Expression of Endogenous Angiotensin-Converting Enzyme 2 in Human Induced Pluripotent Stem Cell-Derived Retinal Organoids
Abstract
1. Introduction
2. Results
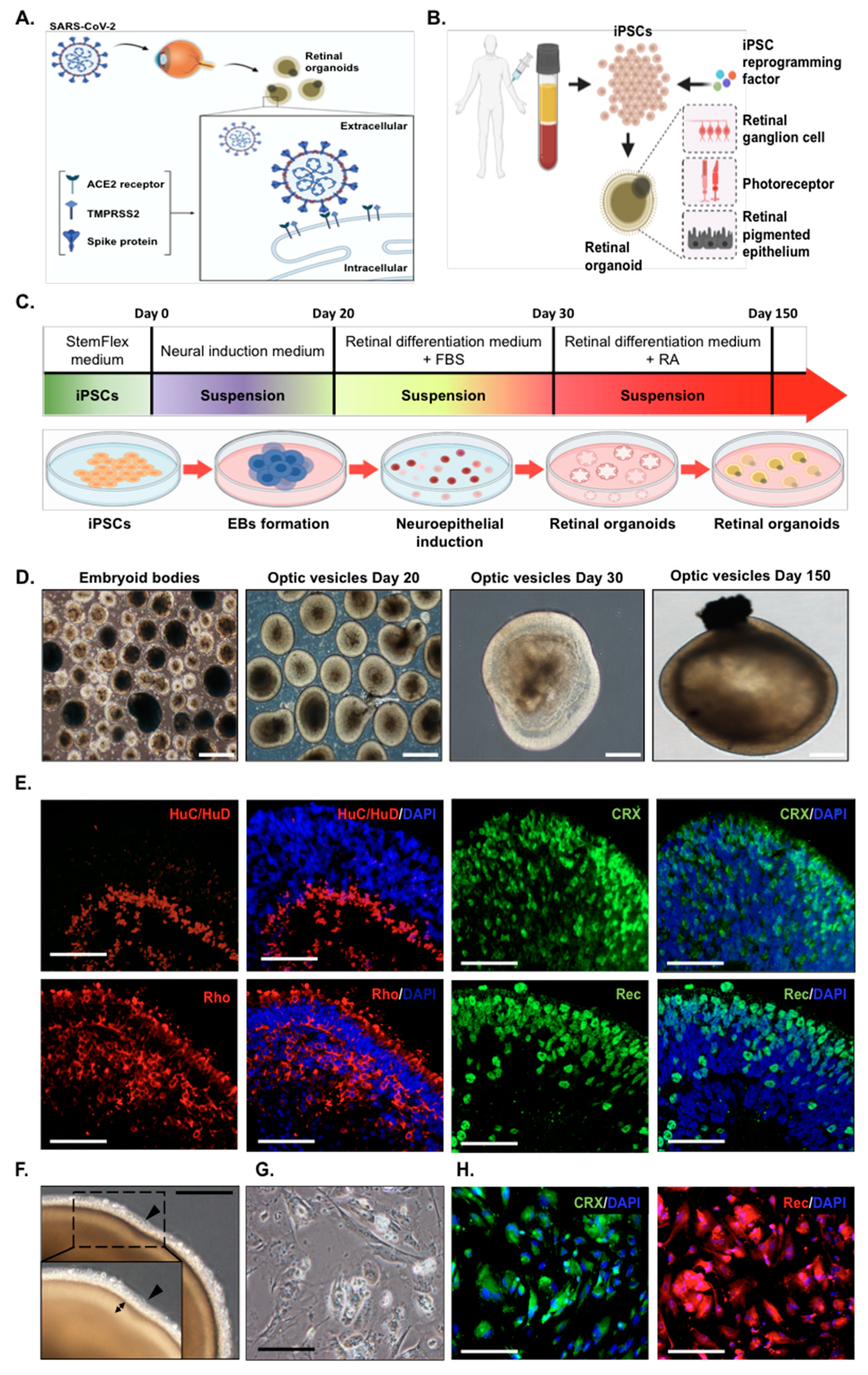
2.1. Generation of Human Induced Pluripotent Stem Cells (hiPSC) into Retinal Organoids and Monolayer Cultures
2.2. Sustained Gene Expression of ACE2 and TMPRSS2 during the Development of Photoreceptor Organoids and Monolayer Cultures
2.3. Modeling Pseudovirus Infection of SARS-CoV-2 with Retinal Organoids and Monolayer Cultures
2.4. Comparative Transcriptome Analysis of the Infected SARS-CoV-2 Pseudovirus
3. Discussion
4. Materials and Methods
4.1. Maintenance and Differentiation of Human-Induced Pluripotent Stem Cells
4.2. Differentiation of Retinal Organoids from hiPSCs
4.3. Infection with SARS-CoV-2 Pseudovirus
4.4. Immunofluorescence
4.5. Quantitative PCR and RT-PCR
4.6. RNA-Seq
4.7. Bioinformatic Analyses
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| dpi | day post-infection |
| MOI | multiplicity of infection |
| COVID-19 | Coronavirus disease 2019 |
| TMPRSS2 | Transmembrane protease serine 2 |
| ACE2 | Angiotensin-converting enzyme 2 |
| qPCR | qualitative polymerase chain reaction |
| SARS-CoV-2 | Severe acute respiratory syndrome-coronavirus-2 |
| iPSC | induced pluripotent stem cell |
| GFP | green fluorescent protein |
References
- Cascella, M.; Rajnik, M.; Cuomo, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation, and Treatment of Coronavirus (COVID-19); StatPearls: Treasure Island, FL, USA, 2020. [Google Scholar]
- Zhou, L.; Xu, Z.; Castiglione, G.M.; Soiberman, U.S.; Eberhart, C.G.; Duh, E.J. ACE2 and TMPRSS2 are expressed on the human ocular surface, suggesting susceptibility to SARS-CoV-2 infection. Ocul. Surf. 2020, 18, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, C.A.; Bergey, C.M.; Jasinska, A.J.; Ramensky, V.; Burt, F.; Svardal, H.; Jorgensen, M.J.; Freimer, N.B.; Grobler, J.P.; Turner, T.R. ACE2 and TMPRSS2 variation in savanna monkeys (Chlorocebus spp.): Potential risk for zoonotic/anthroponotic transmission of SARS-CoV-2 and a potential model for functional studies. PLoS ONE 2020, 15, e0235106. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.J. SARS-CoV-2 infection of kidney organoids prevented with soluble human ACE2. Nat. Rev. Nephrol. 2020, 16, 316. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Han, Y.; Nilsson-Payant, B.E.; Gupta, V.; Wang, P.; Duan, X.; Tang, X.; Zhu, J.; Zhao, Z.; Jaffre, F.; et al. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell 2020, 27, 125–136.e7. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhong, L.; Deng, J.; Peng, J.; Dan, H.; Zeng, X.; Li, T.; Chen, Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral. Sci. 2020, 12, 8. [Google Scholar] [CrossRef]
