Arginine Methylation Regulates Ribosome CAR Function
Abstract
:1. Introduction
2. Results
2.1. Ribosome Subsystems Provide Neighborhoods for Molecular Dynamics
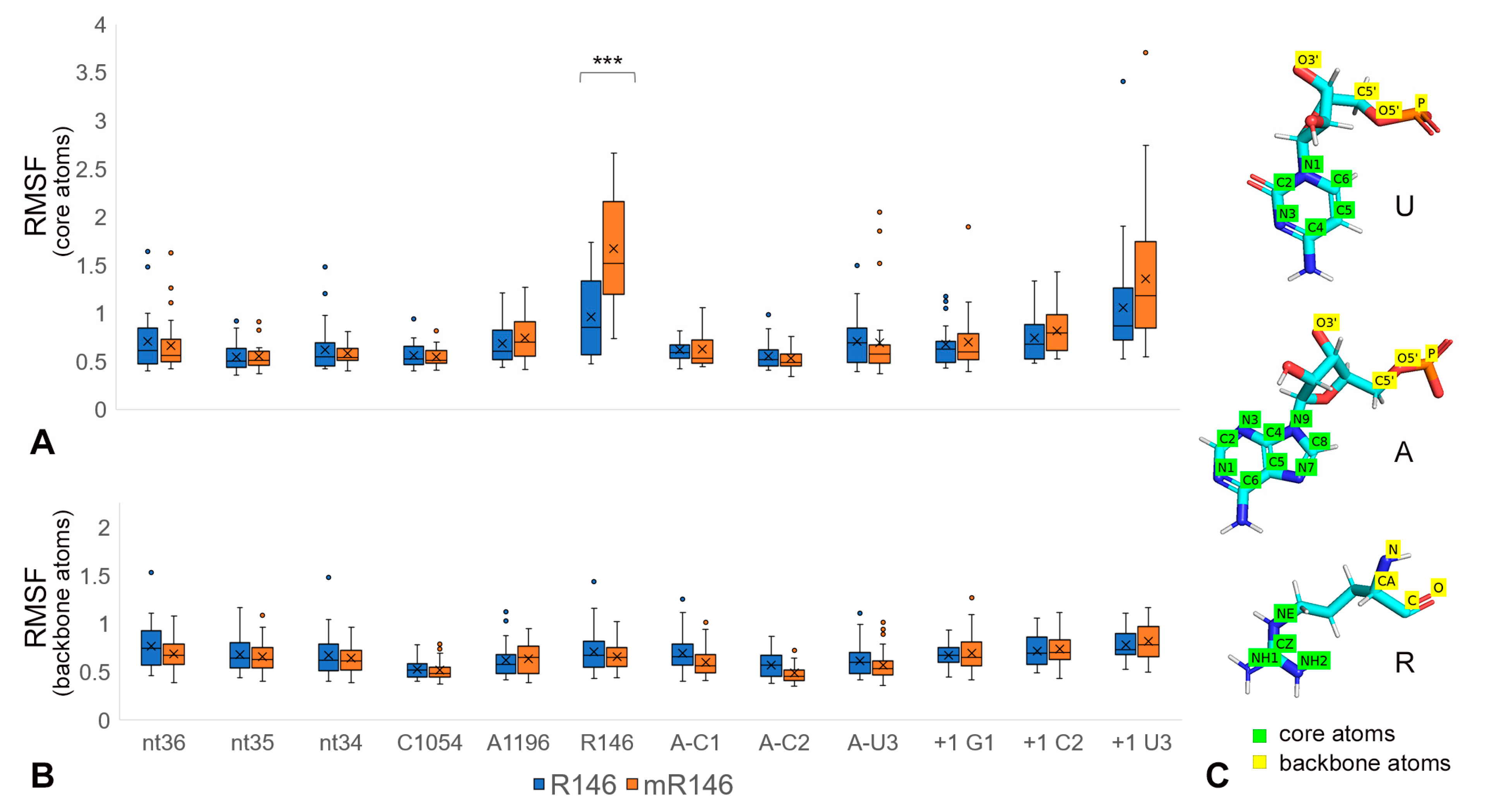
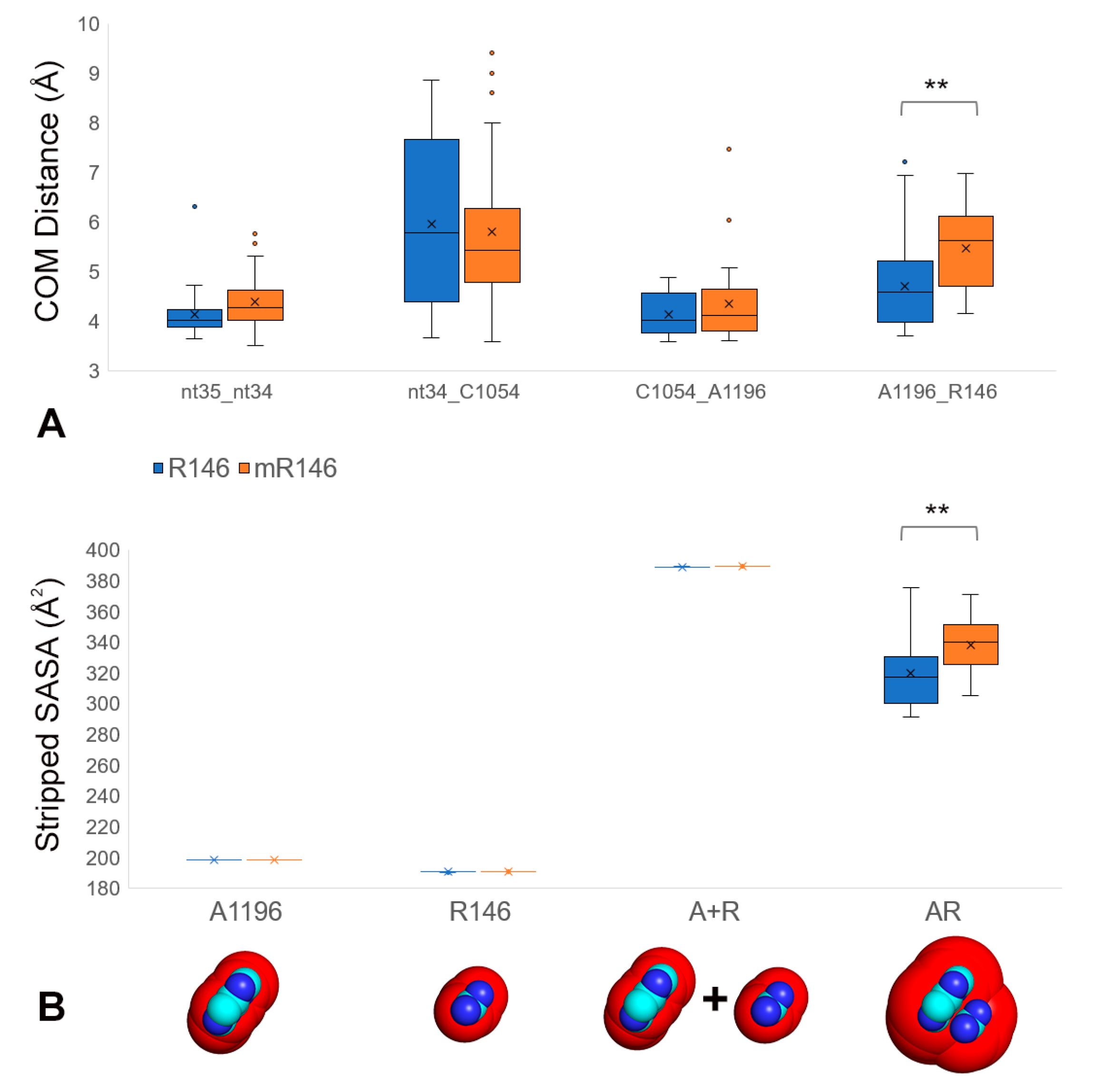
2.2. R146 Methylation Disrupts CAR Integrity
2.3. R146 Methylation Reduces CAR/+1 Codon Hydrogen Bonding
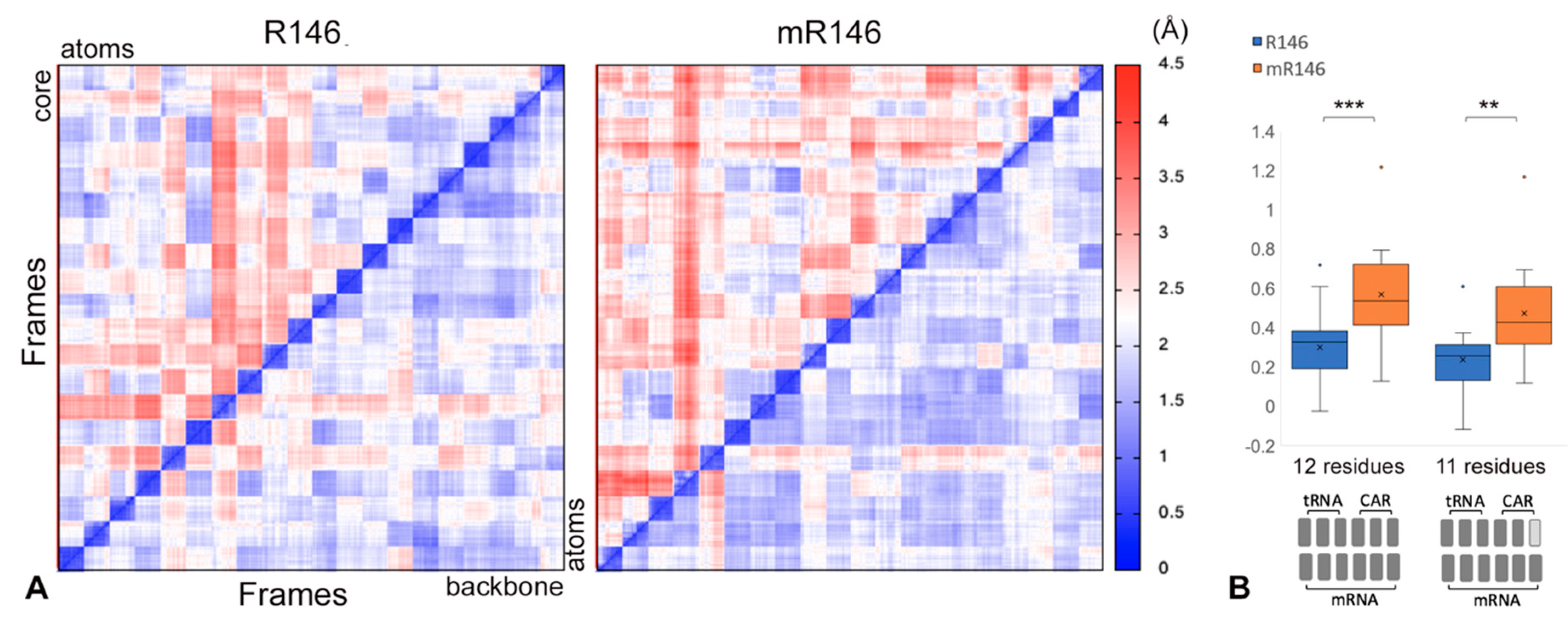
2.4. Cross-Trajectory Comparisons Show Variability of Behavior
3. Discussion
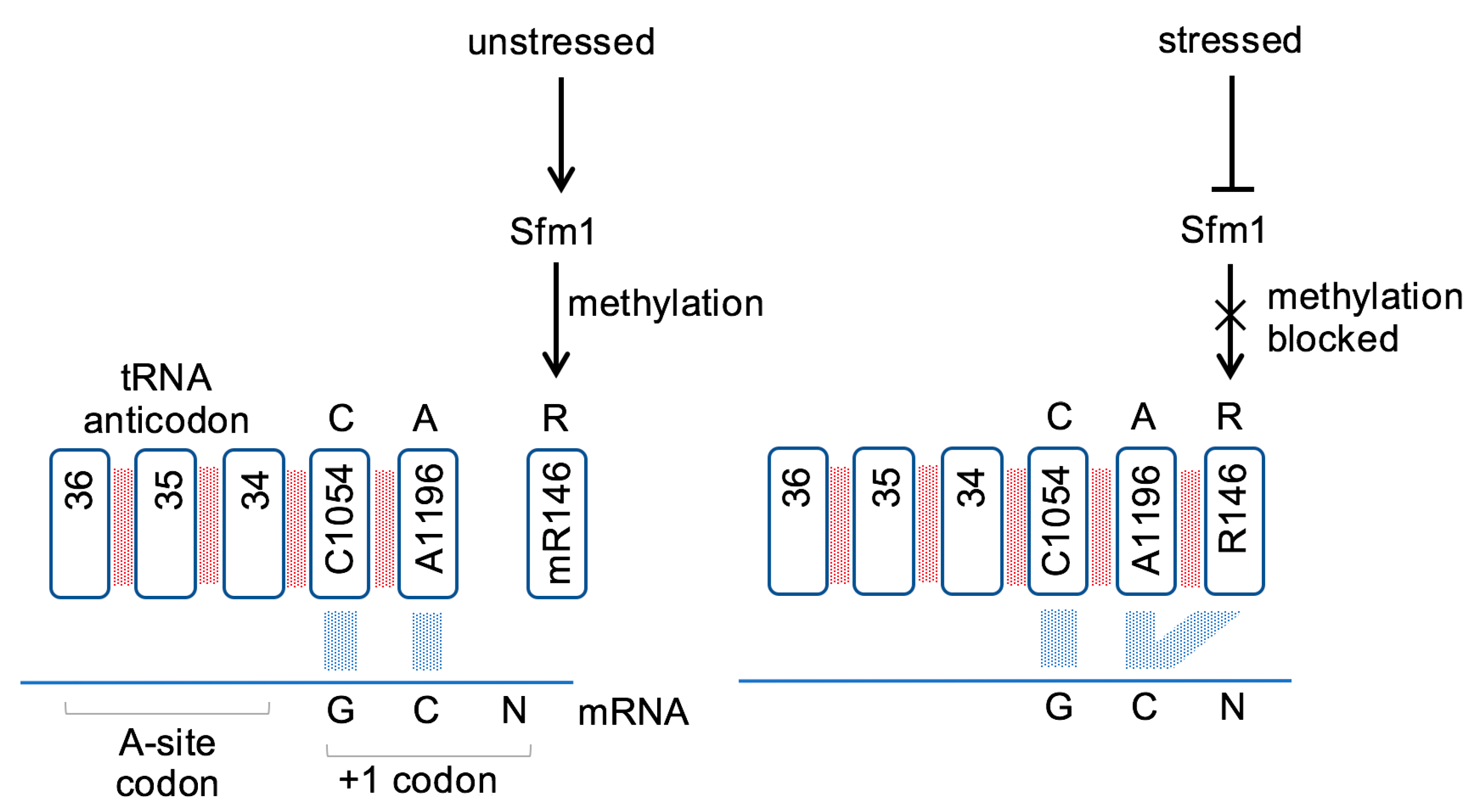
3.1. A Key Role for Arginine Methylation
3.2. Rps3 R146 Methylation Modulates CAR Function
3.3. Conclusions
4. Materials and Methods
4.1. Subsystem Neighborhoods
4.2. Molecular Dynamics Setup
4.3. Trajectory Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CAR | C1054: A1196, R146 interaction surface |
| RMSF | Root mean square fluctuation |
| RMSD | Root mean square deviation |
| RMS2D | Root mean square 2-dimensional deviation |
| SASA | Solvent accessible surface area |
References
- Pavitt, G.D. Regulation of translation initiation factor eIF2B at the hub of the integrated stress response. WIREs RNA 2018, 9, e1491. [Google Scholar] [CrossRef] [PubMed]
- Shenton, D.; Smirnova, J.B.; Selley, J.N.; Carroll, K.; Hubbard, S.J.; Pavitt, G.D.; Ashe, M.P.; Grant, C.M. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 2006, 281, 29011–29021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, M.; Lin, Y.; Yang, C.; McQuary, P.; Campos, A.R.; Blanc, P.A.; Wolf, D.A. Cross talk between eIF2a and eEF2 phosphorylation pathways optimizes translational arrest in response to oxidative stress. iScience 2019, 20, 466–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Back, S.; Gorman, A.W.; Vogel, C.; Silva, G.M. Site-specific