Equilibrative Nucleoside Transporters Mediate the Import of Nicotinamide Riboside and Nicotinic Acid Riboside into Human Cells
Abstract
1. Introduction
2. Results
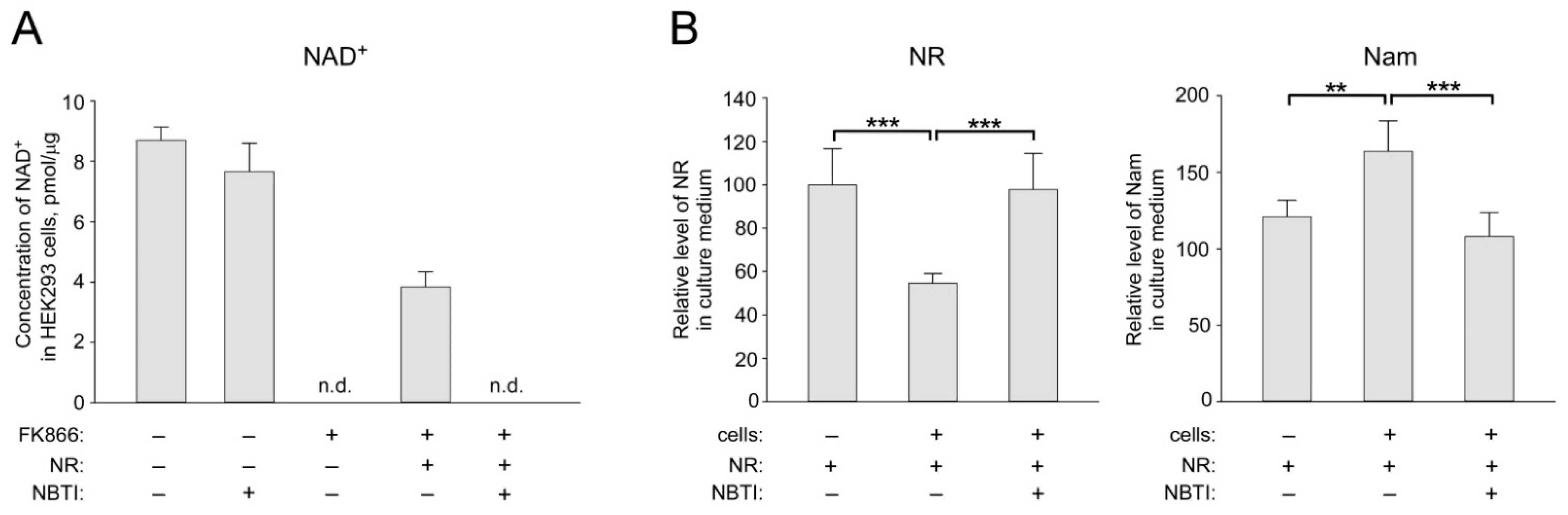
2.1. ENT Inhibition Suppresses NR Utilization by HEK293 Cells for NAD+ Biosynthesis
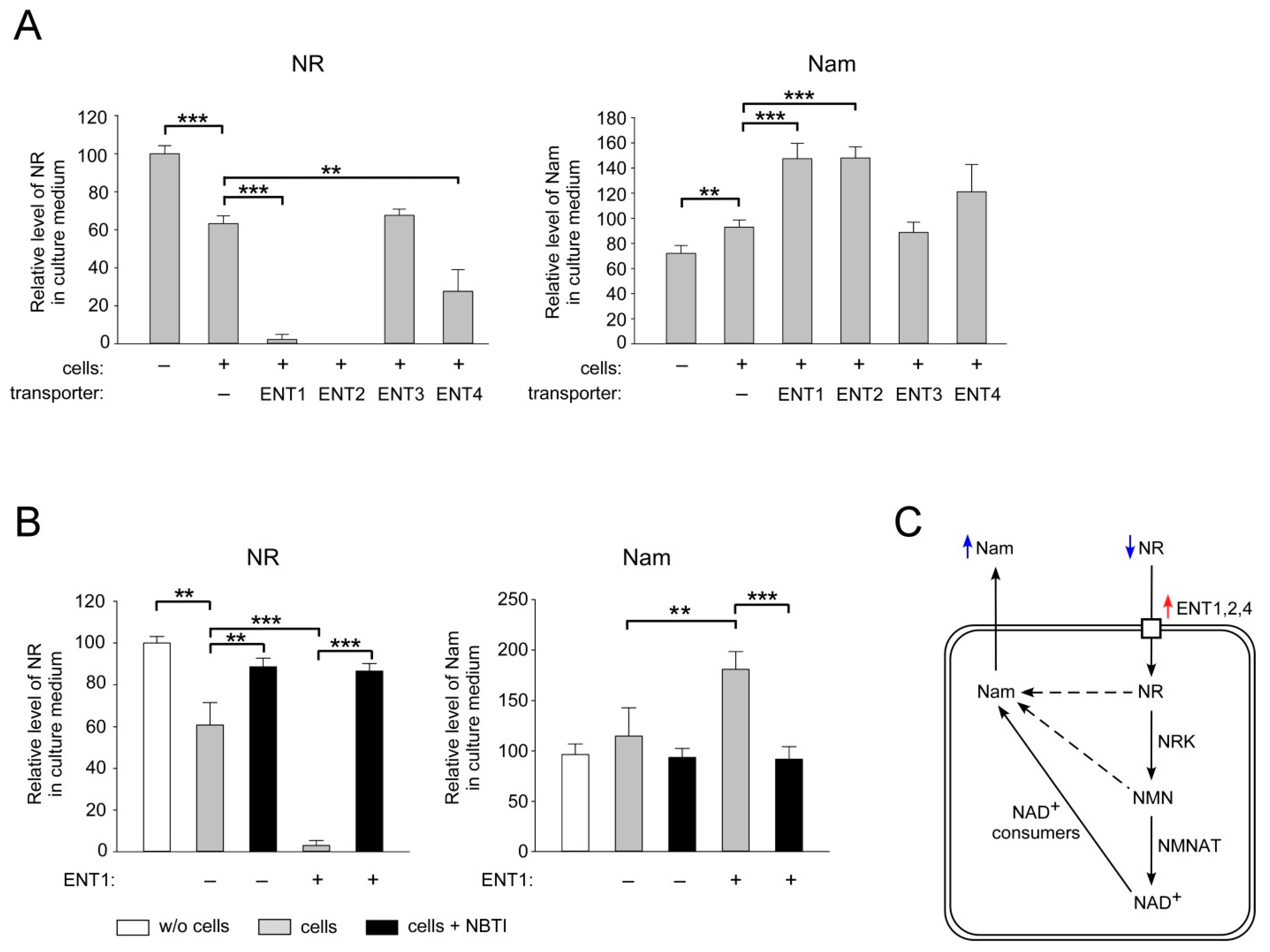
2.2. ENT1, ENT2 and ENT4 Mediate NR Uptake into Human Cells
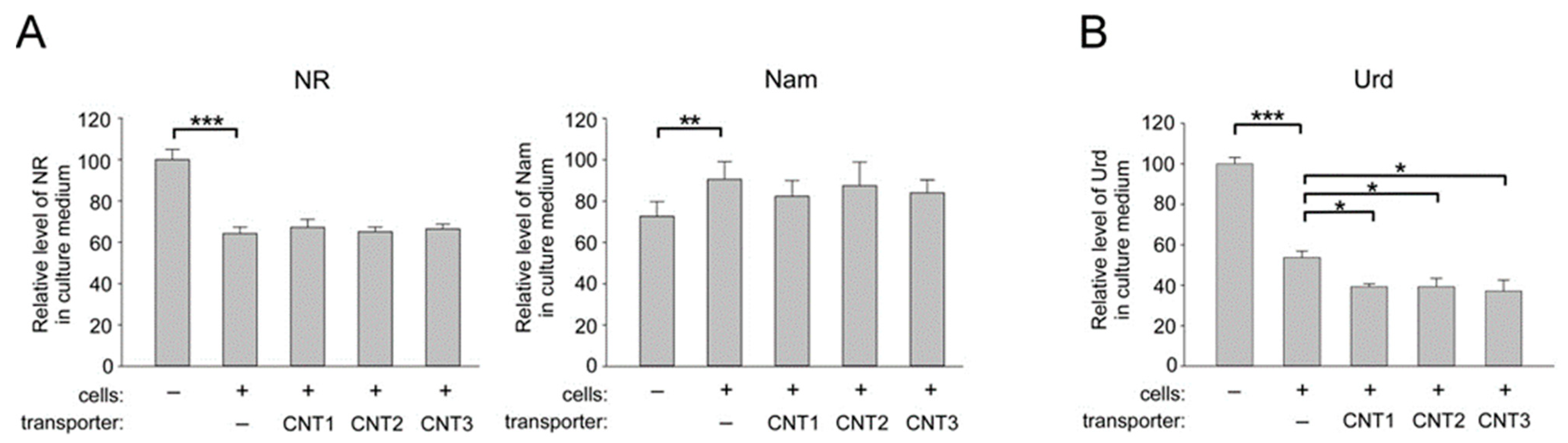
2.3. Overexpression of CNT1, CNT2, and CNT3 in Human Cells does not Affect the Import of NR into HEK293 Cells
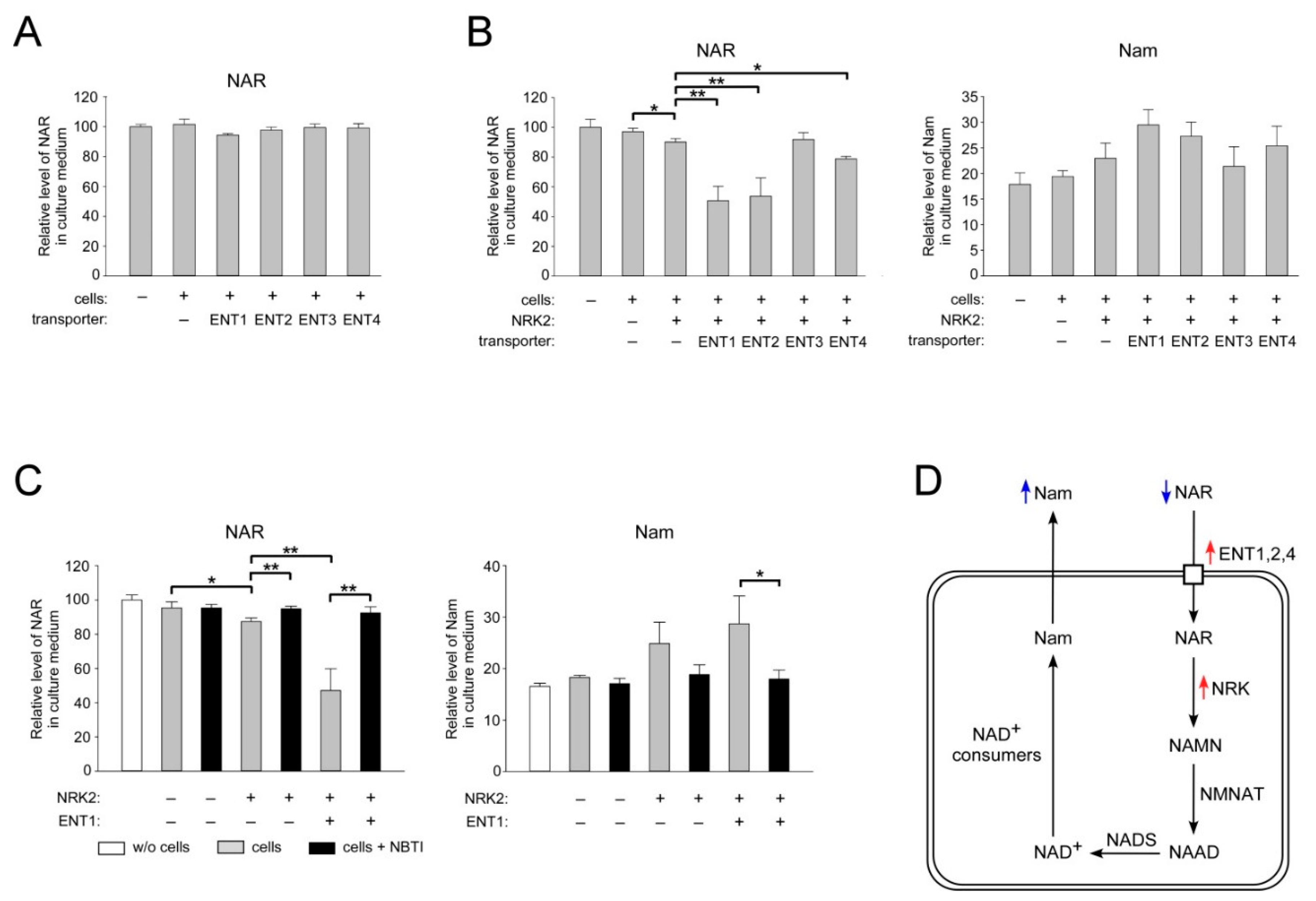
2.4. ENT1, ENT2, and ENT4 Mediate NAR Uptake into Human Cells
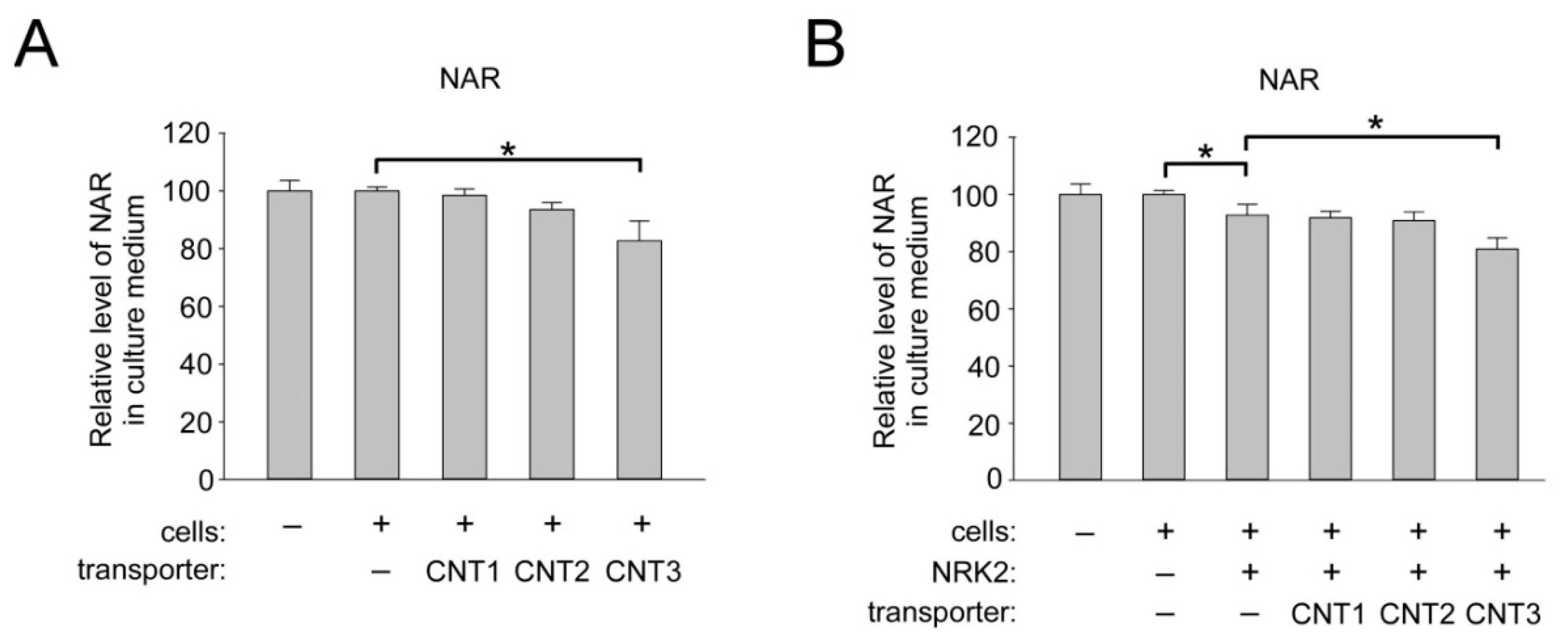
2.5. Overexpression of CNT3 in HEK293 Cells Moderately Stimulates NAR Uptake
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Generation of Eukaryotic Expression Vectors
4.4. Western Blotting
4.5. Metabolite Measurements
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, L.; Su, X.; Quinn, W.J., 3rd; Hui, S.; Krukenberg, K.; Frederick, D.W.; Redpath, P.; Zhan, L.; Chellappa, K.; White, E.; et al. Quantitative Analysis of NAD Synthesis-Breakdown Fluxes. Cell Metab. 2018, 27, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Kulikova, V.A.; Gromyko, D.V.; Nikiforov, A.A. The Regulatory Role of NAD in Human and Animal Cells. Biochemistry 2018, 83, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Stromland, O.; Niere, M.; Nikiforov, A.A.; VanLinden, M.R.; Heiland, I.; Ziegler, M. Keeping the balance in NAD metabolism. Biochem. Soc. Trans. 2019, 47, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sauve, A.A. NAD(+) metabolism: Bioenergetics, signaling and manipulation for therapy. Biochim. Et Biophys. Acta 2016, 1864, 1787–1800. [Google Scholar] [CrossRef]
