase kinase-3 beta (GSK-3β) is a promising target with an overexpressed oncogene in different breast cancers [49], so we evaluated the binding modes of our potent compound 4k as a selective MCF-7 cytotoxic agent against the GSK-3β ATP binding pocket and confirmed the overall prognostic dependency of breast cancer patients on the GSK-3β inhibition effect. We docked the 4k test ligand on our target protein, i.e., GSK-3β (PDB ID: 1UV5), in order to analyze the binding pattern and affinity using the software MOE. Docking poses of the bound and tested compound 4k are depicted in Figure 3A–C. The compound 4k displayed an excellent binding interaction and affinity, i.e., −9.8 kcal/mol toward GSK-3β target compared to the reference drug; 6-Bromoindirubin-3′-oxime with −8.8 kcal/mol. The interaction analysis of the bound ligand; 6-bromoindirubin-3′-oxime, exhibited that the cyclic nitrogen of the pyrrole ring donates a hydrogen bond to the peptide carbonyl oxygen of Val135, while the cyclic nitrogen of the lactam ring donates a hydrogen bond to the carbonyl oxygen of Asp133, and the lactam carbonyl oxygen accepts a hydrogen bond from the backbone amide of Val135 and Tyr134. The molecular docking studies revealed that the compound 4k emerged to be a promising drug candidate; on the account of its lowest binding energy, i.e., −9.8 kcal/mol with the active site residues of the target receptor GSK-3β and this might be one of the reasons for its good in vitro anticancer activity. The putative interaction of compound 4k with GSK3 suggested that the terminal aromatic ring might display two hydrogen bond interactions through the COOH substituent with the backbone carbonyl group of Asp133 and the backbone amide of Val135, respectively. Moreover, another essential linker hydrogen bond through the carbonyl of amide with conserved Cys199 stabilized the compound in the pocket. In addition, the fit of the aromatic systems for the whole compound occurs through a group of aromatic stacking interactions with corresponding residues; Ile62, Tyr134, Asn64 for triazole ring, Val70, and Leu188 that might support selective binding. The 2D and 3D analysis of the target compound 4k mapped to the 6-Bromoindirubin-3′-oxime as a reference is shown in Figure 3. Taken together, these observations might rationalize the observed cytotoxic activity of 4k through the inhibitory effect of overexpressed GSK-3β within the cancer cell lines.
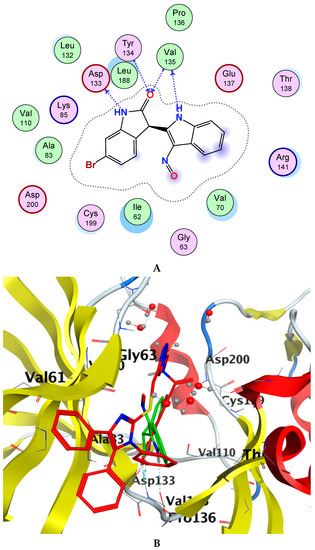
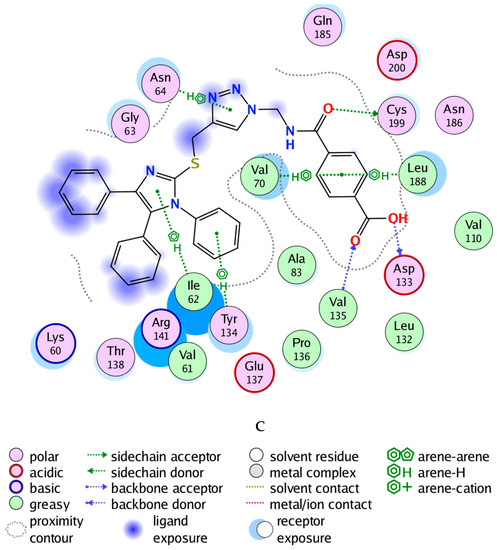
Figure 3.
Docking poses of selected compound 4k onto the active site of glycogen synthase kinase-3 beta (GSK-3β) (PDB ID: 1UV5). (A) Two-dimensional (2D) interaction data and residues of bound ligand 6-bromoindirubin-3′-oxime within the ATP pocket of the target. (B) 2D of the compound within the ATP pocket with the corresponding hydrogen bonding and aromatic interactions with residues. (C) Three-dimensional (3D) analysis of the target compound 4k mapped to the reference drug, i.e., 6-Bromoindirubin-3′-oxime.
3. Experimental Section
The 1H NMR spectra were recorded using an Advance Bruker NMR spectrometer (Bruker, Switzerland) at 400–600 MHz, while 13C NMR spectra were recorded on the same instrument at 100–150 MHz using tetramethylsilane (TMS) (d, ppm) as the internal standard. The EI mass spectra were measured with a Finnigan MAT 95XL spectrometer (Finnigan, Germany). Sonochemical reactions were performed in a Kunshan KQ-250B ultrasound cleaner (50 kHz, 240 W, China).
All the solvents and reagents used in this work were of the highest quality of analytical reagent grade and purchased from Sigma-Aldrich, USA, and were used without further purification. All the reactions were monitored by thin layer chromatography (TLC), using UV fluorescent Silica gel type Merck 60 F254 plates. The spots were visualized using a UV lamp (254 nm). The melting points of the synthesized products were measured using a Stuart Scientific SMP1 (Stuart, UK). The functional groups were identified using a SHIMADZU FTIR-Affinity-1S spectrometer in the range of 400–4000 cm−1 using a Perkin-Elmer 1430 series FT-IR spectrometer (Perkin-Elmer, USA) as potassium bromide pellets. All the synthesized compounds were fully characterized by 1H, 13C NMR, and elemental analysis. The 1H NMR (400 MHz) and 13C NMR (100 MHz) spectra were investigated with an Advance Bruker NMR spectrometer (Bruker, Switzerland) (400 MHz) with TMS as an internal standard to calibrate the chemical shifts (δ) reported in ppm. Elemental analyses were performed using a GmbH, Vario EL III, Elementar Analyzer (HEKAtech GmbH, Germany).
3.1. Synthesis of the Title Compounds
The azides 3a–k and 5a–e used in this study were prepared according to the reported procedures [50,51,52]. The procedures for the synthesis of the reported compounds are described below.
