T Cell Mediated Conversion of a Non-Anti-La Reactive B Cell to an Autoreactive Anti-La B Cell by Somatic Hypermutation †
Abstract
:1. Introduction
2. Results
2.1. Hybridoma Fusion from a Human La Transgenic Mouse after Adoptive Transfer of T Cells from Non-Transgenic Mice Immunized with Human La
2.2. The Hybridomas Originate from the La Transgenic Mouse
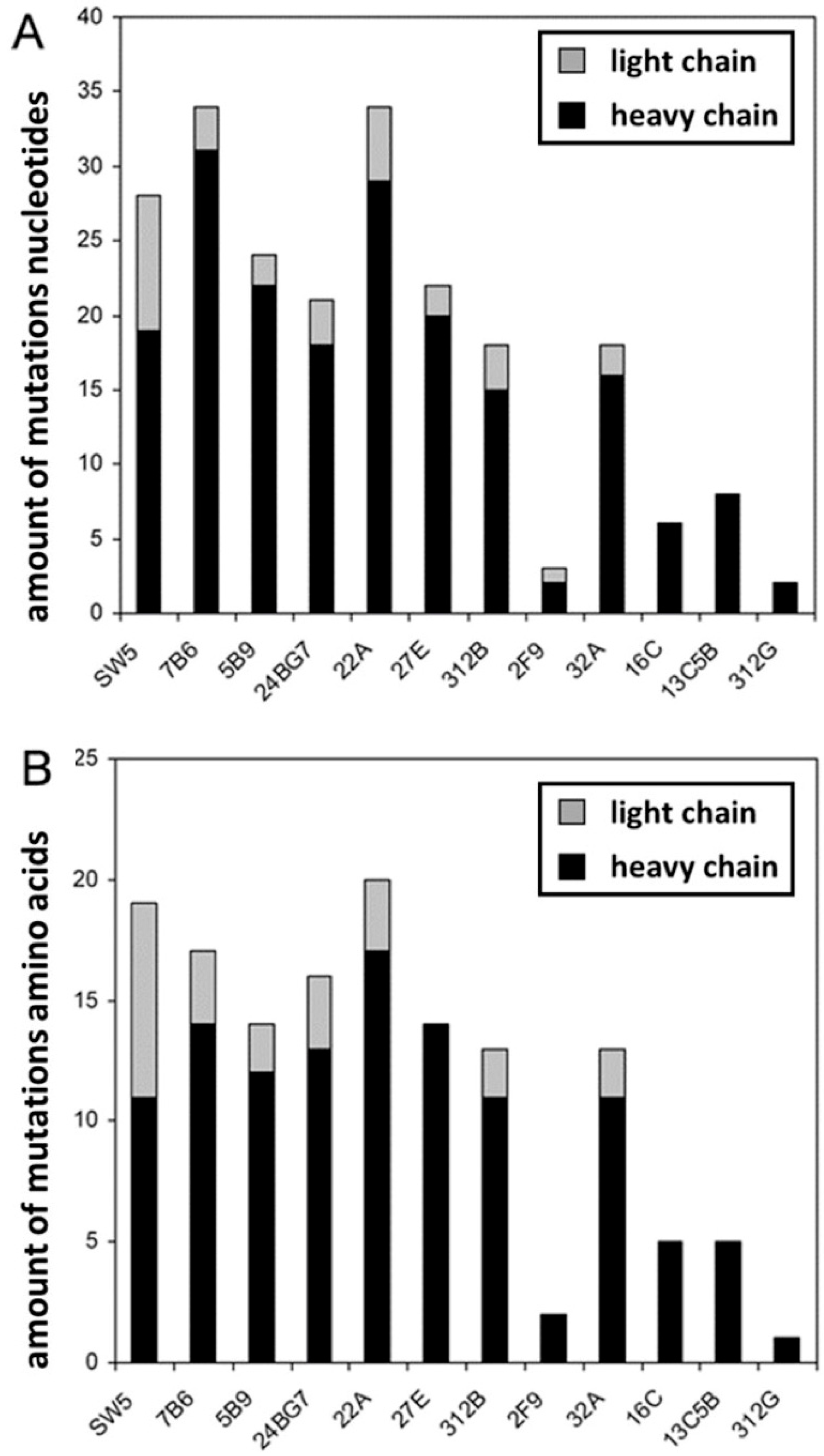
2.3. The Hybridomas Originate from Individual B Cells
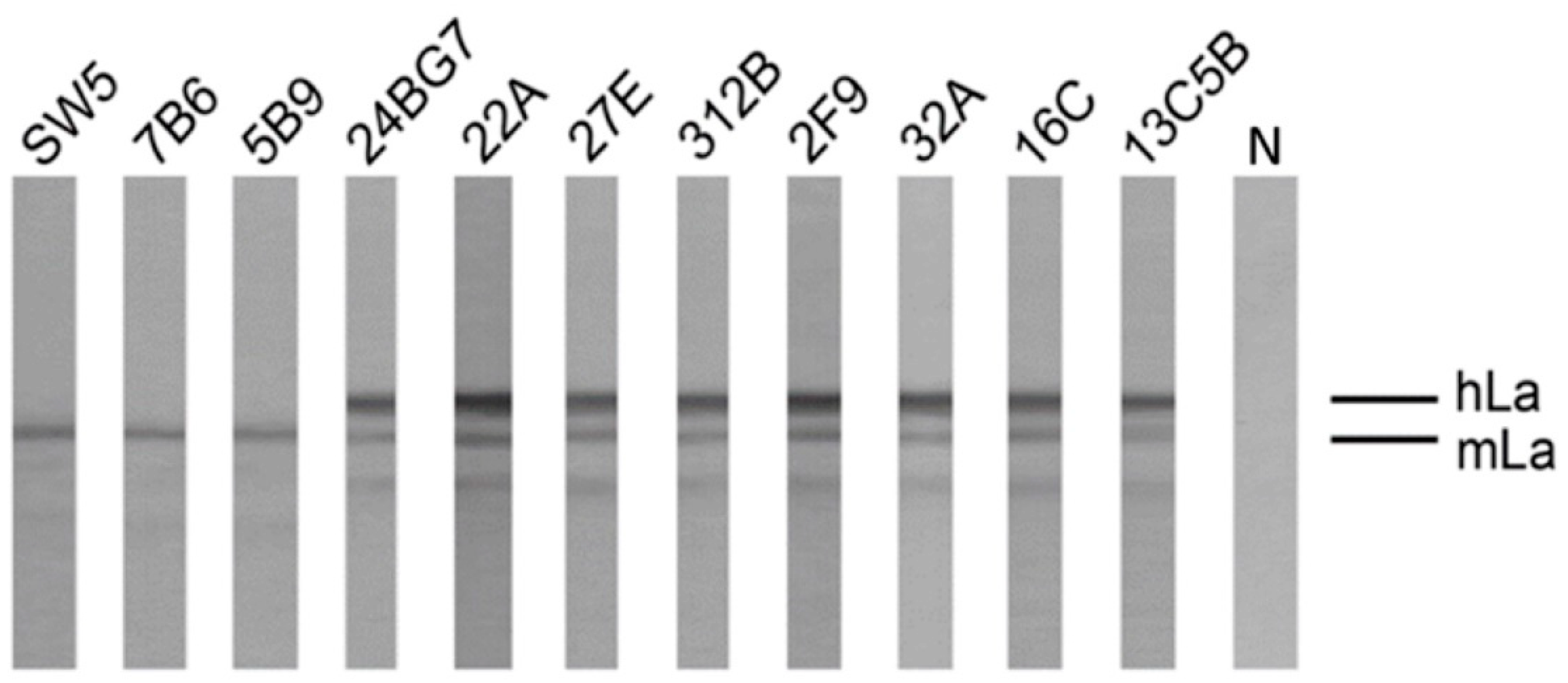
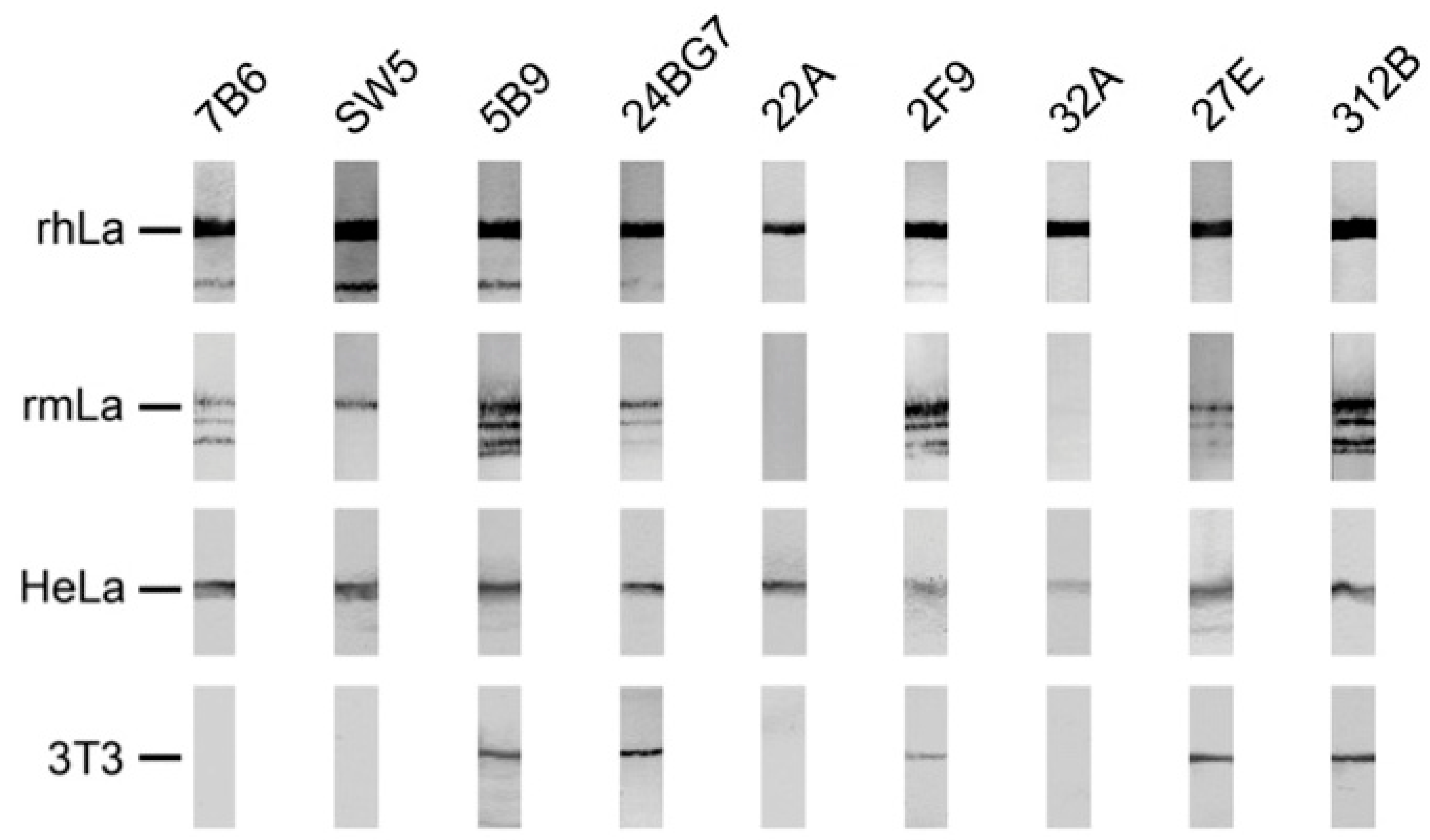
2.4. Reactivity of Anti-La Mabs to Denatured and Native Human and Mouse La Protein
2.4.1. Analysis by SDS-PAGE and Immunoblotting Using La Proteins Recombinantly Expressed in E. coli and Total Extracts from Murine or Human Cell Lines
2.4.2. Coprecipitation Study of Native Human and Mouse La Protein from Total Cell Extracts of a Mouse 3T3 Cell Line Expressing the Human La Transgene
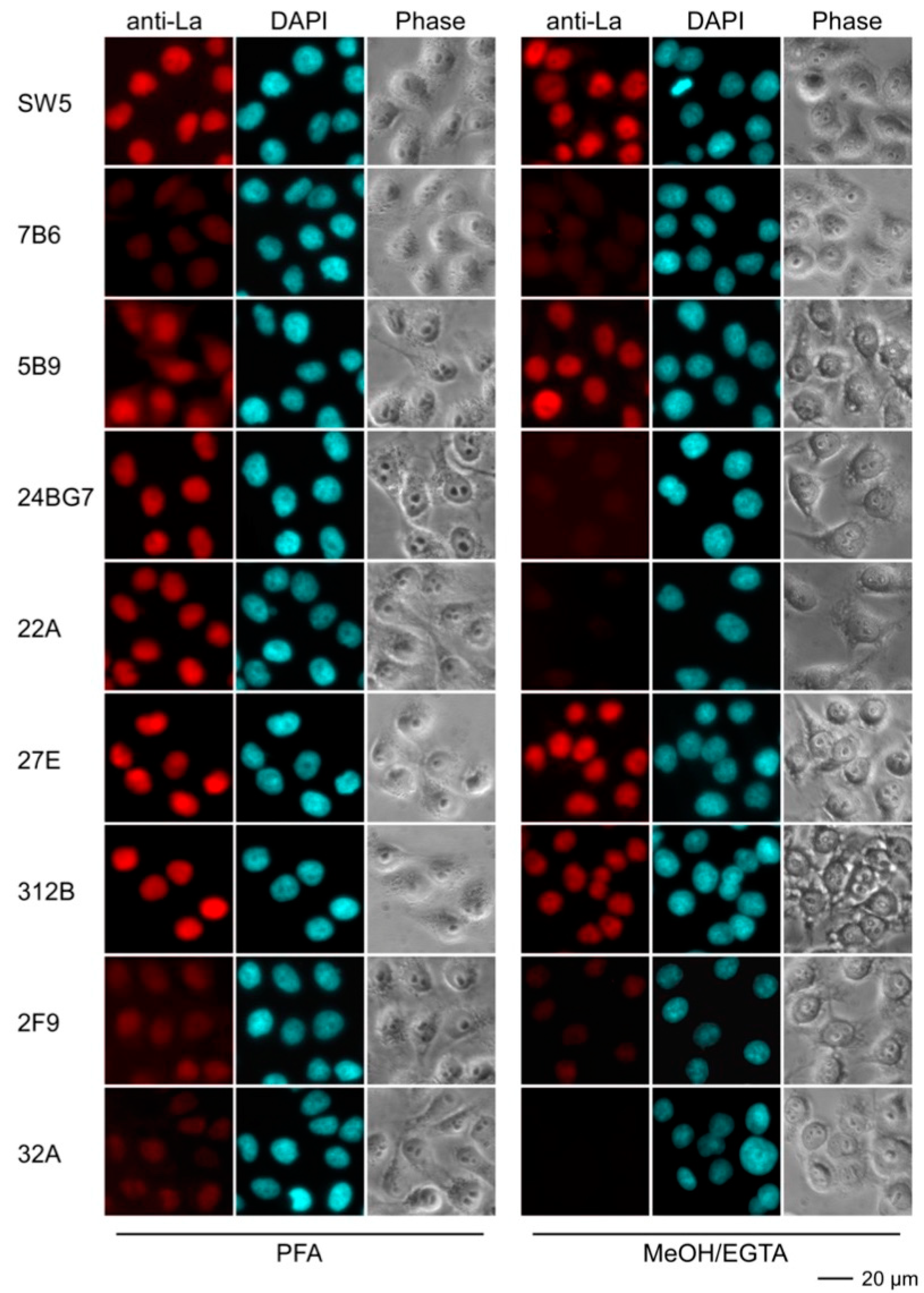
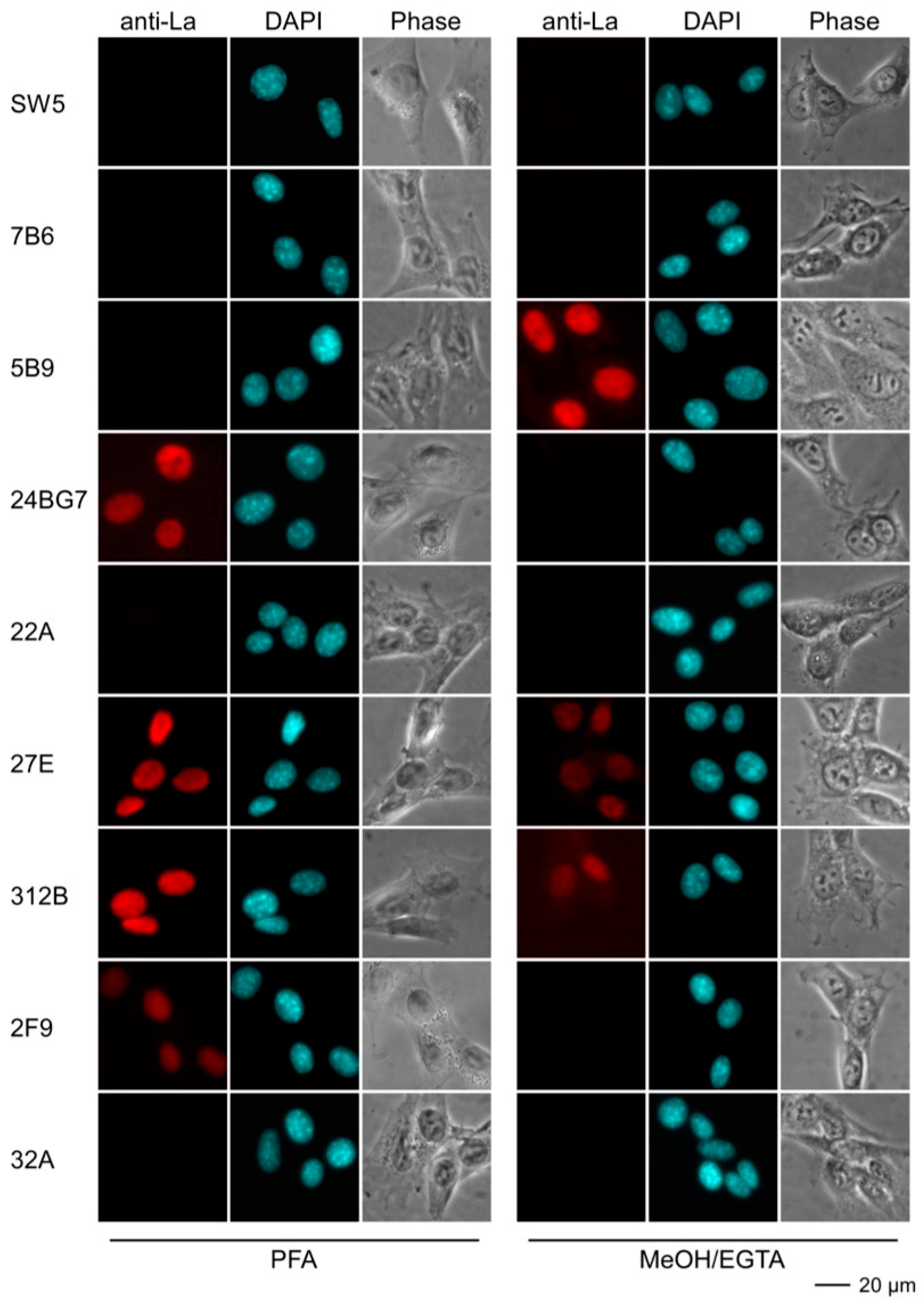
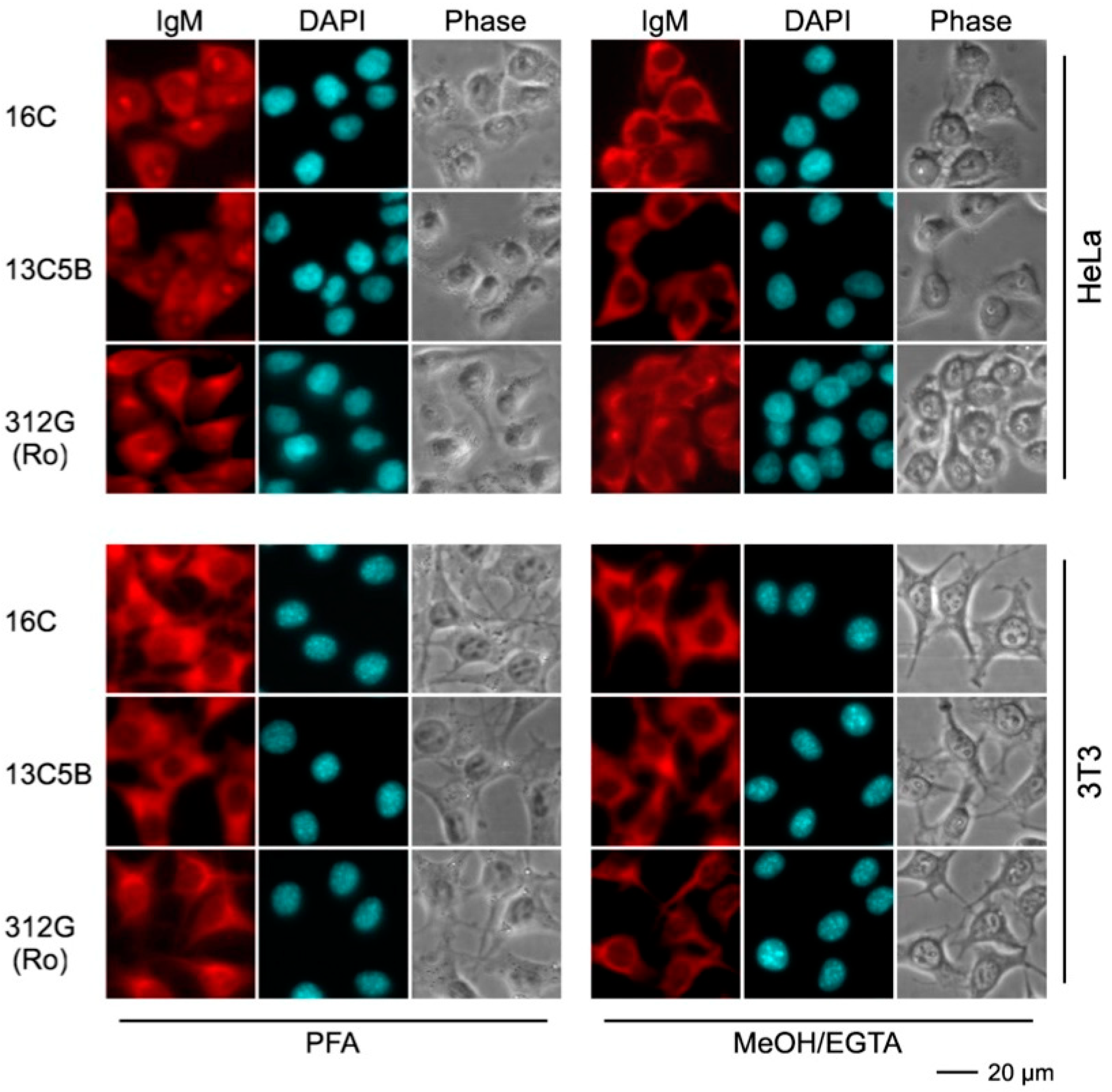
2.4.3. Immunofluorescence Analysis Using Human and Murine Cell Lines
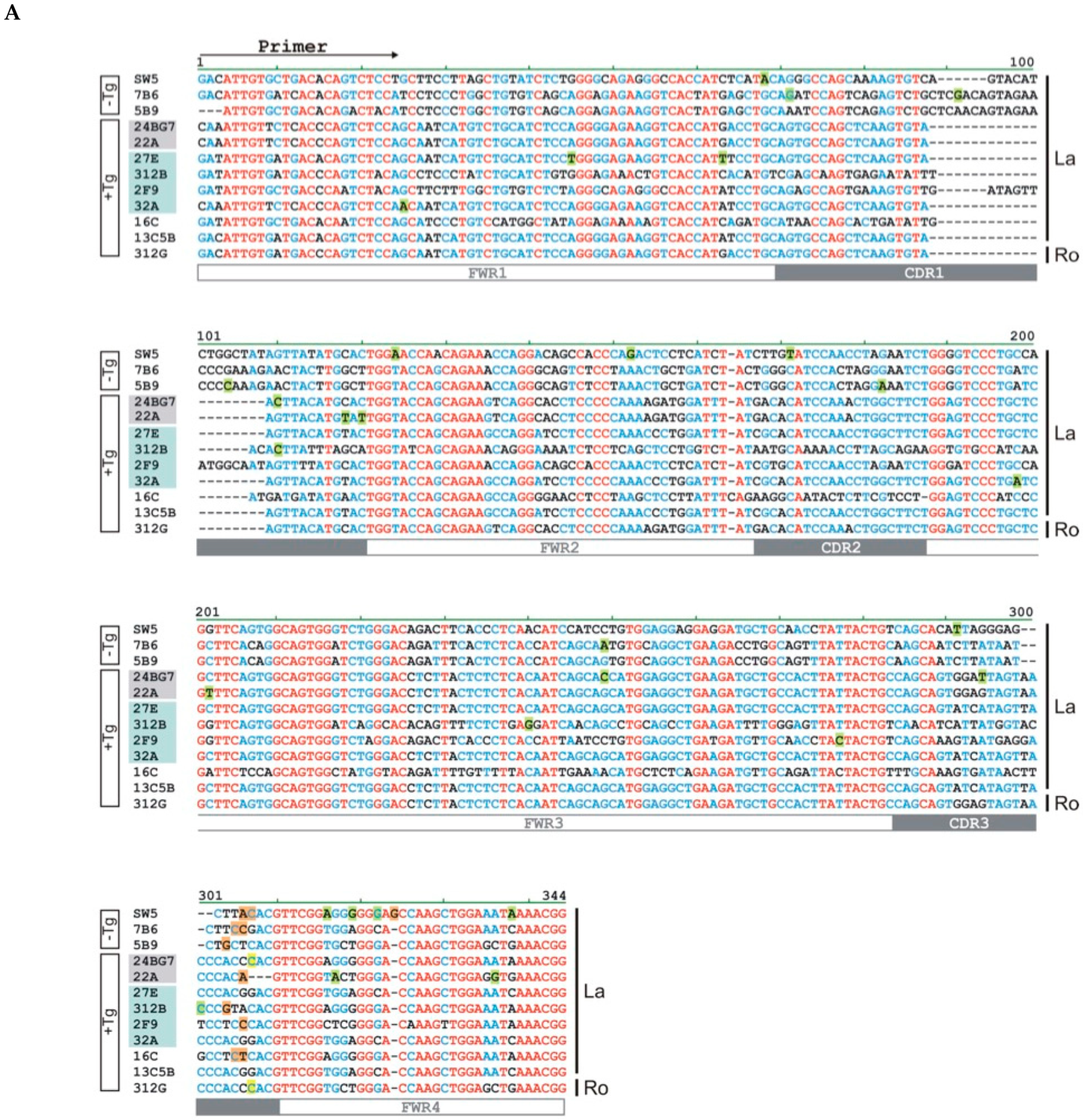
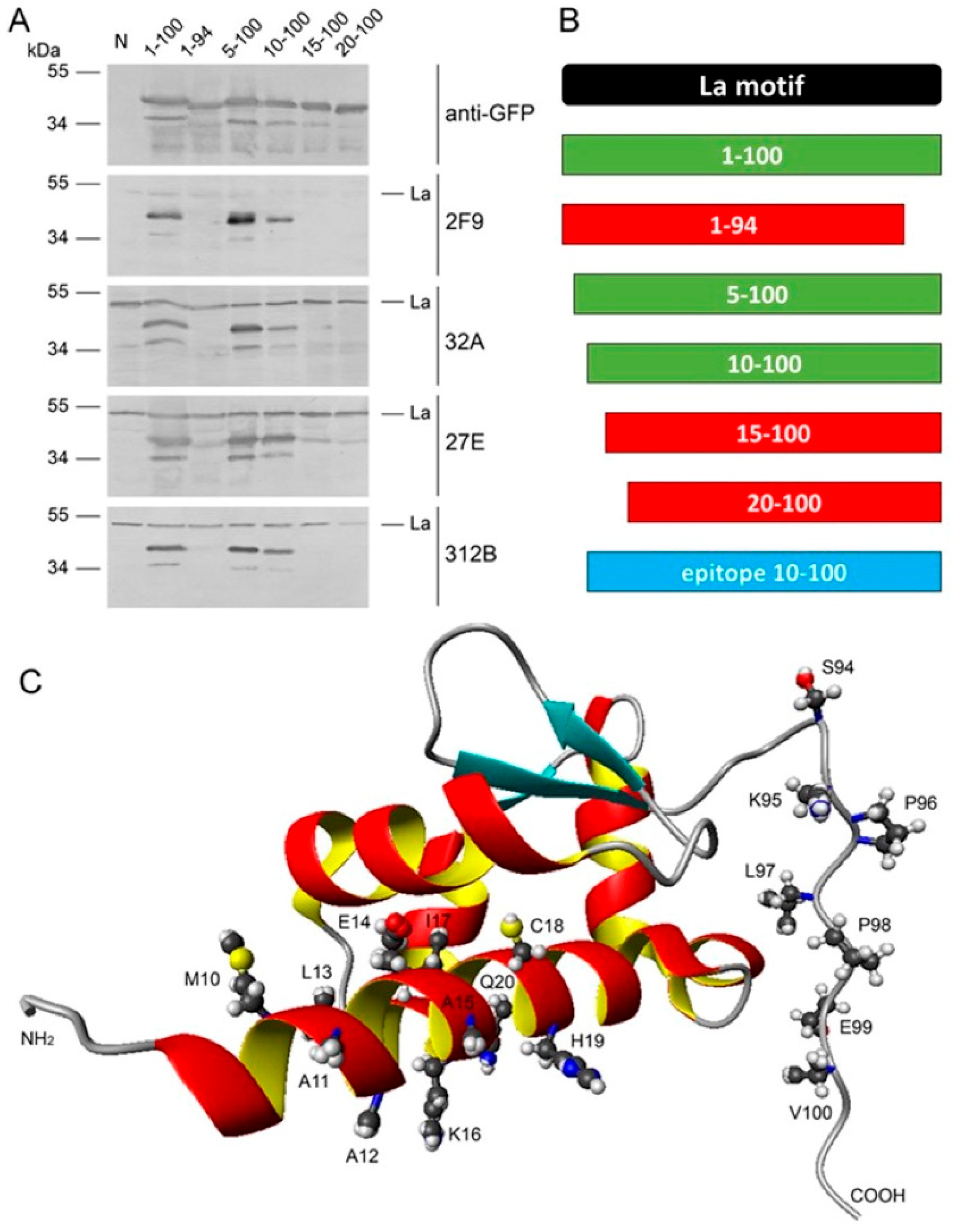
2.5. Epitope Mapping
2.6. Lack of Anti-La Reactivity of the 312B Germline Sequence
3. Discussion
4. Materials and Methods
4.1. Recombinant Human La Protein Expression and Characterization
4.2. Mice
4.3. T Cell Isolation, Adoptive Transfer, and Hybridoma Fusion
4.4. Hybridoma Fusion
4.5. Sequence Analysis
4.6. Construction, Expression, and Purification of Recombinant Antibodies
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tan, E.M. Antinuclear Antibodies: Diagnostic Markers for Autoimmune Diseases and Probes for Cell Biology. Adv. Immunol. 1989, 44, 93–151. [Google Scholar] [CrossRef]
- Harley, J.B.; Alexander, E.L.; Bias, W.B.; Fox, O.F.; Provost, T.T.; Reichlin, M.; Yamagata, H.; Arnett, F.C. Anti-Ro (SS-A) and Anti-La (SS-B) in patients with Sjögren’s syndrome. Arthritis Rheum. 1986, 29, 196–206. [Google Scholar] [CrossRef]
- Buyon, J.P.; Hiebert, R.; Copel, J.; Craft, J.; Friedman, D.; Katholi, M.; Lee, L.A.; Provost, T.T.; Reichlin, M.; Rider, L.; et al. Autoimmune-Associated Congenital Heart Block: Demographics, Mortality, Morbidity and Recurrence Rates Obtained From a National Neonatal Lupus Registry. J. Am. Coll. Cardiol. 1998, 31, 1658–1666. [Google Scholar] [CrossRef]
- Oldstone, M.B. Molecular mimicry and autoimmune disease. Cell 1987, 50, 819–820. [Google Scholar] [CrossRef]
- Dean, G.S.; Tyrrell-Price, J.; Crawley, E.; A Isenberg, D. Cytokines and systemic lupus erythematosus. Ann. Rheum. Dis. 2000, 59, 243–251. [Google Scholar] [CrossRef] [Green Version]
- Flescher, E.; Talal, N. Do Viruses Contribute to the Development of Sjögren’s Syndrome? Am. J. Med. 1991, 90, 283–285. [Google Scholar] [CrossRef]