- Bieganowski, P.; Brenner, C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 2004, 117, 495–502. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, N.; Zhang, G.; Sauve, A.A. NRH salvage and conversion to NAD(+) requires NRH kinase activity by adenosine kinase. Nat. Metab. 2020, 2, 364–379. [Google Scholar] [CrossRef]
- Giroud-Gerbetant, J.; Joffraud, M.; Giner, M.P.; Cercillieux, A.; Bartova, S.; Makarov, M.V.; Zapata-Perez, R.; Sanchez-Garcia, J.L.; Houtkooper, R.H.; Migaud, M.E.; et al. A reduced form of nicotinamide riboside defines a new path for NAD(+) biosynthesis and acts as an orally bioavailable NAD(+) precursor. Mol. Metab. 2019, 30, 192–202. [Google Scholar] [CrossRef]
- Trammell, S.A.; Weidemann, B.J.; Chadda, A.; Yorek, M.S.; Holmes, A.; Coppey, L.J.; Obrosov, A.; Kardon, R.H.; Yorek, M.A.; Brenner, C. Nicotinamide Riboside Opposes Type 2 Diabetes and Neuropathy in Mice. Sci. Rep. 2016, 6, 26933. [Google Scholar] [CrossRef]
- Yoshino, J.; Mills, K.F.; Yoon, M.J.; Imai, S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011, 14, 528–536. [Google Scholar] [CrossRef]
- Fang, E.F.; Kassahun, H.; Croteau, D.L.; Scheibye-Knudsen, M.; Marosi, K.; Lu, H.; Shamanna, R.A.; Kalyanasundaram, S.; Bollineni, R.C.; Wilson, M.A.; et al. NAD(+) Replenishment Improves Lifespan and Healthspan in Ataxia Telangiectasia Models via Mitophagy and DNA Repair. Cell Metab. 2016, 24, 566–581. [Google Scholar] [CrossRef]