3.1.1. Synthesis and Characterization of 1,4,5-triphenyl-2-(prop-2-yn-1-ylthio)-1H-imidazole (2)
A solution of imidazole 1 (12 mmol) in a methanolic solution (30 mL) of sodium methoxide (12 mmol) was stirred, then propargyl bromide (12 mmol) was added under stirring. The mixture was heated under reflux for 1 h until the completion of the reaction (as indicated by TLC; hexane—ethyl acetate 2:1). The solvent was removed by evaporation under reduced pressure; the solid formed was collected by filtration, washed with water, dried, and recrystallized from ethanol to give the desired imidazole-based alkyne 2 as colorless crystals in 90% yield, mp: 103–105 °C. IR (υ, cm−1): 1590 (C=C), 1620 (C=N), 2140 (C≡C), 2888, 2894 (C-H Al), 3050 (C-H Ar), 3300 (≡CH). 1H NMR (400 MHz, DMSO-d6): δH = 3.25 (s, 1H, ≡CH), 3.99 (s, 2H, SCH2), 7.18–7.28 (m, 10H, Ar-H), 7.39–7.46 (m, 5H, Ar-H) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 21.54 (SCH2); 74.81 (C≡CH); 80.47 (C≡CH); 126.72, 127.09, 128.68, 128.73, 128.94, 129.05, 129.53, 129.65, 130.40, 131.17, 131.89, 134.44, 135.59, 138.08, 141.39 (Ar-C, C=N) ppm. Calculated for C24H18N2S: C, 78.66; H, 4.95; N, 7.64. Found: C, 78.59; H, 4.89; N, 7.82.
3.1.2. General Procedure for the Synthesis of 1,4-disubstituted 1,2,3-triazoles Bearing Imidazole Moiety 4a–k and 6a–e
A solution of copper sulfate (0.10 g) and sodium ascorbate (0.15 g) in water (10 mL) was added dropwise to a solution of alkyne 2 (1 mmol) in DMSO (10 mL) under stirring. Then, the appropriate azide 3a–k and/or 5a–e (1 mmol) was added. The stirring was continued for 6-10 h at room temperature. The reaction was monitored via TLC (hexane-ethyl acetate 2:1), and after the completion of the reaction, the mixture was poured onto iced-water. The precipitate thus formed was collected by filtration, washed with a saturated solution of ammonium chloride, and recrystallized from ethanol/DMF to give the targeted 1,2,3-triazoles 4a–k and 6a–e.
4-(((1,4,5-Triphenyl-1H-imidazol-2-yl)thio)methyl)-1-undecyl-1H-1,2,3-triazole (4a)
This compound was obtained as a yellow solid in 90% yield, mp: 99–101 °C. IR (υ, cm−1): 1525 (C=C), 1610 (C=N), 2910-2880 (CH-Al), 3030 (CH-Ar). 1H NMR (400 MHz, DMSO-d6): δH = 0.87 (t, 3H, J = 4.0 Hz, CH3), 1.19-1.28 (m, 16H, 8×CH2), 1.77 (bs, 2H, NCH2CH2), 4.27 (t, 2H, J = 4.0 Hz, NCH2), 4.46 (s, 2H, SCH2), 7.18-7.29 (m, 10H, Ar-H), 7.33-7.46 (m, 5H, Ar-H), 8.08 (s, 1H, CH-1,2,3-triazole) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 14.45 (CH3); 26.15, 28.66, 29.22, 29.30, 29.36, 29.38, 30.14 (CH2); 27.53 (SCH2); 54.87 (NCH2); 125.42, 126.62, 126.77, 127.06, 128.65, 128.68, 128.86, 129.03, 129.45, 129.63, 130.38, 131.13, 131.64, 134.57, 135.59, 142.39, 142.46 (Ar-C, C=N) ppm. Calculated for C27H25N5S: C, 74.56; H, 7.33; N, 12.42. Found: C, 74.78; H, 7.45; N, 12.66.
4-(((1,4,5-Triphenyl-1H-imidazol-2-yl)thio)methyl)-1-hexadecyl-1H-1,2,3-triazole (4b)
This compound was obtained as a light-yellow solid in 88% yield, mp: 90–91 °C. IR (υ, cm−1): 1530 (C=C), 1620 (C=N), 2955 (C-H Al), 3050 (C-H Ar). 1H NMR (400 MHz, DMSO-d6): δH = 0.88 (t, 3H, J = 4.0 Hz, CH3), 1.16-1.26 (m, 26H, 13×CH2), 1.77 (t, 2H, J = 4.0 Hz, NCH2CH2), 4.29 (s, 2H, NCH2), 4.45 (s, 2H, SCH2), 7.20-7.28 (m, 10H, Ar-H), 7.36-7.47 (m, 5H, Ar-H), 8.06 (s, 1H, CH-1,2,3-triazole) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 14.46 (CH3); 22.56, 26.24, 28.79, 29.24, 29.29, 29.39, 29.48, 29.56, 30.13 (CH2); 27.57 (SCH2); 54.81 (NCH2); 125.40, 126.65, 126.76, 127.09, 128.62, 128.65, 128.81, 129.08, 129.48, 129.69, 130.35, 131.10, 131.61, 134.53, 135.55, 142.33, 142.52 (Ar-C, C=N) ppm. Calculated for C27H25N5S: C, 75.79; H, 8.11; N, 11.05. Found: C, 75.58; H, 8.23; N, 11.29.
4-(((1,4,5-Triphenyl-1H-imidazol-2-yl)thio)methyl)-1-octadecyl-1H-1,2,3-triazole (4c)
This compound was obtained as a pale yellow solid in 88% yield, mp: 10–110 °C. IR (υ, cm−1): 1560 (C=C), 1620 (C=N), 2955 (C-H Al), 3050 (C-H Ar). 1H NMR (400 MHz, DMSO-d6): δH = 0.85 (t, 3H, J = 4.0 Hz, CH3), 1.14-1.29 (m, 30H, 15×CH2), 1.74 (t, 2H, J = 4.0 Hz, NCH2CH2), 4.26 (s, 2H, NCH2), 4.49 (s, 2H, SCH2), 7.21-7.30 (m, 10H, Ar-H), 7.35-7.48 (m, 5H, Ar-H), 8.10 (s, 1H, CH-1,2,3-triazole) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 14.43 (CH3); 22.59, 26.27, 26.65, 26.89, 28.72, 29.27, 29.35, 29.48, 29.56, 29.59, 30.15 (CH2); 27.59 (SCH2); 54.89 (NCH2), 125.42, 126.69, 126.79, 127.17, 128.71, 128.69, 128.88, 129.13, 129.42, 129.68, 130.34, 131.13, 131.64, 134.55, 135.58, 142.37, 142.58 (Ar-C, C=N) ppm. Calculated for C27H25N5S: C, 76.20; H, 8.37; N, 10.58. Found: C, 76.48; H, 8.26; N, 10.77.