- Yi, S.A.; Nam, K.H.; Yun, J.; Gim, D.; Joe, D.; Kim, Y.H.; Kim, H.J.; Han, J.W.; Lee, J. Infection of Brain Organoids and 2D Cortical Neurons with SARS-CoV-2 Pseudovirus. Viruses 2020, 12, 1004. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Pather, S.R.; Huang, W.K.; Zhang, F.; Wong, S.Z.H.; Zhou, H.; Cubitt, B.; Fan, W.; Chen, C.Z.; Xu, M.; et al. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 2020, 27, 937–950.e9. [Google Scholar] [CrossRef]
- Casagrande, M.; Fitzek, A.; Puschel, K.; Aleshcheva, G.; Schultheiss, H.P.; Berneking, L.; Spitzer, M.S.; Schultheiss, M. Detection of SARS-CoV-2 in Human Retinal Biopsies of Deceased COVID-19 Patients. Ocul. Immunol. Inflamm. 2020, 28, 721–725. [Google Scholar] [CrossRef]
- Csobonyeiova, M.; Polak, S.; Danisovic, L. Recent Overview of the Use of iPSCs Huntington’s Disease Modeling and Therapy. Int. J. Mol. Sci. 2020, 21, 2239. [Google Scholar] [CrossRef]
- Fleischer, A.; Vallejo-Diez, S.; Martin-Fernandez, J.M.; Sanchez-Gilabert, A.; Castresana, M.; Del Pozo, A.; Esquisabel, A.; Avila, S.; Castrillo, J.L.; Gainza, E.; et al. iPSC-Derived Intestinal Organoids from Cystic Fibrosis Patients Acquire CFTR Activity upon TALEN-Mediated Repair of the p.F508del Mutation. Mol. Ther. Methods Clin. Dev. 2020, 17, 858–870. [Google Scholar] [PubMed]
- Caputo, L.; Granados, A.; Lenzi, J.; Rosa, A.; Ait-Si-Ali, S.; Puri, P.L.; Albini, S. Acute conversion of patient-derived Duchenne muscular dystrophy iPSC into myotubes reveals constitutive and inducible over-activation of TGFbeta-dependent pro-fibrotic signaling. Skelet. Muscle 2020, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhao, A.; Pan, Y.; Cai, S. Patient-Specific Cells for Modeling and Decoding Amyotrophic Lateral Sclerosis: Advances and Challenges. Stem Cell Rev. Rep. 2020, 16, 482–502. [Google Scholar] [CrossRef] [PubMed]
- Pomeshchik, Y.; Klementieva, O.; Gil, J.; Martinsson, I.; Hansen, M.G.; de Vries, T.; Sancho-Balsells, A.; Russ, K.; Savchenko, E.; Collin, A.; et al. Human iPSC-Derived Hippocampal Spheroids: An Innovative Tool for Stratifying Alzheimer Disease Patient-Specific Cellular Phenotypes and Developing Therapies. Stem Cell Rep. 2020, 15, 256–273. [Google Scholar] [PubMed]
- Songstad, A.E.; Wiley, L.A.; Duong, K.; Kaalberg, E.; Flamme-Wiese, M.J.; Cranston, C.M.; Riker, M.J.; Levasseur, D.; Stone, E.M.; Mullins, R.F.; et al. Generating iPSC-Derived Choroidal Endothelial Cells to Study Age-Related Macular Degeneration. Invest. Ophthalmol. Vis. Sci. 2015, 56, 8258–8267. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.P.; Liu, H.; Jin, Z.B. Generation of three human iPSC lines from a retinitis pigmentosa family with SLC7A14 mutation. Stem Cell Res. 2020, 49, 102075. [Google Scholar] [CrossRef]