- Wang, J.; Zhai, Q.; Chen, Y.; Lin, E.; Gu, W.; McBurney, M.W.; He, Z. A local mechanism mediates NAD-dependent protection of axon degeneration. J. Cell Biol. 2005, 170, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.D.; Maqsood, S.; Huang, J.Y.; Pan, Y.; Harkcom, W.; Li, W.; Sauve, A.; Verdin, E.; Jaffrey, S.R. Activation of SIRT3 by the NAD(+) precursor nicotinamide riboside protects from noise-induced hearing loss. Cell Metab. 2014, 20, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.P.; Oka, S.; Shao, D.; Hariharan, N.; Sadoshima, J. Nicotinamide phosphoribosyltransferase regulates cell survival through NAD+ synthesis in cardiac myocytes. Circ. Res. 2009, 105, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Diguet, N.; Trammell, S.A.J.; Tannous, C.; Deloux, R.; Piquereau, J.; Mougenot, N.; Gouge, A.; Gressette, M.; Manoury, B.; Blanc, J.; et al. Nicotinamide Riboside Preserves Cardiac Function in a Mouse Model of Dilated Cardiomyopathy. Circulation 2018, 137, 2256–2273. [Google Scholar] [CrossRef]
- Ryu, D.; Zhang, H.; Ropelle, E.R.; Sorrentino, V.; Mazala, D.A.; Mouchiroud, L.; Marshall, P.L.; Campbell, M.D.; Ali, A.S.; Knowels, G.M.; et al. NAD+ repletion improves muscle function in muscular dystrophy and counters global PARylation. Sci. Transl. Med. 2016, 8, 361ra139. [Google Scholar] [CrossRef]
- Poyan Mehr, A.; Tran, M.T.; Ralto, K.M.; Leaf, D.E.; Washco, V.; Messmer, J.; Lerner, A.; Kher, A.; Kim, S.H.; Khoury, C.C.; et al. De novo NAD(+) biosynthetic impairment in acute kidney injury in humans. Nat. Med. 2018, 24, 1351–1359. [Google Scholar] [CrossRef]
- Braidy, N.; Guillemin, G.J.; Mansour, H.; Chan-Ling, T.; Poljak, A.; Grant, R. Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in wistar rats. PLoS ONE 2011, 6, e19194. [Google Scholar] [CrossRef]
- Gomes, A.P.; Price, N.L.; Ling, A.J.; Moslehi, J.J.; Montgomery, M.K.; Rajman, L.; White, J.P.; Teodoro, J.S.; Wrann, C.D.; Hubbard, B.P.; et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell 2013, 155, 1624–1638. [Google Scholar] [CrossRef]
