Tumor-Associated Microbiome: Where Do We Stand?
Abstract
1. Introduction: Microbiology Meets Oncology
2. Tumor-Associated Microbiome
2.1. Cancers of the Upper Aerodigestive Tract: Head and Neck and Esophageal Tumors
2.2. Hepatocarcinoma, Pancreas, and Biliary Tract Cancers
2.3. Colorectal Cancer
2.4. Genitourinary Cancers
2.5. Other Cancers
3. Microbiome and Antitumor Immunity
3.1. Interplay between the Microbiome, the Immune System, and Response to Anticancer Therapies
3.2. Modulation of Gut Microbiome to Boost Antitumor Responses
4. Microbiome in Oncology: Are We Ready for Prime Time?
4.1. Benchmarks in Standardization of Collection and Preservation Methods
4.2. Microbiome Analysis
4.2.1. Sequencing Techniques
4.2.2. Bioinformatic Analysis
4.2.3. Statistics for Microbiome Analysis
4.2.4. Spatial In Situ Resolution
4.2.5. Pre-Clinical Tools to Study Microbiome in Cancer
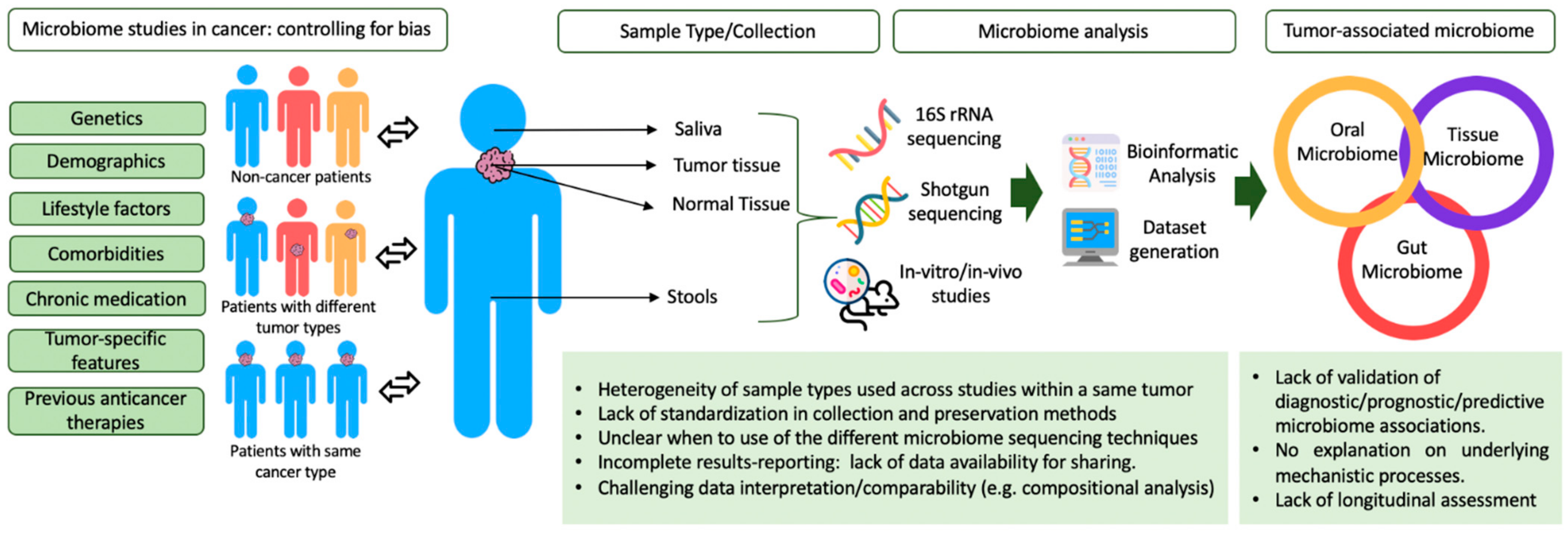
4.3. Challenges in Microbiome Studies in Cancer: Controlling for Bias
5. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Belkaid, Y.; Naik, S. Compartmentalized and systemic control of tissue immunity by commensals. Nat. Immunol. 2013, 14, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Trinchieri, G. Microbiota: A key orchestrator of cancer therapy. Nat. Rev. Cancer 2017, 17, 271–285. [Google Scholar] [CrossRef] [PubMed]
- McQuade, J.L.; Daniel, C.R.; Helmink, B.A.; Wargo, J.A. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 2019, 20, e77–e91. [Google Scholar] [CrossRef]
- Routy, B.; Gopalakrishnan, V.; Daillère, R.; Zitvogel, L.; Wargo, J.A.; Kroemer, G. The gut microbiota influences anticancer immunosurveillance and general health. Nat. Rev. Clin. Oncol. 2018, 15, 382–396. [Google Scholar] [CrossRef]
- Zur Hausen, H. The search for infectious causes of human cancers: Where and why (Nobel lecture). Angewandte Chemie 2009, 48, 5798–5808. [Google Scholar] [CrossRef]
- Peek, R.M.; Blaser, M.J. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2002, 2, 28–37. [Google Scholar] [CrossRef]
- Moore, P.S.; Chang, Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer 2010, 10, 878–889. [Google Scholar] [CrossRef]
- Bonnet, M.; Buc, E.; Sauvanet, P.; Darcha, C.; Dubois, D.; Pereira, B.; Dechelotte, P.; Bonnet, R.; Pezet, D.; Darfeuille-Michaud, A. Colonization of the human gut by E. coli and colorectal cancer risk. Clin. Cancer Res. 2014, 20, 859–867. [Google Scholar] [CrossRef]
- Scott, A.J.; Alexander, J.L.; Merrifield, C.A.; Cunningham, D.; Jobin, C.; Brown, R.; Alverdy, J.; O’Keefe, S.J.; Gaskins, H.R.; Teare, J.; et al. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut 2019, 68, 1624–1632. [Google Scholar] [CrossRef]