4-(((1,4,5-Triphenyl-1H-imidazol-2-yl)thio)methyl)-1-benzyl-1H-1,2,3-triazole (4d)
This compound was obtained as colorless crystals in 93% yield, mp: 239–240 °C. IR (υ, cm−1): 1505 (C=C), 1630 (C=N), 2960 (CH- Al), 3090 (CH-Ar). 1H NMR (400 MHz, DMSO-d6): δH = 4.44 (s, 2H, SCH2), 5.58 (s, 2H, CH2), 7.15-7.21 (m, 5H, Ar-H), 7.23-7.45 (m, 15H, Ar-H), 8.05 (s, 1H, CH-1,2,3-triazole) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 27.85 (SCH2); 53.22 (NCH2); 124.04, 126.71, 127.02, 128.46, 128.61, 128.67, 128.87, 129.02, 129.22, 129.40, 129.57, 130.44, 131.14, 131.65, 134.54, 135.64, 136.45, 137.95, 142.29, 143.93 (Ar-C, C=N). Calculated for C31H25N5S: C, 74.52; H, 5.04; N, 14.02. Found: C, 74.88; H, 5.12; N, 14.24.
Ethyl-2-(4-(((1,4,5-triphenyl-1H-imidazol-2-yl)thio)methyl)-1H-1,2,3-triazol-1-yl)acetate (4e)
This compound was obtained as a yellow solid in 90% yield, mp: 103–105 °C. IR (υ, cm−1): 1255 (C-O), 1509 (C=C), 1610 (C=N), 1725 (C=O), 2889, 2967, (CH-Al), 3088 (CH-Ar). 1H NMR (400 MHz, DMSO-d6): δH 1.20 (t, 3H, J = 4.0 Hz, CH3), 4.14-4.19 (q, 2H, OCH2CH3), 4.50 (s, 2H, SCH2), 5.39 (s, 2H, NCH2), 7.19-7.27 (m, 10H, Ar-H), 7.36-7.48 (m, 5H, Ar-H), 8.04 (s, 1H, CH-1,2,3-triazole) ppm. 13C NMR (100 MHz, DMSO-d6): δC 14.43 (CH3); 27.88 (SCH2); 50.54 (NCH2), 61.34 (OCH2), 125.40, 124.43, 126.63, 126.79, 127.03, 128.63, 128.66, 128.88, 129.02, 129.45, 129.61, 130.36, 130.44, 131.15, 131.62, 131.69, 134.52, 134.58, 135.56, 135.63, 137.96, 142.35, 142.41 (Ar-C, C=N); 167.74 (C=O) ppm. Calculated for C28H25N5O2S: C, 67.86; H, 5.08; N, 14.13. Found: C, 67.58; H, 5.18; N, 14.39.
2-(4-(((1,4,5-Triphenyl-1H-imidazol-2-yl)thio)methyl)-1H-1,2,3-triazol-1-yl)-1-(4-methoxyphenyl) ethanone (4f)
This compound was obtained as a yellow solid in 87% yield, mp: 154–155 °C. IR (υ, cm−1): 1560 (C=C), 1620 (C=N), 1730 (C=O), 2960 (CH-Al), 3030 (CH-Ar). 1H NMR (400 MHz, DMSO-d6): δH = 3.80 (s, 3H, OCH3), 4.52 (s, 2H, SCH2), 6.02 (d, 2H, NCH2), 7.16-7.29 (m, 10H, Ar-H), 7.32-7.48 (m, 5H, Ar-H), 8.06-8.20 (m, 4H, Ar-H), 8.10 (s, 1H, CH-1,2,3-triazole) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 27.80 (SCH2); 52.42 (NCH2); 56.26 (OCH3); 118.87, 122.60, 126.09, 126.62, 128.81, 128.48, 129.65, 131.01, 131.41, 131.83, 134.44, 135.24, 136.24, 137.60, 138.45, 140.23, 141.86, 145.11, 145.29 (Ar-C, C=N); 191.13 (CO). Calculated for C33H27N5O2S: C, 71.07; H, 4.88; N, 12.56. Found: C, 71.32; H, 4.729; N, 12.78.
N-(3,4-Dichlorophenyl)-2-(4-(((1,4,5-triphenyl-1H-imidazol-2-yl)thio)methyl)-1H-1,2,3-triazol-1-yl)acetamide (4g)
This compound was obtained as a yellow solid in 90% yield, mp: 161–162 °C. IR (υ, cm−1): 1565 (C=C), 1628 (C=N), 1745 (C=O), 2899 (CH- Al), 3055 (CH-Ar), 3355 (NH). 1H NMR (400 MHz, DMSO-d6): δH = 4.50 (s, 2H, SCH2), 5.36 (s, 2H, NCH2), 7.11-7.64 (m, 17H, Ar-H), 7.96 (s, 1H, Ar-H), 8.06 (s, 1H, CH-1,2,3-triazole), 10.82 (s, 1H, NH) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 27.50 (SCH2); 52.68 (NCH2); 119.65, 120.87, 125.73, 126.95, 127.67, 128.23, 128.60, 128.99, 129.60, 130.19, 131.05, 131.38, 131.76, 134.45, 135.53, 138.92 (Ar-C, C=N); 165.11 (C=O). Calculated for C32H24Cl2N6OS: C, 62.85; H, 3.96; N, 13.74. Found: C, 62.69; H, 3.90; N, 13.93.
N-(4-Nitrophenyl)-2-(4-(((1,4,5-triphenyl-1H-imidazol-2-yl)thio)methyl)-1H-1,2,3-triazol-1-yl)acetamide(4h)
This compound was obtained as a yellow solid in 87% yield, mp: 144–145 °C. IR (υ, cm−1): 1565 (C=C), 1620 (C=N), 1700 (C=O), 2940 (CH-Al), 3080 (CH-Ar), 3340 (NH). 1H NMR (400 MHz, DMSO-d6): δH = 4.46 (s, 2H, SCH2), 5.36 (s, 2H, NCH2), 7.00-7.49 (m, 15H, Ar-H), 7.59-8.00 (m, 4H, Ar-H), 8.15 (s, 1H, CH-1,2,3-triazole), 10.60 (s, 1H, NH) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 27.53 (SCH2); 52.57 (NCH2); 115.91, 116.13, 121.38, 121.36, 125.69, 127.28, 128.49, 128.92, 129.06, 129.61, 129.71, 129.99, 131.03, 134.14, 135.22, 135.64 (Ar-C, C=N); 164.53 (C=O). Calculated for C32H25N7O3S: C, 65.40; H, 4.29; N, 16.68. Found: C, 65.66; H, 4.33; N, 16.82.