- Fox, R.I.; Pearson, G.; Vaughan, J.H. Detection of Epstein-Barr virus-associated antigens and DNA in salivary gland biopsies from patients with Sjogren’s syndrome. J. Immunol. 1986, 137, 137. [Google Scholar]
- Fox, R.I.; Luppi, M.; Pisa, P.; I Kang, H. Potential role of Epstein-Barr virus in Sjögren’s syndrome and rheumatoid arthritis. J. Rheumatol. Suppl. 1992, 32, 18–24. [Google Scholar]
- Magistrelli, C.; Samoilova, E.; Agarwal, R.K.; Banki, K.; Ferrante, P.; Vladutiu, A.; Phillips, P.E.; Perl, A. Polymorphic genotypes of the HRES-1 human endogenous retrovirus locus correlate with systemic lupus erythematosus and autoreactivity. Immunogenetics 1999, 49, 829–834. [Google Scholar] [CrossRef]
- A James, J.; Gross, T.; Scofield, R.H.; Harley, J.B. Immunoglobulin epitope spreading and autoimmune disease after peptide immunization: Sm B/B’-derived PPPGMRPP and PPPGIRGP induce spliceosome autoimmunity. J. Exp. Med. 1995, 181, 453–461. [Google Scholar] [CrossRef]
- Vaishnaw, A.K.; McNally, J.D.; Elkon, K.B. Apoptosis in the rheumatic diseases. Arthritis Rheum. 1997, 40, 1917–1927. [Google Scholar] [CrossRef]
- Andrade, F.; Casciola-Rosen, L.; Rosen, A. Apoptosis in Systemic Lupus Erythematosus. Rheum. Dis. Clin. N. Am. 2000, 26, 215–227. [Google Scholar] [CrossRef]
- Kurien, B.T.; Hensley, K.; Bachmann, M.; Scofield, R.H. Oxidatively modified autoantigens in autoimmune diseases. Free. Radic. Biol. Med. 2006, 41, 549–556. [Google Scholar] [CrossRef]
- Guo, W.; Smith, D.; Aviszus, K.; Detanico, T.; Heiser, R.A.; Wysocki, L.J. Somatic hypermutation as a generator of antinuclear antibodies in a murine model of systemic autoimmunity. J. Exp. Med. 2010, 207, 2225–2237. [Google Scholar] [CrossRef] [Green Version]
- Smith, P.R.; Williams, D.G.A.; Venables, P.J.; Maini, R.N.; Maimi, R.N. Monoclonal antibodies to the Sjögren’s syndrome associated antigen SS-B (La). J. Immunol. Methods 1985, 77, 63–76. [Google Scholar] [CrossRef]
- Veldhoven, C.H.A.; Pruijn, G.J.M.; Meilof, J.F.; Thijssen, J.P.H.; Kemp, A.W.C.M.; Venrooij, W.J.; Smeenk, R.J.T. Characterization of murine monoclonal antibodies against 60-kD Ro/SS-A and La/SS-B autoantigens. Clin. Exp. Immunol. 1995, 101, 45–54. [Google Scholar] [CrossRef]
- Buskila, D.; Weigl, D.; Shoenfeld, Y. The detection of anti-Ro/SS-A and anti-La/SS-B activity of human serum monoclonal immunoglobulins (monoclonal gammopathies). Hum. Antibodies Hybridomas 1992, 3, 75–80. [Google Scholar] [CrossRef]
- Harley, J.B.; O Rosario, M.; Yamagata, H.; Fox, O.F.; Koren, E. Immunologic and structural studies of the lupus/Sjögren’s syndrome autoantigen, La/SSB, with a monoclonal antibody. J. Clin. Investig. 1985, 76, 801–806. [Google Scholar] [CrossRef] [Green Version]
- Chan, E.K.; Francoeur, A.M.; Tan, E.M. Epitopes, structural domains, and asymmetry of amino acid residues in SS-B/La nu-clear protein. J. Immunol. 1986, 136, 3744–3749. [Google Scholar]