K63 ubiquitinomics provides insights into translation regulation under stress. J. Proteome Res. 2019, 18, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.M.; Daniel; Finley, D.; Vogel, C. K63 polyubiquitination is a new modulator of the oxidative stress response. Nat. Struct. Mol. Biol. 2015, 22, 116–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, C.T.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, C.; Begley, T.J.; Dedon, P.C. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014, 588, 4287–4296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Y.M.; Gamper, H.; Yang, W. Post-transcriptional modifications to tRNA—A response to the genetic code degeneracy. RNA 2015, 21, 642–644. [Google Scholar] [CrossRef]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Schosserer, M.; Minois, N.; Angerer, T.B.; Amring, M.; Dellago, H.; Harreither, E.; Calle-Perez, A.; Pircher, A.; Gerstl, M.P.; Pfeifenberger, S.; et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 2015, 6, 6158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scopino, K.; Williams, E.; Elsayed, A.; Barr, W.A.; Krizanc, D.; Thayer, K.M.; Weir, M.P. A ribosome interaction surface sensitive to mRNA GCN periodicity. Biomolecules 2020, 10, 849. [Google Scholar] [CrossRef] [PubMed]
- Culver, G.M. Meanderings of the mRNA through the ribosome. Structure 2001, 9, 751–758. [Google Scholar] [CrossRef] [Green Version]
- Kozak, M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 2005, 361, 13–37. [Google Scholar] [CrossRef] [PubMed]
- Shine, J.; Dalgarno, L. The 3’-terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 1974, 71, 1342–1346. [Google Scholar] [CrossRef] [Green Version]