N-(2-Fluorophenyl)-2-(4-(((1,4,5-triphenyl-1H-imidazol-2-yl)thio)methyl)-1H-1,2,3-triazol-1-yl) acetamide (4i)
This compound was obtained as a pale yellow solid in 89% yield, mp: 118–119 °C. IR (υ, cm−1): 1555 (C=C), 1630 (C=N), 1693 (C=O), 3070 (CH-Ar), 3330 (NH). 1H NMR (400 MHz, DMSO-d6): δH = 4.58 (s, 2H, SCH2), 5.39 (s, 2H, NCH2), 7.18-7.29 (m, 10H, Ar-H), 7.34-7.48 (m, 5H, Ar-H), 8.07-8.22 (m, 4H, Ar-H), 8.09 (s, 1H, CH-1,2,3-triazole), 10.85 (s, 1H, NH) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 27.59 (SCH2); 52.76 (NCH2); 119.89, 120.77, 125.69, 126.95, 127.70, 128.56, 128.47, 128.80, 129.61, 131.12, 131.21, 132.07, 132.56, 133.98, 135.76, 140.57 (Ar-C, C=N); 165.23 (C=O). Calculated for C32H25FN6OS: C, 68.55; H, 4.49; N, 14.99. Found: C, 68.70; H, 4.60; N, 14.82.
N-(4-Fluorophenyl)-2-(4-(((1,4,5-triphenyl-1H-imidazol-2-yl)thio)methyl)-1H-1,2,3-triazol-1-yl) acetamide (4j)
This compound was obtained as a pale yellow solid in 86% yield, mp: 126–127 °C. IR (υ, cm−1): 1570 (C=C), 1625 (C=N), 1695 (C=O), 2925 (CH-Al), 3085 (CH-Ar), 3310 (NH). 1H NMR (400 MHz, DMSO-d6): δH = 4.51 (s, 2H, SCH2), 5.41 (s, 2H, NCH2), 7.16-7.28 (m, 10H, Ar-H), 7.31-7.45 (m, 5H, Ar-H), 8.02-8.15 (m, 4H, Ar-H), 8.14 (s, 1H, CH-1,2,3-triazole), 10.89 (s, 1H, NH) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 27.59 (SCH2); 52.76 (NCH2); 119.60, 120.82, 125.85, 126.91, 127.76, 128.45, 128.54, 128.87, 129.68, 130.34, 131.11, 131.29, 131.67, 134.51, 135.48, 140.36 (Ar-C, C=N); 165.47 (C=O). Calculated for C32H25FN6OS: C, 68.55; H, 4.49; N, 14.99. Found: C, 68.78; H, 4.58; N, 14.76.
4-(2-(4-(((1,4,5-Triphenyl-1H-imidazol-2-yl)thio)methyl)-1H-1,2,3-triazol-1-yl)acetamido)benzoic acid (4k)
This compound was obtained as a yellow solid in 86% yield, mp: 129–130 °C. IR (υ, cm−1): 1550 (C=C), 1615 (C=N), 1725 (C=O), 2510-3300 (OH, NH). 1H NMR (400 MHz, DMSO-d6): δH = 4.50 (s, 2H, SCH2), 5.39 (s, 2H, NCH2), 7.22-7.30 (m, 10H, Ar-H), 7.38-7.50 (m, 5H, Ar-H), 7.94 (d, 2H, J = 4.0 Hz, Ar-H), 8.14 (d, 2H, J = 4.0 Hz, Ar-H), 8.02 (s, 1H, CH-1,2,3-triazole), 10.78 (s, 1H, NH), 12.45 (s, 1H, COOH) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 27.60 (SCH2); 52.67 (NCH2); 119.76, 120.81, 125.72, 126.85, 127.76, 128.38, 128.55, 128.78, 129.54, 130.54, 131.21, 131.44, 132.09, 134.56, 135.78, 141.57 (Ar-C, C=N); 167.78 (C=O); 168.44 (C=O). Calculated for C33H26N6O3S: C, 67.56; H, 4.47; N, 14.33. Found: C, 67.83; H, 4.33; N, 14.78.
1-(3,4-Dichlorophenyl)-4-(((1,4,5-triphenyl-1H-imidazol-2-yl)thio)methyl)-1H-1,2,3-triazole (6a)
This compound was obtained as a pale yellow solid in 85% yield, mp: 124–125 °C. IR (υ, cm−1): 1525 (C=C), 1625 (C=N), 2925 (CH- Al), 3075 (CH-Ar). 1H NMR (400 MHz, DMSO-d6): δH = 4.54 (s, 2H, SCH2), 7.17-7.27 (m, 10H, Ar-H), 7.35-7.46 (m, 5H, Ar-H), 7.84-7.94 (m, 2H, Ar-H), 8.23 (s, 1H, Ar-H), 8.84 (s, 1H, CH-1,2,3-triazole) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 27.55 (SCH2); 120.33, 122.11, 122.68, 126.70, 127.05, 128.63, 128.97, 129.73, 131.13, 131.30, 132.29, 132.83, 141.86, 144.93 (Ar-C, C=N). Calculated for C30H21Cl2N5S: C, 64.98; H, 3.82; N, 12.63. Found: C, 64.63; H, 3.69; N, 12.88.
1-(4-Nitrophenyl)-4-(((1,4,5-triphenyl-1H-imidazol-2-yl)thio)methyl)-1H-1,2,3-triazole (6b)
This compound was obtained as a colorless solid in 86% yield, mp: 129–130 °C. IR (υ, cm−1): 1540 (C=C), 1605 (C=N), 2930 (CH-Al), 3085 (CH-Ar). 1H NMR (400 MHz, DMSO-d6): δH = 4.51 (s, 2H, SCH2), 7.13-7.25 (m, 10H, Ar-H), 7.38-7.50 (m, 5H, Ar-H), 7.90 (d, 2H, J = 4.0 Hz, Ar-H), 8.04 (d, 2H, J = 4.0 Hz, Ar-H), 8.80 (s, 1H, CH-1,2,3-triazole) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 27.68 (SCH2); 119.21, 121.35, 126.45, 127.21, 128.42, 127.82, 129.52, 130.36, 131.45, 131.28, 132.41, 132.77, 134.62, 135.67, 136.31, 138.14, 141.90, 144.76 (Ar-C, C=N). Calculated for C30H22N6O2S: C, 67.91; H, 4.18; N, 15.84. Found: C, 67.66; H, 4.25; N, 15.72.