- Chan, E.K.; Tan, E.M. Human autoantibody-reactive epitopes of SS-B/La are highly conserved in comparison with epitopes recognized by murine monoclonal antibodies. J. Exp. Med. 1987, 166, 1627–1640. [Google Scholar] [CrossRef] [Green Version]
- Offen, D.; Mendlovic, S.; Fricke, H.; Sperling, J.; Mozes, E.; Sperling, R. Monoclonal anti-la antibody derived from a mouse with experimental SLE is similar to human anti-la antibodies. J. Autoimmun. 1990, 3, 701–713. [Google Scholar] [CrossRef]
- Pruijn, G.J.M.; Thijssen, J.P.H.; Smith, P.R.; Williams, D.G.A.; Venrooij, W.J. Anti-La Monoclonal Antibodies Recognizing Epitopes Within the RNA-Binding Domain of the La Protein Show Differential Capacities to Immunoprecipitate RNA-Associated La Protein. JBIC J. Biol. Inorg. Chem. 1995, 232, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.; Mayet, W.J.; Schroder, H.C.; Pfeifer, K.; Büschenfelde, K.H.M.Z.; Müller, W.E. Association of La and Ro antigens with intracellular structures in HEp-2 carcinoma cells. Proc. Natl. Acad. Sci. USA 1986, 83, 7770–7774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachmann, M.; Pfeifer, K.; Schröder, H.C.; Müller, W.E.; Schrdder, H.-C. Characterization of the autoantigen La as a nucleic acid-dependent ATPase/dATPase with melting properties. Cell 1990, 60, 85–93. [Google Scholar] [CrossRef]
- Tröster, H.; Bartsch, H.; Klein, R.; Metzger, T.E.; Pollak, G.; Semsei, I.; Schwemmle, M.; Pruijn, G.J.; Van Venrooij, W.J.; Bachmann, M. Activation of a murine autoreactive B cell by immunization with human recombinant autoantigen La/SS-B: Characterization of the autoepitope. J. Autoimmun. 1995, 8, 825–842. [Google Scholar] [CrossRef] [Green Version]
- Grölz, D.; Laubinger, J.; Wilmer, F.; Tröster, H.; Bachmann, M. Transfection Analysis of Expression of mRNA Isoforms Encoding the Nuclear Autoantigen La/SS-B. J. Biol. Chem. 1997, 272, 12076–12082. [Google Scholar] [CrossRef] [Green Version]
- Bippes, C.C.; Feldmann, A.; Stamova, S.; Cartellieri, M.; Schwarzer, A.; Wehner, R.; Schmitz, M.; Rieber, E.P.; Zhao, S.; Schäkel, K.; et al. A Novel Modular Antigen Delivery System for Immuno Targeting of Human 6-sulfo LacNAc-Positive Blood Dendritic Cells (SlanDCs). PLoS ONE 2011, 6, e16315. [Google Scholar] [CrossRef]
- Bachmann, M. The UniCAR system: A modular CAR T cell approach to improve the safety of CAR T cells. Immunol. Lett. 2019, 211, 13–22. [Google Scholar] [CrossRef]
- Cartellieri, M.; Koristka, S.; Arndt, C.; Feldmann, A.; Stamova, S.; Von Bonin, M.; Töpfer, K.; Krüger, T.; Geib, M.; Michalk, I.; et al. A Novel Ex Vivo Isolation and Expansion Procedure for Chimeric Antigen Receptor Engrafted Human T Cells. PLoS ONE 2014, 9, e93745. [Google Scholar] [CrossRef]