- Mouchiroud, L.; Houtkooper, R.H.; Moullan, N.; Katsyuba, E.; Ryu, D.; Canto, C.; Mottis, A.; Jo, Y.S.; Viswanathan, M.; Schoonjans, K.; et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell 2013, 154, 430–441. [Google Scholar] [CrossRef]
- Massudi, H.; Grant, R.; Braidy, N.; Guest, J.; Farnsworth, B.; Guillemin, G.J. Age-associated changes in oxidative stress and NAD+ metabolism in human tissue. PLoS ONE 2012, 7, e42357. [Google Scholar] [CrossRef]
- Zhu, X.H.; Lu, M.; Lee, B.Y.; Ugurbil, K.; Chen, W. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc. Natl. Acad. Sci. USA 2015, 112, 2876–2881. [Google Scholar] [CrossRef] [PubMed]
- Rajman, L.; Chwalek, K.; Sinclair, D.A. Therapeutic Potential of NAD-Boosting Molecules: The In Vivo Evidence. Cell Metab. 2018, 27, 529–547. [Google Scholar] [CrossRef] [PubMed]
- Katsyuba, E.; Romani, M.; Hofer, D.; Auwerx, J. NAD(+) homeostasis in health and disease. Nat. Metab. 2020, 2, 9–31. [Google Scholar] [CrossRef] [PubMed]
- Purhonen, J.; Rajendran, J.; Tegelberg, S.; Smolander, O.P.; Pirinen, E.; Kallijarvi, J.; Fellman, V. NAD(+) repletion produces no therapeutic effect in mice with respiratory chain complex III deficiency and chronic energy deprivation. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018, 32, 5913–5926. [Google Scholar] [CrossRef] [PubMed]
- Dall, M.; Trammell, S.A.J.; Asping, M.; Hassing, A.S.; Agerholm, M.; Vienberg, S.G.; Gillum, M.P.; Larsen, S.; Treebak, J.T. Mitochondrial function in liver cells is resistant to perturbations in NAD(+) salvage capacity. J. Biol. Chem. 2019, 294, 13304–13326. [Google Scholar] [CrossRef] [PubMed]