1-(4-(4-(((1,4,5-Triphenyl-1H-imidazol-2-yl)thio)methyl)-1H-1,2,3-triazol-1-yl)phenyl)ethan-1-one (6c)
This compound was obtained as a yellow solid in 85% yield, mp: 161–163 °C. IR (υ, cm−1): 1545 (C=C), 1635 (C=N), 1765 (C=O), 2992 (CH-Al), 3079 (CH-Ar). 1H NMR (400 MHz, DMSO-d6): δH = 2.64 (s, 3H, CH3), 4.56 (s, 2H, SCH2), 7.18-7.28 (m, 10H, Ar-H), 7.36-7.47 (m, 5H, Ar-H), 8.07 (d, 2H, J = 12 Hz, Ar-H), 8.17 (d, 2H, J = 8 Hz, Ar-H), 8.88 (s, 1H, CH-1,2,3-triazole) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 27.16 (SCH2); 27.68 (CH3); 119.62, 122.63, 126.05, 126.68, 128.80, 129.42, 130.43, 131.48, 131.80, 134.49, 135.20, 136.21, 137.57, 137.96, 139.94, 141.92, 145.02, 145.36 (Ar-C, C=N); 197.45 (C=O). Calculated for C32H25N5OS: C, 72.84; H, 4.78; N, 13.27. Found: C, 72.59; H, 4.90; N, 13.46.
Ethyl 4-(4-(((1,4,5-triphenyl-1H-imidazol-2-yl)thio)methyl)-1H-1,2,3-triazol-1-yl)benzoate (6d)
This compound was obtained as a pale yellow solid in 86% yield, mp: 134–135 °C. IR (υ, cm−1): 1210 (C-O), 1565 (C=C), 1610 (C=N), 1740 (C=O), 2975 (CH-Al), 3055 (CH-Ar). 1H NMR (400 MHz, DMSO-d6): δH = 1.30 (t, 3H, J = 4.0 Hz, CH3), 4.27-4.34 (q, 2H, OCH2), 4.52 (s, 2H, SCH2), 7.21-7.30 (m, 10H, Ar-H), 7.39-7.49 (m, 5H, Ar-H), 8.01-8.16 (m, 4H, Ar-H), 8.84 (s, 1H, CH-1,2,3-triazole) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 14.48 (CH3); 27.49 (SCH2); 60.98 (OCH2); 119.78, 122.60, 126.51, 126.66, 128.83, 128.46, 129.47, 130.37, 131.29, 131.86, 134.52, 135.24, 136.29, 137.64, 138.11, 139.89, 141.96, 145.08, 145.42 (Ar-C, C=N); 166.22 (C=O). Calculated for C33H27N5O2S: C, 71.07; H, 4.88; N, 12.56. Found: C, 71.29; H, 4.99; N, 12.72.
4-(4-(((1,4,5-Triphenyl-1H-imidazol-2-yl)thio)methyl)-1H-1,2,3-triazol-1-yl)benzoic acid (6e)
This compound was obtained as a yellow solid in 85% yield, mp: 219–220 °C. IR (υ, cm−1): 1535 (C=C), 1605 (C=N), 1720 (C=O), 2580-3350 (OH). 1H NMR (400 MHz, DMSO-d6): δH = 4.57 (s, 2H, SCH2), 7.15-7.30 (m, 10H, Ar-H), 7.39-7.48 (m, 5H, Ar-H), 7.92 (d, 2H, J = 4.0 Hz, Ar-H), 8.06 (d, 2H, J = 4.0 Hz, Ar-H), 8.82 (s, 1H, CH-1,2,3-triazole), 12.23 (s, 1H, COOH) ppm. 13C NMR (100 MHz, DMSO-d6): δC = 27.74 (SCH2); 119.26, 121.38, 126.38, 127.28, 128.53, 127.77, 129.65, 130.41, 131.37, 131.78, 132.48, 132.80, 134.78, 135.64, 136.29, 138.24, 141.82, 144.89 (Ar-C, C=N); 167.58 (C=O). Calculated for C31H23N5O2S: C, 70.30; H, 4.38; N, 13.22. Found: C, 70.47; H, 4.42; N, 13.34.
3.2. MTT Assay
The cytotoxic activity was assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) colorimetric assay as reported previously [53,54,55]. In brief, the tumor cell lines were suspended in the medium at a concentration of 1 × 104 cell/well in Corning® 96-well tissue culture plates and then incubated for 24 h. The tested compounds with concentrations were then added into 96-well plates to achieve different concentrations for each compound. After incubating for 24 h, the number of viable cells was determined by the MTT test. Briefly, the medium was removed and the MTT solution (50 μL, 0.5 mg/mL in RPMI 1640 without phenol red) was added. After 3 h of incubation, the MTT solution was removed and the acquired formazan was dissolved in an isopropanol:HCl system. The 96-well plates were then incubated at 37 °C and 5% CO2 for 4 h. Finally, the optical density was measured at 570 nm with the microplate reader to determine the number of viable cells and the percentage of viability were calculated as [1-(ODt/ODc)] × 100% where ODt is the mean optical density of wells treated with the tested sample and ODc is the mean optical density of the untreated cells. The experiment was conducted in three independent iterations with four technical repetitions. Tests were conducted at concentrations of tested compounds ranging from 0.01 to 0.8 mM solutions. The 50% inhibitory concentration (IC50), the concentration required to cause toxic effects in 50% of intact cells, was estimated from graphic plots of the dose response curve for each concentration using CalcuSyn.
3.3. In Silico Analysis
Several physicochemical, pharmacokinetic, and pharmacodynamic attributes (i.e., topological polar surface area (TPSA), lipophilicity, absorption, distribution, metabolism, excretion and toxicity (ADMET)), molar refractivity, fragment-based drug-likeness and violations, etc., of the newly synthesized compounds were predicted through an in silico strategy utilizing the Swiss ADME tool from the Swiss Institute of Bioinformatics (http://www.sib.swiss) and ADMET SAR server (http://lmmd.ecust.edu.cn).
