Establishment and Long-Term Expansion of Small Cell Lung Cancer Patient-Derived Tumor Organoids
Abstract
1. Introduction
2. Results
2.1. Establishment of PDTO Culture Systems Optimized for SCLC
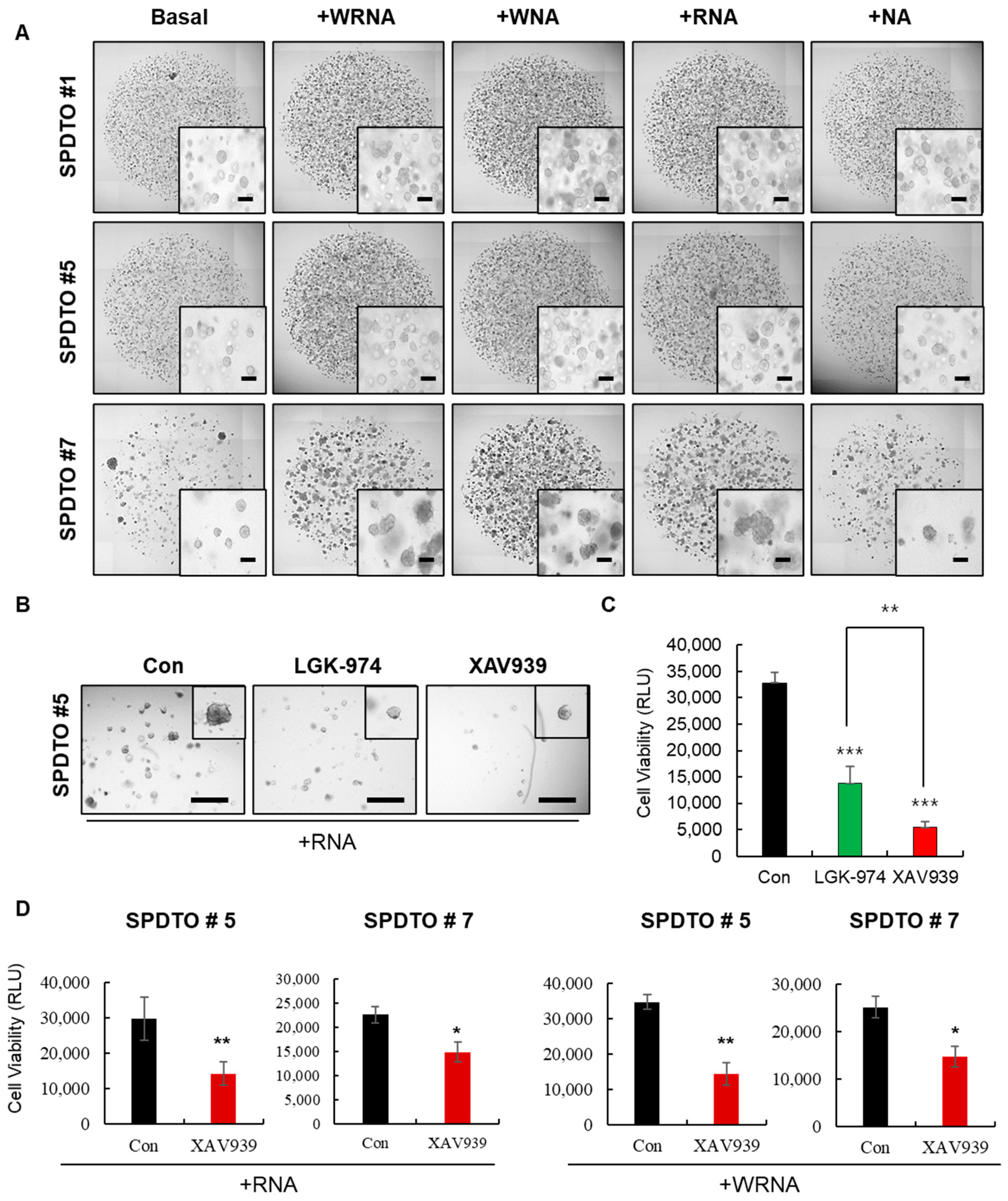
2.2. Optimization of Culture Conditions for Long-Term Viability of SCLC Cells
2.3. Characterization of SCLC Tumor Organoids as a Patient Avatar Model
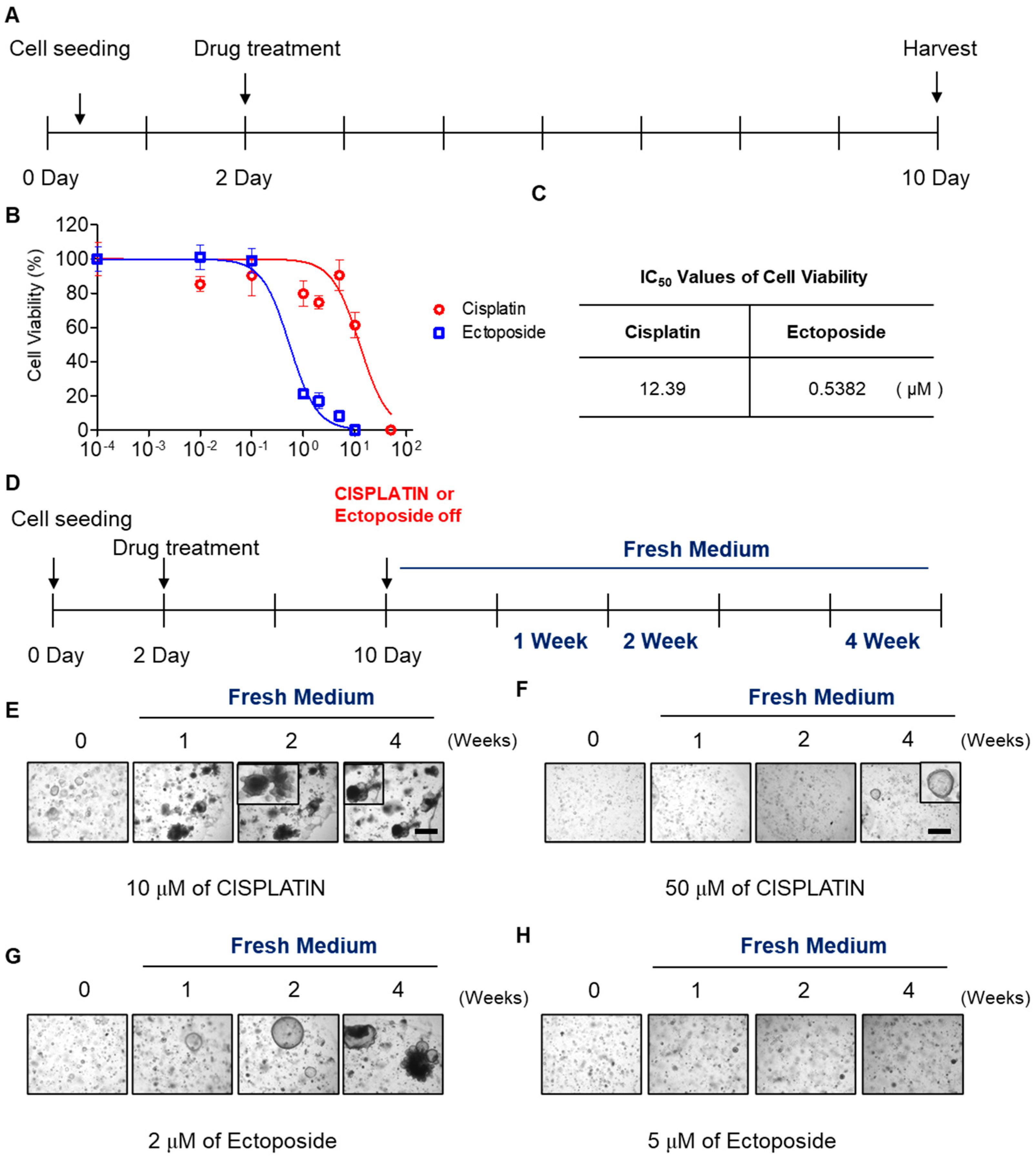
2.4. SCLC Organoid Proof-of-Concept Drug Screen
3. Discussion
4. Materials and Methods
4.1. Tumor Tissue Preparation and Organoid Establishment
4.2. Immunohistochemistry
4.3. Organoid Drug Tests
4.4. DNA Extraction and WES Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008, 83, 584–594. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, Z.; Wang, Q. Emerging therapies for small cell lung cancer. J. Hematol. Oncol. 2019, 12, 47. [Google Scholar] [CrossRef] [PubMed]
- Anbazhagan, R.; Tihan, T.; Bornman, D.M.; Johnston, J.C.; Saltz, J.H.; Weigering, A.; Piantadosi, S.; Gabrielson, E. Classification of small cell lung cancer and pulmonary carcinoid by gene expression profiles. Cancer Res. 1999, 59, 5119–5122. [Google Scholar] [PubMed]
- Demedts, I.K.; Vermaelen, K.Y.; van Meerbeeck, J.P. Treatment of extensive-stage small cell lung carcinoma: Current status and future prospects. Eur. Respir. J. 2010, 35, 202–215. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Johnson, B.E.; Mazor, T.; Hong, C.; Barnes, M.; Aihara, K.; McLean, C.Y.; Fouse, S.D.; Yamamoto, S.; Ueda, H.; Tatsuno, K.; et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014, 343, 189–193. [Google Scholar] [CrossRef]
- Greaves, M. Evolutionary determinants of cancer. Cancer Discov. 2015, 5, 806–820. [Google Scholar] [CrossRef]
- Xue, Y.; Martelotto, L.; Baslan, T.; Vides, A.; Solomon, M.; Mai, T.T.; Chaudhary, N.; Riely, G.J.; Li, B.T.; Scott, K.; et al. An approach to suppress the evolution of resistance in BRAF(V600E)-mutant cancer. Nat. Med. 2017, 23, 929–937. [Google Scholar] [CrossRef]