- Frederick, D.W.; McDougal, A.V.; Semenas, M.; Vappiani, J.; Nuzzo, A.; Ulrich, J.C.; Becherer, J.D.; Preugschat, F.; Stewart, E.L.; Sevin, D.C.; et al. Complementary NAD(+) replacement strategies fail to functionally protect dystrophin-deficient muscle. Skelet. Muscle 2020, 10, 30. [Google Scholar] [CrossRef]
- Gong, B.; Pan, Y.; Vempati, P.; Zhao, W.; Knable, L.; Ho, L.; Wang, J.; Sastre, M.; Ono, K.; Sauve, A.A.; et al. Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-gamma coactivator 1alpha regulated beta-secretase 1 degradation and mitochondrial gene expression in Alzheimer’s mouse models. Neurobiol. Aging 2013, 34, 1581–1588. [Google Scholar] [CrossRef]
- Canto, C.; Houtkooper, R.H.; Pirinen, E.; Youn, D.Y.; Oosterveer, M.H.; Cen, Y.; Fernandez-Marcos, P.J.; Yamamoto, H.; Andreux, P.A.; Cettour-Rose, P.; et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012, 15, 838–847. [Google Scholar] [CrossRef]
- Zhang, H.; Ryu, D.; Wu, Y.; Gariani, K.; Wang, X.; Luan, P.; D’Amico, D.; Ropelle, E.R.; Lutolf, M.P.; Aebersold, R.; et al. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 2016, 352, 1436–1443. [Google Scholar] [CrossRef]
- Martens, C.R.; Denman, B.A.; Mazzo, M.R.; Armstrong, M.L.; Reisdorph, N.; McQueen, M.B.; Chonchol, M.; Seals, D.R. Chronic nicotinamide riboside supplementation is well-tolerated and elevates NAD(+) in healthy middle-aged and older adults. Nat. Commun. 2018, 9, 1286. [Google Scholar] [CrossRef]