- Chang, T.J.; Yang, D.M.; Wang, M.L.; Liang, K.H.; Tsai, P.H.; Chiou, S.H.; Lin, T.H.; Wang, C.T. Genomic analysis and comparative multiple sequences of SARS-CoV2. J. Chin. Med. Assoc. 2020, 83, 537–543. [Google Scholar] [CrossRef]
- Huo, K.G.; D’Arcangelo, E.; Tsao, M.S. Patient-derived cell line, xenograft and organoid models in lung cancer therapy. Transl. Lung Cancer Res. 2020, 9, 2214–2232. [Google Scholar] [CrossRef]
- Sun, G.; Chiuppesi, F.; Chen, X.; Wang, C.; Tian, E.; Nguyen, J.; Kha, M.; Trinh, D.; Zhang, H.; Marchetto, M.C.; et al. Modeling Human Cytomegalovirus-Induced Microcephaly in Human iPSC-Derived Brain Organoids. Cell. Rep. Med. 2020, 1, 100002. [Google Scholar]
- Cakir, B.; Xiang, Y.; Tanaka, Y.; Kural, M.H.; Parent, M.; Kang, Y.J.; Chapeton, K.; Patterson, B.; Yuan, Y.; He, C.S.; et al. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods 2019, 16, 1169–1175. [Google Scholar] [CrossRef]
- Forbes, T.A.; Howden, S.E.; Lawlor, K.; Phipson, B.; Maksimovic, J.; Hale, L.; Wilson, S.; Quinlan, C.; Ho, G.; Holman, K.; et al. Patient-iPSC-Derived Kidney Organoids Show Functional Validation of a Ciliopathic Renal Phenotype and Reveal Underlying Pathogenetic Mechanisms. Am. J. Hum. Genet. 2018, 102, 816–831. [Google Scholar] [CrossRef]
- Huang, K.C.; Wang, M.L.; Chen, S.J.; Kuo, J.C.; Wang, W.J.; Nhi Nguyen, P.N.; Wahlin, K.J.; Lu, J.F.; Tran, A.A.; Shi, M.; et al. Morphological and Molecular Defects in Human Three-Dimensional Retinal Organoid Model of X-Linked Juvenile Retinoschisis. Stem Cell Reports 2019, 13, 906–923. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.X.; Pek, N.M.Q.; Soh, B.-S. Disease Modeling Using 3D Organoids Derived from Human Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2018, 19, 936. [Google Scholar] [CrossRef] [PubMed]
- Strange, D.P.; Zarandi, N.P.; Trivedi, G.; Atala, A.; Bishop, C.E.; Sadri-Ardekani, H.; Verma, S. Human testicular organoid system as a novel tool to study Zika virus pathogenesis. Emerg. Microbes. Infect. 2018, 7, 82. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, C.; Sachs, N.; Chiu, M.C.; Wong, B.H.; Chu, H.; Poon, V.K.; Wang, D.; Zhao, X.; Wen, L.; et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl. Acad. Sci. USA 2018, 115, 6822–6827. [Google Scholar]
- Perez-Bermejo, J.A.; Kang, S.; Rockwood, S.J.; Simoneau, C.R.; Joy, D.A.; Ramadoss, G.N.; Silva, A.C.; Flanigan, W.R.; Li, H.; Nakamura, K.; et al. SARS-CoV-2 infection of human iPSC-derived cardiac cells predicts novel cytopathic features in hearts of COVID-19 patients. BioRxiv 2020. [Google Scholar] [CrossRef]
- Sharma, A.; Garcia, G.; Arumugaswami, V.; Svendsen, C.N. Human iPSC-Derived Cardiomyocytes are Susceptible to SARS-CoV-2 Infection. BioRxiv 2020. [Google Scholar] [CrossRef]