- Gillet, J.P.; Varma, S.; Gottesman, M.M. The clinical relevance of cancer cell lines. J. Natl. Cancer Inst. 2013, 105, 452–458. [Google Scholar] [CrossRef]
- Sachs, N.; Clevers, H. Organoid cultures for the analysis of cancer phenotypes. Curr. Opin. Genet. Dev. 2014, 24, 68–73. [Google Scholar]
- Siolas, D.; Hannon, G.J. Patient-derived tumor xenografts: Transforming clinical samples into mouse models. Cancer Res. 2013, 73, 5315–5319. [Google Scholar] [CrossRef] [PubMed]
- Tentler, J.J.; Tan, A.C.; Weekes, C.D.; Jimeno, A.; Leong, S.; Pitts, T.M.; Arcaroli, J.J.; Messersmith, W.A.; Eckhardt, S.G. Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 2012, 9, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.J.; Chun, S.M.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019, 10, 3991. [Google Scholar] [CrossRef] [PubMed]
- Boonekamp, K.E.; Kretzschmar, K.; Wiener, D.J.; Asra, P.; Derakhshan, S.; Puschhof, J.; Lopez-Iglesias, C.; Peters, P.J.; Basak, O.; Clevers, H. Long-term expansion and differentiation of adult murine epidermal stem cells in 3D organoid cultures. Proc. Natl. Acad. Sci. USA 2019, 116, 14630–14638. [Google Scholar] [CrossRef] [PubMed]
- Broutier, L.; Mastrogiovanni, G.; Verstegen, M.M.; Francies, H.E.; Gavarro, L.M.; Bradshaw, C.R.; Allen, G.E.; Arnes-Benito, R.; Sidorova, O.; Gaspersz, M.P.; et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017, 23, 1424–1435. [Google Scholar] [CrossRef]
- Driehuis, E.; van Hoeck, A.; Moore, K.; Kolders, S.; Francies, H.E.; Gulersonmez, M.C.; Stigter, E.C.A.; Burgering, B.; Geurts, V.; Gracanin, A.; et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl. Acad. Sci. USA 2019, 116, 26580–26590. [Google Scholar] [CrossRef]
- Jung, J.; Seol, H.S.; Chang, S. The Generation and Application of Patient-Derived Xenograft Model for Cancer Research. Cancer Res. Treat. 2018, 50, 1–10. [Google Scholar] [CrossRef]