3.4. Molecular Docking Studies
The newly potent synthesized compound 4k was docked into the active site of 3D crystallographic structure of the GSK-3β (PDB: 1UV5), used as the target model. The AutoDock 3.0 [56] and the MOE software [57] were used for all the docking calculations and molecular representations. The AutoDock Tools package was employed to generate the docking input files and analyze the docking results. A grid box size of 90 × 90 × 90 points with a spacing of 0.375 Å between the grid points was generated that covered almost the entire protein surface. The ligand was built by means of the MOE builder interface, their geometries were optimized with the CHARMm forcefield and then prepared for docking calculations with the python scripts available in the AutoDock package. Fifty runs were performed, and the resulting poses were clustered with 1.8 Å tolerance. Lamarckian GA was used for the conformational space search with the initial population set to 150, and fitness function evaluations set to 2,500,0000. The most abundant low energy clusters were selected for the analysis. The protein-ligand interaction plots were generated, using MOE 2012.10.
4. Conclusions
An efficient copper(I)catalyzed click reaction of imidazole-based alkyne 2 with a variety of organoazides furnished a focused library of imidazole-1,2,3-triazole hybrids carrying different un/functionalized alkyl/aryl side chains 4a–k and 6a–e. The targeted click products were fully characterized using different spectroscopic techniques such as IR, 1H, 13C NMR, and elemental analyses. The resulted adducts were investigated for their anticancer activity against four cancer cell lines (Caco-2, HCT-116, HeLa, and MCF-7) by the MTT assay and showed moderate to significant activity. Of these compounds, 4k displayed potent cytotoxic activity against the cancer cell lines, especially MCF-7 with IC50 0.38 µM. The study of pharmacokinetic parameters of the synthesized compounds revealed that most of the compounds are CYP2C19 and CYP3A4 inhibitors. All the synthesized compounds emerged to be non-carcinogenic in nature. The in silico molecular docking study was also performed recruiting GSK-3β as the promising cancer target receptor. The results of in silico studies were in accordance with the in vitro results. The overall results of this study showed that a fascinating protocol for the production of new anticancer agents targeting the GSK3β enzyme for the treatment of breast cancer was the molecular hybridization of 1,2,3-triazole to biologically interesting imidazole scaffolds with diverse aromatic substituents.
Author Contributions
F.A.-b., S.A.S., and F.A. carried out of the experimental work, data curation and cooperated in the preparation of the manuscript. N.R. and M.R.A. gave the concepts of work, collected data, interpreted the results and prepared the first drat of the manuscript. A.N. and S.I. performed the biological assays and their investigation. All authors discussed the results and commented on the manuscript. F.A.-b., A.N., N.R. and M.R.A. wrote the paper and edited English language. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No data Available.
Conflicts of Interest
Authors declare no conflict of interests.
References
- Jardim, D.L.; Gagliato, D.D.; Nikanjam, M.; Barkauskas, D.A.; Kurzrock, R. Efficacy and safety of anticancer drug combinations: A meta-analysis of randomized trials with a focus on immunotherapeutics and gene-targeted compounds. OncoImmunology 2020, 9, 1710052. [Google Scholar] [CrossRef]
- Meegan, M.J.; O’Boyle, N.M. Anticancer drugs. Pharmaceuticals 2019, 12, 134. [Google Scholar] [CrossRef]
- Kharb, R. Updates on receptors targeted by heterocyclic scaffolds: New horizon in anticancer drug development. Anticancer Agents Med. Chem. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Lone, M.N.; Al-Othman, Z.A.; Al-Warthan, A.; Sanagi, M.M. Heterocyclic scaffolds: Centrality in anticancer drug development. Curr. Drug Targets 2015, 16, 711–734. [Google Scholar] [CrossRef]
- Kerru, N.; Gummidi, L.; Maddila, S.; Gangu, K.K.; Jonnalagadda, S.B. A review on recent advances in nitrogen-containing molecules and their biological applications. Molecules 2020, 25, 1909. [Google Scholar] [CrossRef]
- Irannejad, H. Nitrogen rich heterocycles as a privileged fragment in lead discovery. Med. Analy. Chem. Int. J. 2018, 2, 1–6. [Google Scholar] [CrossRef]
- Gaba, M.; Mohan, C. Development of drugs based on imidazole and benzimidazole bioactive heterocycles: Recent advances and future directions. Med. Chem. Res. 2016, 25, 173–210. [Google Scholar] [CrossRef]
- Agarwal, S.; Sasane, S.; Kumar, J.; Darji, B.; Bhayani, H.; Soman, S.; Kulkarni, N.; Jain, M. Novel 2-mercapto imidazole and triazole derivatives as potent TGR5 receptor agonists. Med. Drug Discov. 2019, 1, 100002. [Google Scholar] [CrossRef]
- Fassihi, A.; Azadpour, Z.; Delbari, N.; Saghaie, L.; Memarian, H.R.; Sabet, R.; Alborzi, A.; Miri, R.; Pourabbas, B.; Mardaneh, J.; et al. Synthesis and antitubercular screening of imidazole derivatives. Eur. J. Med. Chem. 2009, 44, 3253–3258. [Google Scholar] [CrossRef]
- Zhang, X.; Sui, Z.; Kauffman, J.; Hou, C.; Chen, C.; Du, F.; Kirchner, T.; Liang, Y.; Johnson, D.; Murray, W.V.; et al. Evaluation of anti-diabetic effect and gall bladder function with 2-thio5-thiomethyl substituted imidazoles as TGR5 receptor agonists. Bioorg. Med. Chem. Lett. 2017, 27, 1760–1764. [Google Scholar] [CrossRef]