- Arndt, C.; Bachmann, M.; Bergmann, R.; Berndt, N.; Feldmann, A.; Koristka, S. Theranostic CAR T cell targeting: A brief review. J. Label. Compd. Radiopharm. 2019, 62, 533–540. [Google Scholar] [CrossRef]
- Jureczek, J.; Bergmann, R.; Berndt, N.; Koristka, S.; Kegler, A.; Puentes-Cala, E.; Soto, J.A.; Arndt, C.; Bachmann, M.; Feldmann, A. An oligo-His-tag of a targeting module does not influence its biodistribution and the retargeting capabilities of UniCAR T cells. Sci. Rep. 2019, 9, 10547. [Google Scholar] [CrossRef] [PubMed]
- Albert, S.; Arndt, C.; Feldmann, A.; Bergmann, R.; Bachmann, D.; Koristka, S.; Ludwig, F.; Ziller-Walter, P.; Kegler, A.; Gärtner, S.; et al. A novel nanobody-based target module for retargeting of T lymphocytes to EGFR-expressing cancer cells via the modular UniCAR platform. OncoImmunology 2017, 6, e1287246. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, A.; Hoffmann, A.; Bergmann, R.; Koristka, S.; Berndt, N.; Arndt, C.; Loureiro, L.R.; Kittel-Boselli, E.; Mitwasi, N.; Kegler, A.; et al. Versatile chimeric antigen receptor platform for controllable and combinatorial T cell therapy. OncoImmunology 2020, 9, 1785608. [Google Scholar] [CrossRef] [PubMed]
- Arndt, C.; Fasslrinner, F.; Loureiro, L.R.; Koristka, S.; Feldmann, A.; Bachmann, M. Adaptor CAR Platforms—Next Generation of T Cell-Based Cancer Immunotherapy. Cancers 2020, 12, 1302. [Google Scholar] [CrossRef] [PubMed]
- Koristka, S.; Cartellieri, M.; Feldmann, A.; Arndt, C.; Loff, S.; Michalk, I.; Aliperta, R.; von Bonin, M.; Bornhäuser, M.; Ehninger, A.; et al. Flexible antigen-specific redirection of human regulatory T cells via a novel universal chimeric antigen receptor system. Blood 2014, 124, 3494. [Google Scholar] [CrossRef]
- Cartellieri, M.; Loff, S.; von Bonin, M.; Bejestani, E.P.; Ehninger, A.; Feldmann, A.; Koristka, S.; Arndt, C.; Ehninger, G.; Bachmann, M.P.; et al. Unicar: A novel modular retargeting platform technology for CAR T cells. Blood 2015, 126, 5549. [Google Scholar] [CrossRef]
- Koristka, S.; Cartellieri, M.; Arndt, C.; Bippes, C.C.; Feldmann, A.; Michalk, I.; Wiefel, K.; Stamova, S.; Schmitz, M.; Ehninger, G. Retargeting of regulatory T cells to surface-inducible autoantigen La/SS-B. J. Autoimmun. 2013, 42, 105–116. [Google Scholar] [CrossRef]
- Koristka, S.; Kegler, A.; Bergmann, R.; Arndt, C.; Feldmann, A.; Albert, S.; Cartellieri, M.; Ehninger, A.; Ehninger, G.; Middeke, J.M.; et al. Engrafting human regulatory T cells with a flexible modular chimeric antigen receptor technology. J. Autoimmun. 2018, 90, 116–131. [Google Scholar] [CrossRef]
- Feldmann, A.; Arndt, C.; Bergmann, R.; Loff, S.; Cartellieri, M.; Bachmann, D.; Aliperta, R.; Hetzenecker, M.; Ludwig, F.; Albert, S.; et al. Retargeting of T lymphocytes to PSCA- or PSMA positive prostate cancer cells using the novel modular chimeric antigen receptor platform technology “UniCAR”. Oncotarget 2017, 8, 31368–31385. [Google Scholar] [CrossRef] [Green Version]