- Dong, X.; Guan, J.; English, J.C.; Flint, J.; Yee, J.; Evans, K.; Murray, N.; Macaulay, C.; Ng, R.T.; Gout, P.W.; et al. Patient-derived first generation xenografts of non-small cell lung cancers: Promising tools for predicting drug responses for personalized chemotherapy. Clin. Cancer Res. 2010, 16, 1442–1451. [Google Scholar] [CrossRef]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef]
- Seino, T.; Kawasaki, S.; Shimokawa, M.; Tamagawa, H.; Toshimitsu, K.; Fujii, M.; Ohta, Y.; Matano, M.; Nanki, K.; Kawasaki, K.; et al. Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell 2018, 22, 454–467.e6. [Google Scholar] [CrossRef]
- Drost, J.; Karthaus, W.R.; Gao, D.; Driehuis, E.; Sawyers, C.L.; Chen, Y.; Clevers, H. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 2016, 11, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, Y.; Cen, J.; Ma, X.; Cui, L.; Qiu, Z.; Zhang, Z.; Li, H.; Yang, R.Z.; Wang, C.; et al. Modelling liver cancer initiation with organoids derived from directly reprogrammed human hepatocytes. Nat. Cell Biol. 2019, 21, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Rosenbluth, J.M.; Schackmann, R.C.J.; Gray, G.K.; Selfors, L.M.; Li, C.M.; Boedicker, M.; Kuiken, H.J.; Richardson, A.; Brock, J.; Garber, J.; et al. Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages. Nat. Commun. 2020, 11, 1711. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.P.; Lapi, E.; Martinez de Villarreal, J.; Alvaro-Espinosa, L.; Fernandez-Barral, A.; Barbachano, A.; Dominguez, O.; Laughney, A.M.; Megias, D.; Munoz, A.; et al. Urothelial organoids originating from Cd49f(high) mouse stem cells display Notch-dependent differentiation capacity. Nat. Commun. 2019, 10, 4407. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Qian, Y.; Li, W.; Liu, L.; Yu, L.; Liu, X.; Wu, G.; Wang, Y.; Luo, W.; Fang, F.; et al. Human Lung Adenocarcinoma-Derived Organoid Models for Drug Screening. iScience 2020, 23, 101411. [Google Scholar] [CrossRef]