- Ramani, A.; Muller, L.; Ostermann, P.N.; Gabriel, E.; Abida-Islam, P.; Muller-Schiffmann, A.; Mariappan, A.; Goureau, O.; Gruell, H.; Walker, A.; et al. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020, 39, e106230. [Google Scholar] [CrossRef]
- Surendran, H.; Nandakumar, S.; Pal, R. Human Induced Pluripotent Stem Cell-Derived Lung Epithelial System for SARS-CoV-2 Infection Modeling and Its Potential in Drug Repurposing. Stem Cells Dev. 2020, 29, 1365–1369. [Google Scholar] [CrossRef]
- Kase, Y.; Okano, H. Expression of ACE2 and a viral virulence-regulating factor CCN family member 1 in human iPSC-derived neural cells: Implications for COVID-19-related CNS disorders. Inflamm. Regen. 2020, 40, 32. [Google Scholar] [CrossRef]
- Deng, C.; Yang, Y.; Chen, H.; Chen, W.; Chen, Z.; Ma, K.; Wang, J. Low risk of SARS-CoV-2 transmission through the ocular surface. Acta Ophthalmol. 2020, 98, e926–e927. [Google Scholar]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Moller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Ohlemacher, S.K.; Sridhar, A.; Xiao, Y.; Hochstetler, A.E.; Sarfarazi, M.; Cummins, T.R.; Meyer, J.S. Stepwise Differentiation of Retinal Ganglion Cells from Human Pluripotent Stem Cells Enables Analysis of Glaucomatous Neurodegeneration. Stem Cells 2016, 34, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Collin, J.; Queen, R.; Zerti, D.; Dorgau, B.; Georgiou, M.; Djidrovski, I.; Hussain, R.; Coxhead, J.M.; Joseph, A.; Rooney, P.; et al. Co-expression of SARS-CoV-2 entry genes in the superficial adult human conjunctival, limbal and corneal epithelium suggests an additional route of entry via the ocular surface. Ocul. Surf. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Quinn, P.M.; Buck, T.M.; Ohonin, C.; Mikkers, H.M.M.; Wijnholds, J. Production of iPS-Derived Human Retinal Organoids for Use in Transgene Expression Assays. Methods Mol. Biol. 2018, 1715, 261–273. [Google Scholar] [PubMed]
- Tanaka, T.; Yokoi, T.; Tamalu, F.; Watanabe, S.; Nishina, S.; Azuma, N. Generation of Retinal Ganglion Cells With Functional Axons From Mouse Embryonic Stem Cells and Induced Pluripotent Stem Cells. Investig. Ophthalmol. Vis. Sci. 2016, 57, 3348–3359. [Google Scholar] [CrossRef] [PubMed]
- Mellough, C.B.; Collin, J.; Khazim, M.; White, K.; Sernagor, E.; Steel, D.H.; Lako, M. IGF-1 Signaling Plays an Important Role in the Formation of Three-Dimensional Laminated Neural Retina and Other Ocular Structures From Human Embryonic Stem Cells. Stem Cells 2015, 33, 2416–2430. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar]
- Davidovic, L.; Durand, N.; Khalfallah, O.; Tabet, R.; Barbry, P.; Mari, B.; Sacconi, S.; Moine, H.; Bardoni, B. A novel role for the RNA-binding protein FXR1P in myoblasts cell-cycle progression by modulating p21/Cdkn1a/Cip1/Waf1 mRNA stability. PLoS Genet. 2013, 9, e1003367. [Google Scholar] [CrossRef]