- Trammell, S.A.; Schmidt, M.S.; Weidemann, B.J.; Redpath, P.; Jaksch, F.; Dellinger, R.W.; Li, Z.; Abel, E.D.; Migaud, M.E.; Brenner, C. Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat. Commun. 2016, 7, 12948. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Chan, N.Y.; Sauve, A.A. Syntheses of nicotinamide riboside and derivatives: Effective agents for increasing nicotinamide adenine dinucleotide concentrations in mammalian cells. J. Med. Chem. 2007, 50, 6458–6461. [Google Scholar] [CrossRef] [PubMed]
- Kulikova, V.; Shabalin, K.; Nerinovski, K.; Dolle, C.; Niere, M.; Yakimov, A.; Redpath, P.; Khodorkovskiy, M.; Migaud, M.E.; Ziegler, M.; et al. Generation, Release, and Uptake of the NAD Precursor Nicotinic Acid Riboside by Human Cells. J. Biol. Chem. 2015, 290, 27124–27137. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.W.; Smith, C.B.; Schmidt, M.S.; Cambronne, X.A.; Cohen, M.S.; Migaud, M.E.; Brenner, C.; Goodman, R.H. Pharmacological bypass of NAD(+) salvage pathway protects neurons from chemotherapy-induced degeneration. Proc. Natl. Acad. Sci. USA 2018, 115, 10654–10659. [Google Scholar] [CrossRef]
- Young, J.D.; Yao, S.Y.; Baldwin, J.M.; Cass, C.E.; Baldwin, S.A. The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Mol. Asp. Med. 2013, 34, 529–547. [Google Scholar] [CrossRef]
- Young, J.D. The SLC28 (CNT) and SLC29 (ENT) nucleoside transporter families: A 30-year collaborative odyssey. Biochem. Soc. Trans. 2016, 44, 869–876. [Google Scholar] [CrossRef]
- Nikiforov, A.; Dolle, C.; Niere, M.; Ziegler, M. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: From entry of extracellular precursors to mitochondrial NAD generation. J. Biol. Chem. 2011, 286, 21767–21778. [Google Scholar] [CrossRef]
- Hasmann, M.; Schemainda, I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003, 63, 7436–7442. [Google Scholar]