- Al-Warhi, T.; Said, M.A.; El Hassab, M.A.; Aljaeed, N.; Ghabour, H.A.; Almahli, H.; Eldehna, W.M.; Abdel-Aziz, H.A. Unexpected synthesis, single-crystal x-ray structure, anticancer activity, and molecular docking studies of certain 2–((imidazole/benzimidazol–2–yl)thio)– 1–arylethanones. Crystals 2020, 10, 446. [Google Scholar] [CrossRef]
- Yurttaş, L.; Ertaş, M.; Çiftçi, G.A.; Temel, H.E.; Demirayak, Ş. Novel benzothiazole based imidazole derivatives as new cytotoxic agents against glioma (C6) and liver (HepG2) cancer cell lines. Acta Pharm. Sci. 2017, 55, 39–47. [Google Scholar] [CrossRef][Green Version]
- Ali, I.; Lonea, M.N.; Aboul-Enein, H.Y. Imidazoles as potential anticancer agents. Med. Chem. Comm. 2017, 8, 1742–1773. [Google Scholar] [CrossRef] [PubMed]
- Rani, N.; Singh, R. Design, synthesis, antimicrobial evaluation and molecular modeling study of new 2-mercaptoimidazoles (series-iii). Lett. Drug Des. Disco. 2019, 16, 512–521. [Google Scholar] [CrossRef]
- Aouad, M.R.; Messali, M.; Rezki, N.; Ali, A.A.; Lesimple, A. Synthesis and characterization of some novel 1,2,4-triazoles, 1,3,4-thiadiazoles and Schiff bases incorporating imidazole moiety as potential antimicrobial agents. Acta Pharm. 2015, 65, 117–132. [Google Scholar] [CrossRef] [PubMed]
- Salman, A.S.; Abdel-Aziem, A.; Alkubbat, M.J. Design, synthesis of some new thio-substituted imidazole and their biological activity. Am. J. Org. Chem. 2015, 5, 57–72. [Google Scholar] [CrossRef]
- Rani, A.; Singh, G.; Singh, A.; Maqbool, U.; Kaur, G.; Singh, J. CuAAC-ensembled 1,2,3-triazole-linked isosteres as pharmacophores in drug discovery: Review. RSC Adv. 2020, 10, 5610–5635. [Google Scholar] [CrossRef]
- Bozorova, K.; Zhaoa, J.; Aisa, H.A. 1,2,3-Triazole-containing hybrids as leads in medicinal chemistry: A recent overview. Bioorg. Med. Chem. 2019, 27, 3511–3531. [Google Scholar] [CrossRef]
- Smit, F.J.; Seldon, R.; Aucamp, J.; Jordaan, A.; Warner, D.F.; N’Da, D.D. Synthesis and antimycobacterial activity of disubstituted benzyltriazoles. Med. Chem. Res. 2019, 28, 2279–2293. [Google Scholar] [CrossRef]
- Ali, A.A.; Gogoi, D.; Chaliha, A.K.; Buragohain, A.K.; Trivedi, P.; Saikia, P.J.; Gehlot, P.S.; Kumar, A.; Chaturvedi, V.; Sarma, D. Synthesis and biological evaluation of novel 1,2,3-triazole derivatives as anti-tubercular agents. Bioorg. Med. Chem. Lett. 2017, 27, 3698–3703. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, Z.; Gao, C.; Ren, Q.-C.; Chang, L.; Lv, Z.-S.; Feng, L.-S. Triazole derivatives and their anti-tubercular activity. Eur. J. Med. Chem. 2017, 138, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Abbasia, M.A.; Ramzana, M.S.; ur-Rehmana, A.; Siddiquia, S.Z.; Hassanb, M.; Shahc, S.A.A.; Ashrafe, M.; Shahidf, M.; Seo, S.-Y. Novel bi-heterocycles as potent inhibitors of urease and less cytotoxic agents: 3-({5-((2-amino-1,3-thiazol-4-yl)methyl)-1,3,4-oxadiazol-2-yl}sulfanyl)-N-(un/substituted-phenyl) propanamides. Iran. J. Pharm. Res. 2020, 19, 487–506. [Google Scholar] [CrossRef]
- Erşatır, M.; Yıldırım, M.; Giray, E.S.; Yalın, S. Synthesis and antiproliferative evaluation of novel biheterocycles based on coumarin and 2-aminoselenophene-3-carbonitrile unit. Mon. Chem. Chem. Mon. 2020, 151, 625–636. [Google Scholar] [CrossRef]
- Mantenuto, S.; Lucarini, S.; De Santi, M.; Piersanti, G.; Brandi, G.; Favi, G.; Mantellini, F. One-pot synthesis of biheterocycles based on indole and azole scaffolds using tryptamines and 1,2-diaza-1,3-dienes as building blocks. Eur. J. Org. Chem. 2016, 9, 3193–3199. [Google Scholar] [CrossRef]
- Reddy, K.H.V.; Brion, J.-D.; Messaoudi, S.; Alami, M. Synthesis of biheterocycles based on quinolinone, chromone and coumarin scaffolds by palladium-catalyzed decarboxylative couplings. Org. Chem. 2016, 81, 424–432. [Google Scholar] [CrossRef]
- Maiti, B.; Chanda, K. Diversity oriented synthesis of benzimidazole based biheterocyclic molecules by combinatorial approach: A critical review. RSC Adv. 2016, 6, 50384–50413. [Google Scholar] [CrossRef]
- Woodgett, J. Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J. 1990, 9, 2431–2438. [Google Scholar] [CrossRef]
- Welsh, G.; Proud, C. Glycogen synthase kinase-3 is rapidly inactivated in response to insulin and phosphorylates eukaryotic initiation factor eIF-2B. Biochem J. 1993, 294, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Ougolkov, A.; Fernandez-Zapico, M.; Savoy, D.; Urrutia, R.; Billadeau, D. Glycogen synthase kinase-3beta participates in nuclear factor kappaB-mediated gene transcription and cell survival in pancreatic cancer cells. Cancer Res. 2005, 65, 2076–2081. [Google Scholar] [CrossRef]
- Shakoori, A.; Ougolkov, A.; Zhang, B.; Modarressi, M.; Billadeau, D.; Mai, M.; Takahashi, Y.; Minamoto, T. Deregulated GSK3beta activity in colorectal cancer: Its association with tumor cell survival and proliferation. Biochem. Biophys. Res. Commun. 2005, 334, 1365–1373. [Google Scholar] [CrossRef]
- Gaisina, I.; Gallier, F.; Ougolkov, A.; Kim, K.; Kurome, T.; Guo, S.; Holzle, D.; Luchini, D.N.; Blond, S.Y.; Billadeau, D.D.; et al. From a natural product lead to the identification of potent and selective benzofuran-3-yl-(indol-3-yl)maleimides as glycogen synthase kinase 3beta inhibitors that suppress proliferation and survival of pancreatic cancer cells. J. Med. Chem. 2009, 52, 1853–1863. [Google Scholar] [CrossRef] [PubMed]
- Ougolkov, A.; Fernandez-Zapico, M.; Bilim, V.; Smyrk, T.; Chari, S.; Billadeau, D. Aberrant nuclear accumulation of glycogen synthase kinase-3beta in human pancreatic cancer: Association with kinase activity and tumor dedifferentiation. Clin. Cancer Res. 2006, 12, 5074–5081. [Google Scholar] [CrossRef]