- Yusupova, G.Z.; Yusupov, M.M.; Cate, J.H.D.; Noller, H.F. The path of messenger RNA through the ribosome. Cell 2001, 106, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Abeyrathne, P.D.; Koh, C.S.; Grant, T.; Grigorieff, N.; Korostelev, A.A. Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome. Elife 2016, 5, e14874. [Google Scholar] [CrossRef]
- Barr, W.A.; Sheth, R.B.; Kwon, J.; Cho, J.; Glickman, J.W.; Hart, F.; Chatterji, O.K.; Scopino, K.; Voelkel-Meiman, K.; Krizanc, D.; et al. GCN sensitive protein translation in yeast. PLoS ONE 2020, 15, e0233197. [Google Scholar] [CrossRef]
- Young, B.D.; Weiss, D.I.; Zurita-Lopez, C.I.; Webb, K.J.; Clarke, S.G.; McBride, A.E. Identification of methylated proteins in the yeast small ribosomal subunit: A role for SPOUT methyltransferases in protein arginine methylation. Biochemistry 2012, 51, 5091–5104. [Google Scholar] [CrossRef] [Green Version]
- Lv, F.; Zhang, T.; Zhou, Z.; Gao, S.; Wong, C.C.; Zhou, J.Q.; Ding, J. Structural basis for Sfm1 functioning as a protein arginine methyltransferase. Cell Discov. 2015, 1, 15037. [Google Scholar] [CrossRef] [Green Version]
- Gerashchenko, M.V.; Gladyshev, V.N. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014, 42, e134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanbonmatsu, K.Y. Energy landscape of the ribosomal decoding center. Biochimie 2006, 88, 1053–1059. [Google Scholar] [CrossRef] [PubMed]
- Sanbonmatsu, K.Y. Computational studies of molecular machines: The ribosome. Curr. Opin. Struct. Biol. 2012, 22, 168–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogle, J.M.; Brodersen, D.E.; Clemons, W.M., Jr.; Tarry, M.J.; Carter, A.P.; Ramakrishnan, V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 2001, 292, 897–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogle, J.M.; Murphy, F.V.; Tarry, M.J.; Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 2002, 111, 721–732. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, C.T.; Mason, P.E.; Anderson, J.L.; Dempsey, C.E. Arginine side chain interactions and the role of arginine as a gating charge carrier in voltage sensitive ion channels. Sci. Rep. 2016, 6, 21759. [Google Scholar] [CrossRef] [Green Version]
- Kumar, K.; Woo, S.M.; Siu, T.; Cortopassi, W.A.; Duarte, F.; Paton, R.S. Cation-pi interactions in protein-ligand binding: Theory and data-mining reveal different roles for lysine and arginine. Chem. Sci. 2018, 9, 2655–2665. [Google Scholar] [CrossRef] [Green Version]
- Vernon, R.M.; Chong, P.A.; Tsang, B.; Kim, T.H.; Bah, A.; Farber, P.; Lin, H.; Forman-Kay, J.D. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife 2018, 7, e31486. [Google Scholar] [CrossRef]
- Amaro, R.E.; Cheng, X.; Ivanov, I.; Xu, D.; McCammon, J.A. Characterizing loop dynamics and ligand recognition in human- and avian-type influenza neuraminidases via generalized born molecular dynamics and end-point free energy calculations. J. Am. Chem. Soc. 2009, 131, 4702–4709. [Google Scholar] [CrossRef] [Green Version]
- Weiser, J.; Shenkin, P.; Still, W. Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO). J. Comput. Chem. 1999, 20, 217–230. [Google Scholar] [CrossRef]
- Shao, J.; Tanner, S.W.; Thompson, N.; Cheatham, T.E. Clustering molecular dynamics trajectories: 1. characterizing the performance of different clustering algorithms. J. Chem. Theory Comput. 2007, 3, 2312–2334. [Google Scholar] [CrossRef] [PubMed]