- Vonaesch, P.; Anderson, M.; Sansonetti, P.J. Pathogens, microbiome and the host: Emergence of the ecological Koch’s postulates. FEMS Microbiol. Rev. 2018, 42, 273–292. [Google Scholar] [CrossRef]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, K.; Izumi, D.; Kandimalla, R.; Sonohara, F.; Baba, Y.; Yoshida, N.; Kodera, Y.; Baba, H.; Goel, A. Intratumoral Fusobacterium nucleatum levels predict therapeutic response to neoadjuvant chemotherapy in esophageal squamous cell carcinoma. Clin. Cancer Res. 2019, 25, 6170–6179. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef]
- Araujo, D.V.; Watson, G.A.; Oliva, M.; Heirali, A.; Coburn, B.; Spreafico, A.; Siu, L.L. Bugs as drugs: The role of microbiome in cancer focusing on immunotherapeutics. Cancer Treat. Rev. 2020, 92, 102125. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef]
- Hayes, R.B.; Ahn, J.; Fan, X.; Peters, B.A.; Ma, Y.; Yang, L.; Agalliu, I.; Burk, R.D.; Ganly, I.; Purdue, M.P.; et al. Association of oral microbiome with risk for incident head and neck squamous cell cancer. JAMA Oncol. 2018, 4, 358–365. [Google Scholar] [CrossRef]
- Le Bars, P.; Matamoros, S.; Montassier, E.; Le Vacon, F.; Potel, G.; Soueidan, A.; Jordana, F.; de La Cochetière, M.F. The oral cavity microbiota: Between health, oral disease, and cancers of the aerodigestive tract. Can. J. Microbiol. 2017, 63, 475–492. [Google Scholar] [CrossRef] [PubMed]
- Huybrechts, I.; Zouiouich, S.; Loobuyck, A.; Vandenbulcke, Z.; Vogtmann, E.; Pisanu, S.; Iguacel, I.; Scalbert, A.; Indave, I.; Smelov, V.; et al. The human microbiome in relation to cancer risk: A Systematic review of epidemiologic studies. Cancer Epidemiol. Biomark. Prev. 2020, 29, 1856–1868. [Google Scholar] [CrossRef] [PubMed]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Mei, Q.X.; Huang, C.L.; Luo, S.Z.; Zhang, X.M.; Zeng, Y.; Lu, Y.Y. Characterization of the duodenal bacterial microbiota in patients with pancreatic head cancer vs. healthy controls. Pancreatology 2018, 18, 438–445. [Google Scholar] [CrossRef]
- Pushalkar, S.; Ji, X.; Li, Y.; Estilo, C.; Yegnanarayana, R.; Singh, B.; Li, X.; Saxena, D. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 2012, 12, 144. [Google Scholar] [CrossRef]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A.; et al. Tumor Microbiome diversity and composition influence pancreatic cancer outcomes. Cell 2019, 178, 795–806.e12. [Google Scholar] [CrossRef]
- Vogelmann, R.; Amieva, M.R. The role of bacterial pathogens in cancer. Curr. Opin. Microbiol. 2007, 10, 76–81. [Google Scholar] [CrossRef]
- Barrett, M.; Hand, C.K.; Shanahan, F.; Murphy, T.; O’Toole, P.W. Mutagenesis by microbe: The role of the microbiota in shaping the cancer genome. Trends Cancer 2020, 6, 277–287. [Google Scholar] [CrossRef]
- Kadosh, E.; Snir-Alkalay, I.; Venkatachalam, A.; May, S.; Lasry, A.; Elyada, E.; Zinger, A.; Shaham, M.; Vaalani, G.; Mernberger, M.; et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature 2020, 586, 133–138. [Google Scholar] [CrossRef]