- Aouad, M.R. Efficient eco-friendly solvent-free click synthesis and antimicrobial evaluation of new fluorinated 1,2,3-triazoles and their conversion into Schiff Bases. J. Braz. Chem. Soc. 2015, 10, 2105–2115. [Google Scholar] [CrossRef]
- Rezki, N. A Green ultrasound synthesis, characterization and antibacterial evaluation of 1,4-disubstituted 1,2,3-triazoles tethering bioactive benzothiazole nucleus. Molecules 2016, 21, 505. [Google Scholar] [CrossRef]
- Rezki, N.; Mayaba, M.M.; Al-blewi, F.F.; El Ashry, E.H.; Aouad, M.R. Click 1,4-regioselective synthesis, characterization, and antimicrobial screening of novel 1,2,3-triazoles tethering fluorinated 1,2,4-triazole and lipophilic side chain. Res. Chem. Intermed. 2017, 43, 995–1011. [Google Scholar] [CrossRef]
- Aouad, M.R. Synthesis and antimicrobial screening of novel thioglycosides analogs carrying 1,2,3-triazole and 1,3,4-oxadiazole moieties. Nucleotides Nucleosides Nucleic Acids 2016, 35, 1–15. [Google Scholar] [CrossRef]
- Aouad, M.R. Click synthesis and antimicrobial screening of novel isatin-1,2,3-triazoles with piperidine, morpholine or piperazine moieties. Org. Prep. Proced. Int. 2017, 49, 216–227. [Google Scholar] [CrossRef]
- Al-blewi, F.F.; Almehmadi, M.A.; Aouad, M.R.; Bardaweel, S.K.; Sahu, P.K.; Messali, M.; Rezki, N.; El Ashry, E.H. Design, synthesis, ADME prediction and pharmacological evaluation of novel benzimidazole-1,2,3-triazole-sulfonamide hybrids as antimicrobial and antiproliferative agents. Chem. Cent. J. 2018, 12, 110–123. [Google Scholar] [CrossRef]
- Aouad, M.R.; Mayaba, M.M.; Naqvi, A.; Bardaweel, S.K.; Al-blewi, F.F.; Messali, M.; Rezki, N. Design, synthesis, in-silico and In-Vitro antimicrobial screenings of novel 1,2,4-triazoles carrying 1,2,3-triazole scaffold with lipophilic side chain tether. Chem. Cent. J. 2017, 11, 117–129. [Google Scholar] [CrossRef]
- Aouad, M.R.; Almehmadi, M.A.; Rezki, N.; Al-blewi, F.F.; Messali, M.; Ali, I. Design, click synthesis, anticancer screening and docking studies of novel benzothiazole-1,2,3-triazoles appended with some bioactive benzofused heterocycles. J. Mol. Struct. 2019, 1188, 153–164. [Google Scholar] [CrossRef]
- Rezki, N. Green microwave synthesis and antimicrobial evaluation of novel triazoles. Org. Prep. Proced. Int. 2017, 49, 525–541. [Google Scholar] [CrossRef]
- Aouad, M.R.; Soliman, M.A.; Alharbi, M.O.; Bardaweel, S.K.; Sahu, P.K.; Ali, A.A.; Messali, M.; Rezki, N.; Al-Soud, Y.A. Design, Synthesis and Anticancer Screening of Novel Benzothiazole-Piperazine-1,2,3-Triazole Hybrids. Molecules 2018, 23, 2788. [Google Scholar] [CrossRef] [PubMed]
- Rezki, N.; Almehmadi, M.A.; Ihmaid, S.; Shehata, A.M.; Omar, A.M.; Ahmed, H.E.A.; Aouad, M.R. Novel scaffold hopping of potent benzothiazole and isatin analogues linked to 1,2,3-triazole fragment that mimics quinazoline epidermal growth factor receptor inhibitors: Synthesis, antitumor and mechanistic analyses. Bioorg. Chem. 2020, 103, 104133. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Del. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. Prediction of hydrophobic (lipophilic) properties of small organic molecules using fragmental methods: An analysis of ALOGP and CLOGP methods. J. Phys. Chem. A. 1998, 102, 3762–3772. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Egan, W.J.; Lauri, G. Prediction of intestinal permeability. Adv. Drug Del. Rev. 2002, 54, 273–289. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Ugolkov, A.; Gaisina, I.; Zhang, J.-S.; Billadeau, D.D.; White, K.; Kozikowski, A.; Jain, S.; Cristofanilli, M.; Giles, F.; O’Halloran, T.; et al. GSK-3 inhibition overcomes chemoresistance in human breast cancer. Cancer Lett. 2016, 380, 384–392. [Google Scholar] [CrossRef]
- Minvielle, M.J.; Bunders, C.A.; Melander, C. Indole/triazole conjugates are selective inhibitors and inducers of bacterial biofilms. Med. Chem. Comm. 2013, 4, 916–919. [Google Scholar] [CrossRef]
- Devender, N.; Gunjan, S.; Tripathi, R.; Tripathi, R.P. Synthesis and antiplasmodial activity of novel indoleamide derivatives bearing sulfonamide and triazole pharmacophores. Eur. J. Med. Chem. 2017, 131, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Peng, Z.; Wang, J.; Li, X.; Li, J. Synthesis, In Vitro evaluation and molecular docking studies of novel triazine-triazole derivatives as potential α-glucosidase inhibitors. Eur. J. Med. Chem. 2017, 12, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Demirci, F.; Başer, K.H.C. Bioassay techniques for drug development by Atta-ur-Rahman, M. Iqbal Choudhary (HEJRIC, University of Karachi, Pakistan), William J. Thomsen (Areana Pharmaceuticals, San Diego, CA). Harwood Academic Publishers, Amsterdam, The Netherlands. 2001. xii + 223 pp. 15.5 × 23.5 cm. $79.00. ISBN 90-5823-051-1. J. Nat. Prod. 2002, 65, 1086–1087. [Google Scholar]
- Omar, A.M.; Bajorath, J.; Ihmaid, S.; Mohamed, H.M.; El-Agrody, A.M.; Mora, A.; El-Araby, M.E.; Ahmed, H.E.A. Novel molecular discovery of promising amidine-based thiazole analogues as potent dual Matrix Metalloproteinase-2 and 9 inhibitors: Anticancer activity data with prominent cell cycle arrest and DNA fragmentation analysis effects. Bioorg. Chem. 2020, 101, 103992. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Chemical Computing Group. Molecular Operating Environment (MOE); Chemical Computing Group: Montreal, QC, Canada, 2012; Available online: http://www.chemcomp.com (accessed on 28 February 2013).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).