- Jung, D.J.; Shin, T.H.; Kim, M.; Sung, C.O.; Jang, S.J.; Jeong, G.S. A one-stop microfluidic-based lung cancer organoid culture platform for testing drug sensitivity. Lab Chip 2019, 19, 2854–2865. [Google Scholar] [CrossRef]


| CASE # | ORGANOID ID | SEX | AGE | LUNG CANCER TYPE | EP RESPONSE | THAWING TEST |
|---|---|---|---|---|---|---|
| 1 | SPDTO #1 | Male | 67 | SCLC | ⃝ | ⃝ |
| 2 | SPDTO #2 | Male | 66 | SCLC | ⃝ | ⃝ |
| 3 | SPDTO #3 | Male | 67 | SCLC | ⃝ | ⃝ |
| 4 | SPDTO #4 | Male | 71 | SCLC | ⃝ | X |
| 5 | SPDTO #5 | Male | 53 | SCLC | ⃝ | ⃝ |
| 6 | SPDTO #6 | Male | 65 | SCLC | ⃝ | ⃝ |
| 7 | SPDTO #7 | Male | 69 | SCLC | X | ⃝ |
| 8 | SPDTO #8 | Male | 64 | SCLC | ⃝ | X |
| 9 | SPDTO #9 | Male | 73 | SCLC | ⃝ | ⃝ |
| 10 | SPDTO #10 | Male | 35 | SCLC | X | ⃝ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, S.Y.; Cho, Y.-H.; Kim, D.-S.; Ji, W.; Choi, C.-M.; Lee, J.C.; Rho, J.K.; Jeong, G.S. Establishment and Long-Term Expansion of Small Cell Lung Cancer Patient-Derived Tumor Organoids. Int. J. Mol. Sci. 2021, 22, 1349. https://doi.org/10.3390/ijms22031349
Choi SY, Cho Y-H, Kim D-S, Ji W, Choi C-M, Lee JC, Rho JK, Jeong GS. Establishment and Long-Term Expansion of Small Cell Lung Cancer Patient-Derived Tumor Organoids. International Journal of Molecular Sciences. 2021; 22(3):1349. https://doi.org/10.3390/ijms22031349
Chicago/Turabian StyleChoi, Seon Young, Yong-Hee Cho, Da-Som Kim, Wonjun Ji, Chang-Min Choi, Jae Cheol Lee, Jin Kyung Rho, and Gi Seok Jeong. 2021. "Establishment and Long-Term Expansion of Small Cell Lung Cancer Patient-Derived Tumor Organoids" International Journal of Molecular Sciences 22, no. 3: 1349. https://doi.org/10.3390/ijms22031349
APA StyleChoi, S. Y., Cho, Y.-H., Kim, D.-S., Ji, W., Choi, C.-M., Lee, J. C., Rho, J. K., & Jeong, G. S. (2021). Establishment and Long-Term Expansion of Small Cell Lung Cancer Patient-Derived Tumor Organoids. International Journal of Molecular Sciences, 22(3), 1349. https://doi.org/10.3390/ijms22031349