- Hsin, J.; Arkhipov, A.; Yin, Y.; Stone, J.E.; Schulten, K. Using VMD: An introductory tutorial. Curr. Protoc. Bioinform. 2008, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Molec. Graph. 1986, 14, 33–38. [Google Scholar] [CrossRef]
- Low, J.K.K.; Wilkins, M.R. Protein arginine methylation in Saccharomyces cerevisiae. FEBS J. 2012, 279, 4423–4443. [Google Scholar] [CrossRef] [PubMed]
- Evich, M.; Stroeva, E.; Zheng, Y.G.; Germann, M.W. Effect of methylation on the side-chain pKa value of arginine. Protein Sci. 2016, 25, 479–486. [Google Scholar] [PubMed] [Green Version]
- Szczepinska, T.; Kutner, J.; Kopczynski, M.; Pawłowski, K.; Dziembowski, A.; Kudlicki, A.; Ginalski, K.; Rowicka, M. Probabilistic approach to predicting substrate specificity of methyltransferases. PLoS Comp. Biol. 2014, 10, e1003514. [Google Scholar] [CrossRef] [Green Version]
- Clarke, S.G. The ribosome: A hot spot for the identification of new types of protein methyltransferases. J. Biol. Chem. 2018, 293, 10438–10446. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Zeng, J.; Xie, W. A flexible cofactor-binding loop in the novel arginine methyltransferase Sfm1. FEBS Lett. 2017, 591, 433–441. [Google Scholar]
- Dong, J.; Aitken, C.E.; Thakur, A.; Shin, B.S.; Lorsch, J.R.; Hinnebusch, A.G. Rps3/uS3 promotes mRNA binding at the 40S ribosome entry channel and stabilizes preinitiation complexes at start codons. Proc. Natl. Acad. Sci. USA 2017, 114, E2126–E2135. [Google Scholar] [CrossRef] [Green Version]
- Llacer, J.L.; Hussain, T.; Marler, L.; Aitken, C.E.; Thakur, A.; Lorsch, J.R.; Hinnebusch, A.G.; Ramakrishnan, V. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell 2015, 59, 399–412. [Google Scholar] [CrossRef] [Green Version]