- Kallman, A.; Capowski, E.E.; Wang, J.; Kaushik, A.M.; Jansen, A.D.; Edwards, K.L.; Chen, L.; Berlinicke, C.A.; Joseph Phillips, M.; Pierce, E.A.; et al. Investigating cone photoreceptor development using patient-derived NRL null retinal organoids. Commun. Biol. 2020, 3, 82. [Google Scholar] [CrossRef]
- Cora, V.; Haderspeck, J.; Antkowiak, L.; Mattheus, U.; Neckel, P.H.; Mack, A.F.; Bolz, S.; Ueffing, M.; Pashkovskaia, N.; Achberger, K.; et al. A Cleared View on Retinal Organoids. Cells 2019, 8, 391. [Google Scholar] [CrossRef] [PubMed]
- Hanke, L.; Vidakovics Perez, L.; Sheward, D.J.; Das, H.; Schulte, T.; Moliner-Morro, A.; Corcoran, M.; Achour, A.; Karlsson Hedestam, G.B.; Hallberg, B.M.; et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020, 11, 4420. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, S.J.R.; Silva, C.; Guarines, K.M.; Mendes, R.P.G.; Pardee, K.; Kohl, A.; Pena, L. Clinical and Laboratory Diagnosis of SARS-CoV-2, the Virus Causing COVID-19. ACS Infect. Dis. 2020, 6, 2319–2336. [Google Scholar] [CrossRef] [PubMed]
- Guler, N.; Siddiqui, F.; Fareed, J. Is the Reason of Increased D-Dimer Levels in COVID-19 Because of ACE-2-Induced Apoptosis in Endothelium? Clin. Appl. Thromb. Hemost. 2020, 26, 1076029620935526. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, W.; Yang, L.; You, R. Physiological and pathological regulation of ACE2, the SARS-CoV-2 receptor. Pharmacol. Res. 2020, 157, 104833. [Google Scholar] [CrossRef]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkruys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado Del Pozo, C.; Prosper, F.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181, 905–913.e7. [Google Scholar] [CrossRef]
- Napoli, P.E.; Nioi, M.; d’Aloja, E.; Fossarello, M. The Ocular Surface and the Coronavirus Disease 2019: Does a Dual ’Ocular Route’ Exist? J. Clin. Med. 2020, 9, 1269. [Google Scholar] [CrossRef]
- Hong, N.; Yu, W.; Xia, J.; Shen, Y.; Yap, M.; Han, W. Evaluation of ocular symptoms and tropism of SARS-CoV-2 in patients confirmed with COVID-19. Acta Ophthalmol. 2020, 26, 10–1111. [Google Scholar] [CrossRef]
- Makovoz, B.; Moeller, R.; Zebitz Eriksen, A.; tenOever, B.R.; Blenkinsop, T.A. SARS-CoV-2 Infection of Ocular Cells from Human Adult Donor Eyes and hESC-Derived Eye Organoids. SSRN 2020, 15, 3650574. [Google Scholar]
- Zhang, Q.; Jeppesen, D.K.; Higginbotham, J.N.; Franklin, J.L.; Crowe, J.E., Jr.; Coffey, R.J. ACE2-containing extracellular vesicles and exomeres bind the SARS-CoV-2 spike protein. Gastroenterology 2020, 11, 876. [Google Scholar] [CrossRef]
- Salahudeen, A.A.; Choi, S.S.; Rustagi, A.; Zhu, J.; de la, O.S.; Flynn, R.A.; Margalef-Catala, M.; Santos, A.J.M.; Ju, J.; Batish, A.; et al. Progenitor identification and SARS-CoV-2 infection in long-term human distal lung organoid cultures. BioRxiv 2020. [Google Scholar]