- Arndt, C.; Feldmann, A.; Koristka, S.; Schäfer, M.; Bergmann, R.; Mitwasi, N.; Berndt, N.; Bachmann, D.; Kegler, A.; Schmitz, M.; et al. A theranostic PSMA ligand for PET imaging and retargeting of T cells expressing the universal chimeric antigen receptor UniCAR. OncoImmunology 2019, 8, 1659095. [Google Scholar] [CrossRef] [Green Version]
- Jureczek, J.; Feldmann, A.; Bergmann, R.; Arndt, C.; Berndt, N.; Koristka, S.; Loureiro, L.R.; Mitwasi, N.; Hoffmann, A.; Kegler, A.; et al. Highly Efficient Targeting of EGFR-Expressing Tumor Cells with UniCAR T Cells via Target Modules Based on Cetuximab®. OncoTargets Ther. 2020, 13, 5515–5527. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, D.; Aliperta, R.; Bergmann, R.; Feldmann, A.; Koristka, S.; Arndt, C.; Loff, S.; Welzel, P.; Albert, S.; Kegler, A.; et al. Retargeting of UniCAR T cells with an in vivo synthesized target module directed against CD19 positive tumor cells. Oncotarget 2017, 9, 7487–7500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loureiro, L.R.; Feldmann, A.; Bergmann, R.; Koristka, S.; Berndt, N.; Arndt, C.; Pietzsch, J.; Novo, C.; Videira, P.A.; Bachmann, M. Development of a novel target module redirecting UniCAR T cells to Sialyl Tn-expressing tumor cells. Blood Cancer J. 2018, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, L.R.; Feldmann, A.; Bergmann, R.; Koristka, S.; Berndt, N.; Máthé, D.; Hegedüs, N.; Szigeti, K.; Videira, P.A.; Bachmann, M.; et al. Extended half-life target module for sustainable UniCAR T-cell treatment of STn-expressing cancers. J. Exp. Clin. Cancer Res. 2020, 39, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mitwasi, N.; Feldmann, A.; Arndt, C.; Koristka, S.; Berndt, N.; Jureczek, J.; Loureiro, L.R.; Bergmann, R.; Máthé, D.; Hegedüs, N.; et al. “UniCAR”-modified off-the-shelf NK-92 cells for targeting of GD2-expressing tumour cells. Sci. Rep. 2020, 10, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Cartellieri, M.; Feldmann, A.; Koristka, S.; Arndt, C.; Loff, S.; Ehninger, A.; Von Bonin, M.; Bejestani, E.P.; Bachmann, M. Switching CAR T cells on and off: A novel modular platform for retargeting of T cells to AML blasts. Blood Cancer J. 2016, 6, e458. [Google Scholar] [CrossRef] [Green Version]
- Feldmann, A.; Arndt, C.; Koristka, S.; Berndt, N.; Bergmann, R.; Bachmann, M. Conventional CARs versus modular CARs. Cancer Immunol. Immunother. 2019, 68, 1713–1719. [Google Scholar] [CrossRef] [Green Version]
- Koristka, S.; Ziller-Walter, P.; Bergmann, R.; Arndt, C.; Feldmann, A.; Kegler, A.; Cartellieri, M.; Ehninger, A.; Ehninger, G.; Bornhäuser, M.; et al. Anti-CAR-engineered T cells for epitope-based elimination of autologous CAR T cells. Cancer Immunol. Immunother. 2019, 68, 1401–1415. [Google Scholar] [CrossRef] [Green Version]