- Tuller, T.; Carmi, A.; Vestsigian, K.; Navon, S.; Dorfan, Y.; Zaborske, J.; Pan, T.; Dahan, O.; Furman, I.; Pilpel, Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 2010, 141, 344–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagunez-Otero, J.; Trifonov, E.N. mRNA periodical infrastructure complementary to the proof-reading site in the ribosome. J. Biomol. Struct. Dyn. 1992, 10, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, L.; Mondragon, M.; Lagunez-Otero, J. Interaction of the 530 ribosomal site with regions of mRNA. Biosystems 1998, 46, 293–298. [Google Scholar] [CrossRef]
- Liang, X.; Liu, Q.; Fournier, M.J. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 2009, 15, 1716–1728. [Google Scholar] [CrossRef] [Green Version]
- Huber, S.M.; Leonardi, A.; Dedon, P.C.; Begley, T.J. The Versatile Roles of the tRNA Epitranscriptome during cellular responses to toxic exposures and environmental stress. Toxics 2019, 7, 17. [Google Scholar] [CrossRef] [Green Version]
- Phizicky, E.M.; Hopper, A.K. tRNA biology charges to the front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [Green Version]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., I.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K.; et al. AMBER 18; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar]
- Wang, J.; Cieplak, P.; Kollman, P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000, 21, 1049–1074. [Google Scholar] [CrossRef]
- Zgarbova, M.; Otyepka, M.; Sponer, J.; Mladek, A.; Banas, P.; Cheatham, T.E., 3rd; Jurecka, P. Refinement of the Cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theory Comput. 2011, 7, 2886–2902. [Google Scholar] [CrossRef]