- Griffith, D.A.; Jarvis, S.M. Nucleoside and nucleobase transport systems of mammalian cells. Biochim. Et Biophys. Acta 1996, 1286, 153–181. [Google Scholar] [CrossRef]
- Kulikova, V.; Shabalin, K.; Nerinovski, K.; Yakimov, A.; Svetlova, M.; Solovjeva, L.; Kropotov, A.; Khodorkovskiy, M.; Migaud, M.E.; Ziegler, M.; et al. Degradation of Extracellular NAD(+) Intermediates in Cultures of Human HEK293 Cells. Metabolites 2019, 9, 293. [Google Scholar] [CrossRef]
- Govindarajan, R.; Leung, G.P.; Zhou, M.; Tse, C.M.; Wang, J.; Unadkat, J.D. Facilitated mitochondrial import of antiviral and anticancer nucleoside drugs by human equilibrative nucleoside transporter-3. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 296, G910–G922. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, S.A.; Yao, S.Y.; Hyde, R.J.; Ng, A.M.; Foppolo, S.; Barnes, K.; Ritzel, M.W.; Cass, C.E.; Young, J.D. Functional characterization of novel human and mouse equilibrative nucleoside transporters (hENT3 and mENT3) located in intracellular membranes. J. Biol. Chem. 2005, 280, 15880–15887. [Google Scholar] [CrossRef] [PubMed]
- Burnstock, G. Purinergic Signalling: Therapeutic Developments. Front. Pharmacol. 2017, 8, 661. [Google Scholar] [CrossRef] [PubMed]
- Latini, S.; Pedata, F. Adenosine in the central nervous system: Release mechanisms and extracellular concentrations. J. Neurochem. 2001, 79, 463–484. [Google Scholar] [CrossRef]
- Makarov, M.V.; Harris, N.W.; Rodrigues, M.; Migaud, M.E. Scalable syntheses of traceable ribosylated NAD(+) precursors. Org. Biomol. Chem. 2019, 17, 8716–8720. [Google Scholar] [CrossRef]