- Arndt, C.; Loureiro, L.R.; Feldmann, A.; Jureczek, J.; Bergmann, R.; Máthé, D.; Hegedüs, N.; Berndt, N.; Koristka, S.; Mitwasi, N.; et al. UniCAR T cell immunotherapy enables efficient elimination of radioresistant cancer cells. OncoImmunology 2020, 9, 1743036. [Google Scholar] [CrossRef] [Green Version]
- Keech, C.L.; Farris, A.D.; Beroukas, D.; Gordon, T.P.; McCluskey, J. Cognate T cell help is sufficient to trigger anti-nuclear autoantibodies in naive mice. J. Immunol. 2001, 166, 5826–5834. [Google Scholar] [CrossRef] [Green Version]
- Fouraux, M.A.; Van Der Heijden, A.; Van Venrooij, W.J.; Pruijn, G.J.M. Cross-reactivity of the anti-La monoclonal antibody SW5 with early endosome antigen 2. Immunology 2002, 106, 336–342. [Google Scholar] [CrossRef] [PubMed]
- E Lewis, J.; Fu, S.M.; Gaskin, F. Autoimmunity, end organ damage, and the origin of autoantibodies and autoreactive T cells in systemic lupus erythematosus. Discov. Med. 2013, 15, 85–92. [Google Scholar]
- Fu, S.M.; Deshmukh, U.S.; Gaskin, F. Pathogenesis of systemic lupus erythematosus revisited 2011: End organ resistance to damage, autoantibody initiation and diversification, and HLA-DR. J. Autoimmun. 2011, 37, 104–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Z.J.; Davis, K.; Maier, S.; Bachmann, M.P.; Kim-Howard, X.R.; Keech, C.; Gordon, T.P.; McCluskey, J.; Farris, A.D. Neo-epitopes are required for immuno-genicity of the La/SS-B nuclear antigen in the context of late apoptotic cells. Clin. Exp. Immunol. 2006, 143, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Massicotte, H.; Harley, J.B.; Bell, D.A. Characterization of human-human hybridoma monoclonal anti-Ro(SS-A) autoanti-bodies derived from normal tonsil lymphoid cells. J. Autoimmun. 1992, 5, 771–785. [Google Scholar] [CrossRef]
- Bachmann, M.; Grölz, D.; Bartsch, H.; Klein, R.R.; Tröster, H. Analysis of expression of an alternative La (SS-B) cDNA and localization of the encoded N- and C-terminal peptides. Biochim. Biophys. Acta (BBA) Bioenerg. 1997, 1356, 53–63. [Google Scholar] [CrossRef] [Green Version]
- Grölz, D.; Bachmann, M. An Altered Intracellular Distribution of the Autoantigen La/SS-B When Translated from a La mRNA Isoform. Exp. Cell Res. 1997, 234, 329–335. [Google Scholar] [CrossRef]
- Arndt, C.; Koristka, S.; Feldmann, A.; Bachmann, M. Native Polyacrylamide Gels. Methods Mol. Biol. 2019, 1855, 87–91. [Google Scholar] [CrossRef]
- Bachmann, M.P.; Bartsch, H.; Gross, J.K.; Maier, S.M.; Gross, T.F.; Workman, J.L.; James, J.A.; Farris, A.D.; Jung, B.; Franke, C.; et al. Autoimmunity as a Result of Escape from RNA Surveillance. J. Immunol. 2006, 177, 1698–1707. [Google Scholar] [CrossRef]
- Wu, T.T.; Kabat, E.A. An analysis of the sequences of the variable regions of bence jones proteins and myeloma light chains and their implications for antibody complementarity. J. Exp. Med. 1970, 132, 211–250. [Google Scholar] [CrossRef] [Green Version]