- Khoury, G.A.; Thompson, J.P.; Smadbeck, J.; Kieslich, C.A.; Floudas, C.A. Forcefield_PTM: Ab initio charge and AMBER forcefield parameters for frequently occurring post-translational modifications. J. Chem. Theory Comput. 2013, 9, 5653–5674. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, W.L.; Jenson, C. Temperature dependence of TIP3P, SPC, and TIP4P water from NPT Monte Carlo simulations: Seeking temperatures of maximum density. J. Comput. Chem. 1998, 19, 1179–1186. [Google Scholar] [CrossRef]
- Rozov, A.; Khusainov, I.; El Omari, K.; Duman, R.; Mykhaylyk, V.; Yusupov, M.; Westhof, E.; Wagner, A.; Yusupova, G. Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction. Nat. Commun. 2019, 10, 2519. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular synamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scopino, K.; Dalgarno, C.; Nachmanoff, C.; Krizanc, D.; Thayer, K.M.; Weir, M.P. Arginine Methylation Regulates Ribosome CAR Function. Int. J. Mol. Sci. 2021, 22, 1335. https://doi.org/10.3390/ijms22031335
Scopino K, Dalgarno C, Nachmanoff C, Krizanc D, Thayer KM, Weir MP. Arginine Methylation Regulates Ribosome CAR Function. International Journal of Molecular Sciences. 2021; 22(3):1335. https://doi.org/10.3390/ijms22031335
Chicago/Turabian StyleScopino, Kristen, Carol Dalgarno, Clara Nachmanoff, Daniel Krizanc, Kelly M. Thayer, and Michael P. Weir. 2021. "Arginine Methylation Regulates Ribosome CAR Function" International Journal of Molecular Sciences 22, no. 3: 1335. https://doi.org/10.3390/ijms22031335
APA StyleScopino, K., Dalgarno, C., Nachmanoff, C., Krizanc, D., Thayer, K. M., & Weir, M. P. (2021). Arginine Methylation Regulates Ribosome CAR Function. International Journal of Molecular Sciences, 22(3), 1335. https://doi.org/10.3390/ijms22031335