- Zhou, J.; Li, C.; Liu, X.; Chiu, M.C.; Zhao, X.; Wang, D.; Wei, Y.; Lee, A.; Zhang, A.J.; Chu, H.; et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020, 26, 1077–1083. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.Z.; Chu, H.; Han, S.; Shuai, H.; Deng, J.; Hu, Y.F.; Gong, H.R.; Lee, A.C.; Zou, Z.; Yau, T.; et al. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020, 30, 928–931. [Google Scholar] [CrossRef]
- Salick, M.R.; Wells, M.F.; Eggan, K.; Kaykas, A. Modelling Zika Virus Infection of the Developing Human Brain In Vitro Using Stem Cell Derived Cerebral Organoids. J. Vis. Exp. 2017, 56404. [Google Scholar] [CrossRef]
- Bordi, L.; Castilletti, C.; Falasca, L.; Ciccosanti, F.; Calcaterra, S.; Rozera, G.; Di Caro, A.; Zaniratti, S.; Rinaldi, A.; Ippolito, G.; et al. Bcl-2 inhibits the caspase-dependent apoptosis induced by SARS-CoV without affecting virus replication kinetics. Arch. Virol. 2006, 151, 369–377. [Google Scholar] [CrossRef]
- Garcia, L.F. Immune Response, Inflammation, and the Clinical Spectrum of COVID-19. Front. Immunol. 2020, 11, 1441. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad Mulyadi Lai, H.I.; Chou, S.-J.; Chien, Y.; Tsai, P.-H.; Chien, C.-S.; Hsu, C.-C.; Jheng, Y.-C.; Wang, M.-L.; Chiou, S.-H.; Chou, Y.-B.; et al. Expression of Endogenous Angiotensin-Converting Enzyme 2 in Human Induced Pluripotent Stem Cell-Derived Retinal Organoids. Int. J. Mol. Sci. 2021, 22, 1320. https://doi.org/10.3390/ijms22031320
Ahmad Mulyadi Lai HI, Chou S-J, Chien Y, Tsai P-H, Chien C-S, Hsu C-C, Jheng Y-C, Wang M-L, Chiou S-H, Chou Y-B, et al. Expression of Endogenous Angiotensin-Converting Enzyme 2 in Human Induced Pluripotent Stem Cell-Derived Retinal Organoids. International Journal of Molecular Sciences. 2021; 22(3):1320. https://doi.org/10.3390/ijms22031320
Chicago/Turabian StyleAhmad Mulyadi Lai, Henkie Isahwan, Shih-Jie Chou, Yueh Chien, Ping-Hsing Tsai, Chian-Shiu Chien, Chih-Chien Hsu, Ying-Chun Jheng, Mong-Lien Wang, Shih-Hwa Chiou, Yu-Bai Chou, and et al. 2021. "Expression of Endogenous Angiotensin-Converting Enzyme 2 in Human Induced Pluripotent Stem Cell-Derived Retinal Organoids" International Journal of Molecular Sciences 22, no. 3: 1320. https://doi.org/10.3390/ijms22031320
APA StyleAhmad Mulyadi Lai, H. I., Chou, S.-J., Chien, Y., Tsai, P.-H., Chien, C.-S., Hsu, C.-C., Jheng, Y.-C., Wang, M.-L., Chiou, S.-H., Chou, Y.-B., Hwang, D.-K., Lin, T.-C., Chen, S.-J., & Yang, Y.-P. (2021). Expression of Endogenous Angiotensin-Converting Enzyme 2 in Human Induced Pluripotent Stem Cell-Derived Retinal Organoids. International Journal of Molecular Sciences, 22(3), 1320. https://doi.org/10.3390/ijms22031320