- Alfano, C.; Sanfelice, D.; Babon, J.; Kelly, G.; Jacks, A.; Curry, S.; Conte, M.R. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 2004, 11, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Jacks, A.; Babon, J.; Kelly, G.; Manolaridis, I.; Cary, P.D.; Curry, S.H.; Conte, M.R. Structure of the C-Terminal Domain of Human La Protein Reveals a Novel RNA Recognition Motif Coupled to a Helical Nuclear Retention Element. Structure 2003, 11, 833–843. [Google Scholar] [CrossRef] [Green Version]





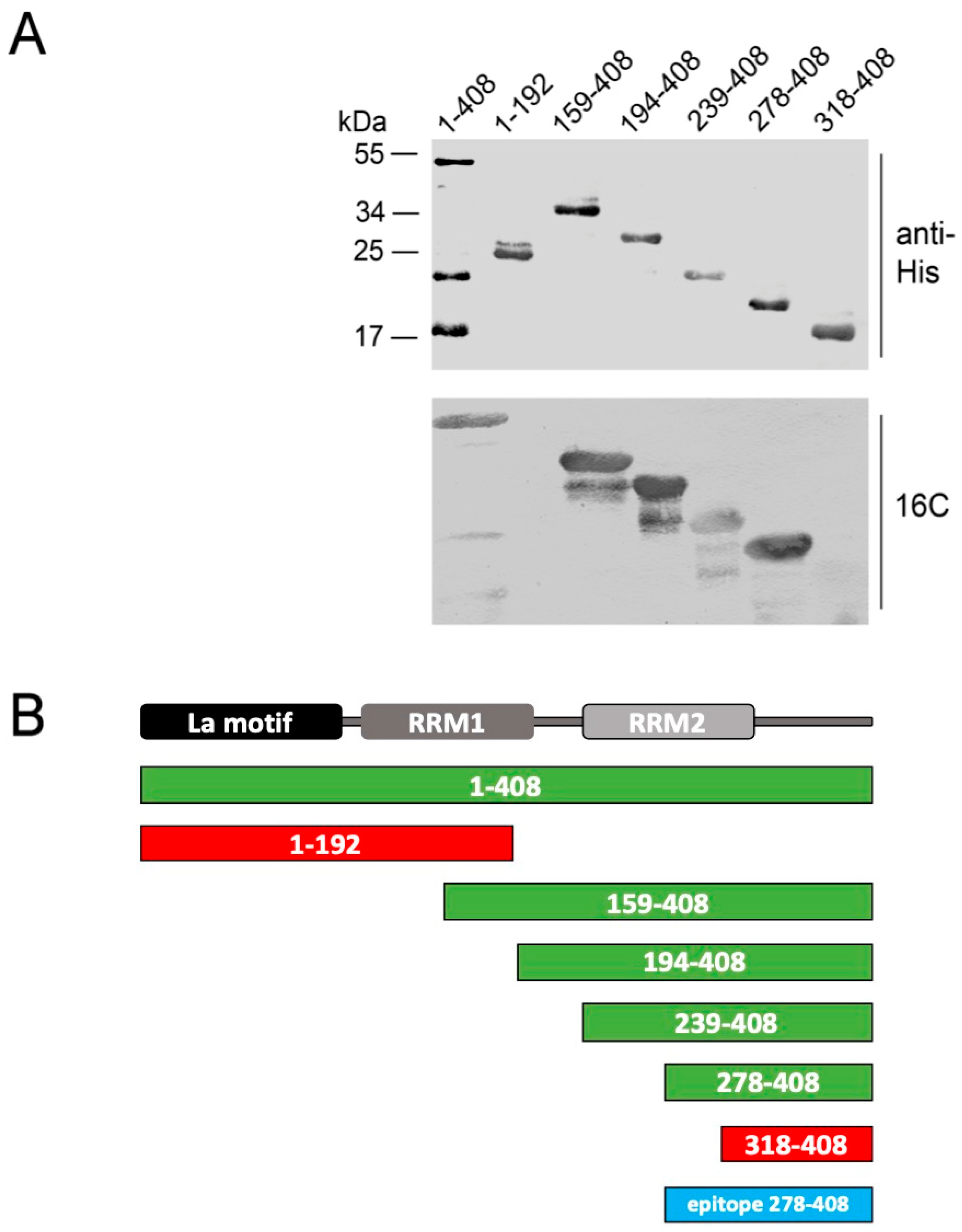
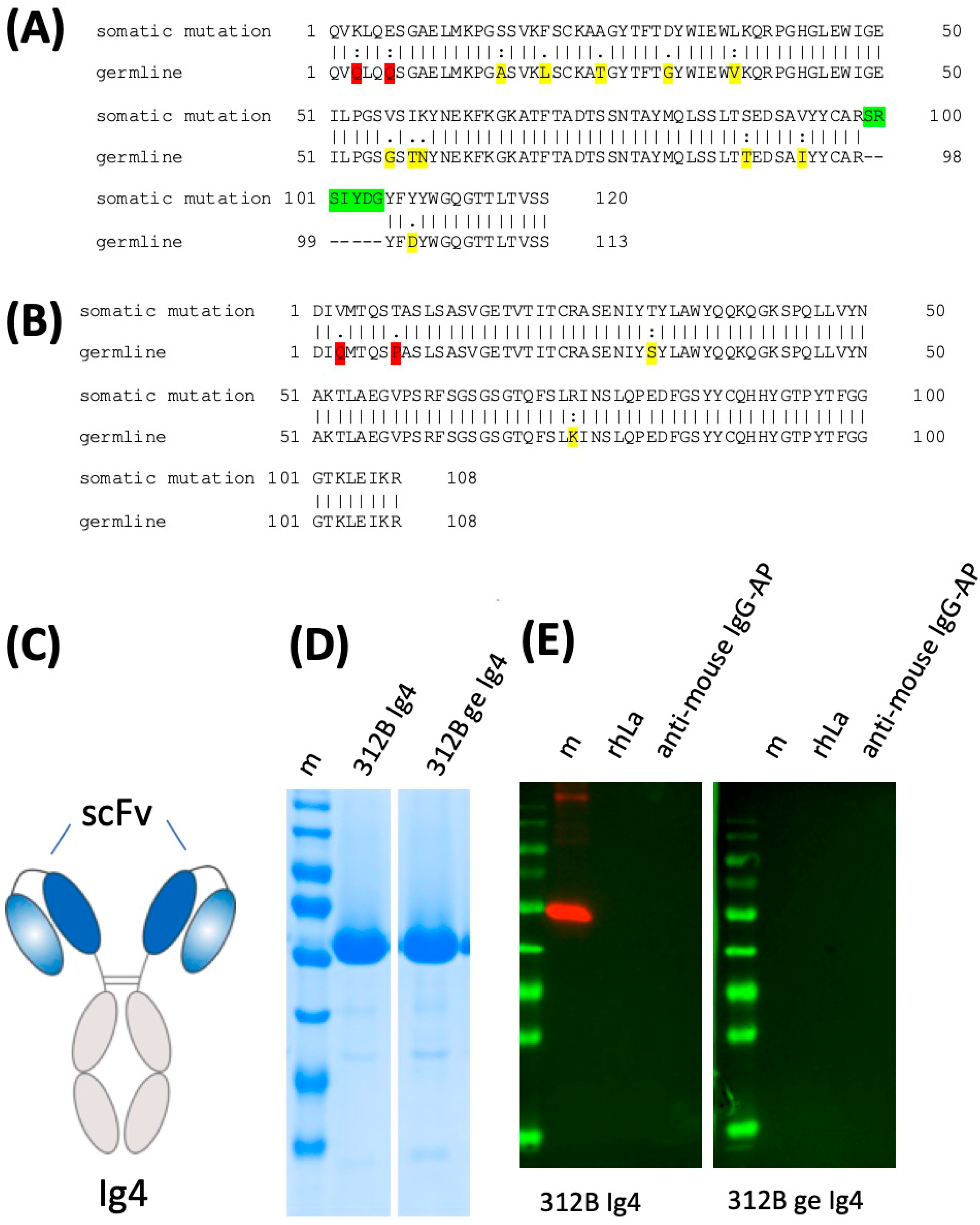
| mab | Isotype | Vl | Jl | Vh | Dh | Jh | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| E | %ID | E | %ID | E | %ID | E | %ID | E | %ID | ||
| SW5 | IgG2b | 21–12 | 98% | JK1 | 86% | VH7183.a7.10 | 94% | DSP2.7 DSP2.5 | 91% | JH3 | 98% |
| 7B6 | IgG1 | 8–21 | 98% | JK1 | 100% | VHJ606.a6.127 | 88% | DSP2.8 DSP2.7 DSP2.5 | 100% | JH3 | 100% |
| 5B9 | IgG2a | 8–21 | 98% | JK5 | 100% | J558.33 | 91% | DSP2.2 | 100% | JH3 | 100% |
| 24BG7 | IgG1 | kk4 | 99% | JK2 | 100% | J558.22 | 94% | DST4.3 DST4.2 | 100% | JH3 | 100% |
| 22A | IgG1 | kk4 | 99% | JK5 | 94% | J558.80.186 | 89% | DSP2.9 | 89% | JH2 | 100% |
| 27E | IgG2b | aa4 | 97% | JK1 | 100% | J558.22 | 93% | DSP2.3 | 100% | JH2 | 100% |
| 312B | IgG1 | 12–44 | 98% | JK2 | 100% | J558.6.96 | 94% | DSP2.9 | 100% | JH2 | 96% |
| 2F9 | IgG1 | 21–5 | 99% | JK4 | 100% | 3609.21.174 | 96% | DST4.3 DST4.2 | 100% | JH2 | 100% |
| 32A | IgG1 | aa4 | 99% | JK1 | 100% | J558.84.190 | 93% | DSP2.x | 100% | JH2 | 100% |
| 16C | IgM | bt20 | 99% | JK2 | 100% | J558.16.106 | 96% | DFL16.3 | 100% | JH4 | 100% |
| 13C5B | IgM | aa4 | 98% | JK1 | 100% | J558.47 | 97% | DSP2.11 DSP2.10 | 100% | JH4 | 98% |
| 312G | IgM | kk4 | 98% | JK5 | 100% | VHQ52.a19.61 | 98% | DFL16.3 | 100% | JH4 | 100% |
| mab | Number of Mutations | n-Nucleotides | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FWR1 | CDR1 | FWR2 | CDR2 | FWR3 | CDR3 | FWR4 | |||||||||
| N | A | N | A | N | A | N | A | N | A | N | A | N | A | Upstream Jl | |
| SW5 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 4 | 3 | +3 | ||||
| 7B6 | 2 | 2 | 1 | 1 | +2 | ||||||||||
| 5B9 | 1 | 1 | 1 | 1 | +1 | ||||||||||
| 24BG7 | 1 | 1 | 1 | 1 | 1 | 1 | −1 | ||||||||
| 22A | 2 | 1 | 1 | 2 | 2 | +1 | |||||||||
| 27E | 2 | ||||||||||||||
| 312B | 1 | 1 | 1 | 1 | 1 | +1 | |||||||||
| 2F9 | 1 | +1 | |||||||||||||
| 32A | 1 | 1 | 1 | 1 | |||||||||||
| 16C | +2 | ||||||||||||||
| 13C5B | |||||||||||||||
| 312G | −1 | ||||||||||||||
| mab | Number of Mutations | n-Nucleotides | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FWR1 | CDR1 | FWR2 | CDR2 | FWR3 | CDR3 | |||||||||
| N | A | N | A | N | A | N | A | N | A | N | A | Upstream Dh | Downstream Dh | |
| SW5 | 1 | 1 | 8 | 5 | 8 | 3 | 2 | 2 | +2 | +1 | ||||