- Shabalin, K.; Nerinovski, K.; Yakimov, A.; Kulikova, V.; Svetlova, M.; Solovjeva, L.; Khodorkovskiy, M.; Gambaryan, S.; Cunningham, R.; Migaud, M.E.; et al. NAD Metabolome Analysis in Human Cells Using (1)H NMR Spectroscopy. Int. J. Mol. Sci. 2018, 19, 3906. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kropotov, A.; Kulikova, V.; Nerinovski, K.; Yakimov, A.; Svetlova, M.; Solovjeva, L.; Sudnitsyna, J.; Migaud, M.E.; Khodorkovskiy, M.; Ziegler, M.; et al. Equilibrative Nucleoside Transporters Mediate the Import of Nicotinamide Riboside and Nicotinic Acid Riboside into Human Cells. Int. J. Mol. Sci. 2021, 22, 1391. https://doi.org/10.3390/ijms22031391
Kropotov A, Kulikova V, Nerinovski K, Yakimov A, Svetlova M, Solovjeva L, Sudnitsyna J, Migaud ME, Khodorkovskiy M, Ziegler M, et al. Equilibrative Nucleoside Transporters Mediate the Import of Nicotinamide Riboside and Nicotinic Acid Riboside into Human Cells. International Journal of Molecular Sciences. 2021; 22(3):1391. https://doi.org/10.3390/ijms22031391
Chicago/Turabian StyleKropotov, Andrey, Veronika Kulikova, Kirill Nerinovski, Alexander Yakimov, Maria Svetlova, Ljudmila Solovjeva, Julia Sudnitsyna, Marie E. Migaud, Mikhail Khodorkovskiy, Mathias Ziegler, and et al. 2021. "Equilibrative Nucleoside Transporters Mediate the Import of Nicotinamide Riboside and Nicotinic Acid Riboside into Human Cells" International Journal of Molecular Sciences 22, no. 3: 1391. https://doi.org/10.3390/ijms22031391
APA StyleKropotov, A., Kulikova, V., Nerinovski, K., Yakimov, A., Svetlova, M., Solovjeva, L., Sudnitsyna, J., Migaud, M. E., Khodorkovskiy, M., Ziegler, M., & Nikiforov, A. (2021). Equilibrative Nucleoside Transporters Mediate the Import of Nicotinamide Riboside and Nicotinic Acid Riboside into Human Cells. International Journal of Molecular Sciences, 22(3), 1391. https://doi.org/10.3390/ijms22031391