| 7B6 | 5 | 2 | 2 | 2 | 5 | 13 | 8 | 6 | 2 | +4 | ||||
| 5B9 | 3 | 1 | 6 | 4 | 4 | 1 | 6 | 5 | 3 | 1 | +6 | +6 | ||
| 24BG7 | 9 | 6 | 3 | 2 | 3 | 3 | 3 | 2 | +3 | +3 | ||||
| 22A | 3 | 3 | 3 | 2 | 2 | 12 | 7 | 8 | 4 | 1 | +12 | −3 | ||
| 27E | 8 | 6 | 4 | 3 | 3 | 3 | 5 | 2 | +5 | +6 | ||||
| 312B | 3 | 3 | 2 | 1 | 1 | 1 | 3 | 3 | 4 | 2 | 2 | 1 | +10 | −4 |
| 2F9 | 1 | 1 | 1 | 1 | −5 | +13 | ||||||||
| 32A | 2 | 3 | 3 | 6 | 6 | 5 | 2 | +6 | ||||||
| 16C | 3 | 2 | 2 | 2 | 1 | 1 | +7 | +6 | ||||||
| 13C5B | 1 | 1 | 3 | 2 | 1 | 1 | 2 | 1 | 1 | +1 | +5 | |||
| 312G | 2 | 1 | +4 | +5 | ||||||||||
| mab | Reactivity Recombinant Proteins Immunoblotting | Reactivity m3T3 hLa Immunoblotting | Immuno-Precipitation m3T3 hLa | Immuno-Fluorescence PFA | Immuno-Fluorescence MeOH | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rhLa | rmLa | HeLa | 3T3 | hLa | mLa | hLa | mLa | HeLa | 3T3 | HeLa | 3T3 | |
| SW5 | +++ | ++ | +++ | - | +++ | - | ++ | - | +++ | - | ++ | - |
| 7B6 | +++ | ++ | +++ | - | ++ | - | - | - | (+) | - | (+) | - |
| 5B9 | +++ | +++ | +++ | +++ | +++ | +++ | - | - | ++ | - | +++ | +++ |
| 24BG7 | +++ | + | +++ | + | ++ | + | + | - | ++ | ++ | - | - |
| 22A | +++ | - | +++ | - | + | - | ++ | - | ++ | - | - | - |
| 27E | +++ | + | +++ | +++ | ++ | ++ | + | + | +++ | +++ | +++ | + |
| 312B | +++ | +++ | +++ | +++ | + | ++ | +++ | +++ | +++ | +++ | ++ | + |
| 2F9 | +++ | +++ | ++ | ++ | + | + | - | - | + | + | (+) | - |
| 32A | +++ | - | ++ | - | (+) | - | - | - | (+) | - | - | - |
| 16C | + | * | * | * | * | * | * | * | C | C | C | C |
| 13C5B | + | * | * | * | * | * | * | * | C | C | C | C |
| 312G | - | * | * | * | * | * | * | * | C | C | C | C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bachmann, M.P.; Bartsch, T.; Bippes, C.C.; Bachmann, D.; Puentes-Cala, E.; Bachmann, J.; Bartsch, H.; Arndt, C.; Koristka, S.; Loureiro, L.R.; et al. T Cell Mediated Conversion of a Non-Anti-La Reactive B Cell to an Autoreactive Anti-La B Cell by Somatic Hypermutation. Int. J. Mol. Sci. 2021, 22, 1198. https://doi.org/10.3390/ijms22031198
Bachmann MP, Bartsch T, Bippes CC, Bachmann D, Puentes-Cala E, Bachmann J, Bartsch H, Arndt C, Koristka S, Loureiro LR, et al. T Cell Mediated Conversion of a Non-Anti-La Reactive B Cell to an Autoreactive Anti-La B Cell by Somatic Hypermutation. International Journal of Molecular Sciences. 2021; 22(3):1198. https://doi.org/10.3390/ijms22031198
Chicago/Turabian StyleBachmann, Michael P., Tabea Bartsch, Claudia C. Bippes, Dominik Bachmann, Edinson Puentes-Cala, Jennifer Bachmann, Holger Bartsch, Claudia Arndt, Stefanie Koristka, Liliana R. Loureiro, and et al. 2021. "T Cell Mediated Conversion of a Non-Anti-La Reactive B Cell to an Autoreactive Anti-La B Cell by Somatic Hypermutation" International Journal of Molecular Sciences 22, no. 3: 1198. https://doi.org/10.3390/ijms22031198