- Moritani, K.; Takeshita, T.; Shibata, Y.; Ninomiya, T.; Kiyohara, Y.; Yamashita, Y. Acetaldehyde production by major oral microbes. Oral Dis. 2015, 21, 748–754. [Google Scholar] [CrossRef]
- Gao, S.; Li, S.; Ma, Z.; Liang, S.; Shan, T.; Zhang, M.; Zhu, X.; Zhang, P.; Liu, G.; Zhou, F.; et al. Presence of Porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect. Agent Cancer 2016, 11, 3. [Google Scholar] [CrossRef]
- Guerrero-Preston, R.; Robert White, J.; Godoy-Vitorino, F.; Rodríguez Hilario, A.; Navarro, K.; González, H.; Michailidi, C.; Jedlicka, A.; Canapp, S.; Bondy, J.; et al. High-resolution microbiome profiling uncovers Fusobacterium nucleatum, Lactobacillus gasseri/johnsonii, and Lactobacillus vaginalis associated to oral and oropharyngeal cancer in saliva from HPV positive and HPV negative patients treated with surgery and chemo-radiation. Oncotarget 2017, 8, 110931–110948. [Google Scholar]
- Oliva, M.; Schneeberger, P.H.H.; Rey, V.; Cho, M.; Taylor, R.; Hansen, A.R.; Taylor, K.; Abdalaty, A.H.; Bayley, A.; Hope, A.J.; et al. Transitions in oral and gut microbiome of HPV+ oropharyngeal squamous cell carcinoma following definitive chemoradiotherapy (ROMA LA-OPSCC study). Br. J. Cancer 2020. [Google Scholar] [CrossRef]
- Blackett, K.L.; Siddhi, S.S.; Cleary, S.; Steed, H.; Miller, M.H.; Macfarlane, S.; Macfarlane, G.T.; Dillon, J.F. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett’s and oesophageal carcinoma: Association or causality? Aliment. Pharmacol. Ther. 2013, 37, 1084–1092. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Francois, F.; Pei, Z. Molecular pathways: Pathogenesis and clinical implications of microbiome alteration in esophagitis and Barrett esophagus. Clin. Cancer Res. 2012, 18, 2138–2144. [Google Scholar] [CrossRef] [PubMed]
- Gall, A.; Fero, J.; McCoy, C.; Claywell, B.C.; Sanchez, C.A.; Blount, P.L.; Li, X.; Vaughan, T.L.; Matsen, F.A.; Reid, B.J.; et al. Bacterial Composition of the human upper gastrointestinal tract microbiome is dynamic and associated with genomic instability in a Barrett’s esophagus cohort. PLoS ONE 2015, 10, e0129055. [Google Scholar] [CrossRef] [PubMed]
- Karpinski, T.M. The Microbiota and pancreatic cancer. Gastroenterol. Clin. N. Am. 2019, 48, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156. [Google Scholar] [CrossRef]
- Nosho, K.; Sukawa, Y.; Adachi, Y.; Ito, M.; Mitsuhashi, K.; Kurihara, H.; Kanno, S.; Yamamoto, I.; Ishigami, K.; Igarashi, H.; et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J. Gastroenterol. 2016, 22, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Vilar, E.; Tabernero, J.; Gruber, S.B. Micromanaging the classification of colon cancer: The role of the microRNAome. Clin. Cancer Res. 2011. [Google Scholar] [CrossRef]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef] [PubMed]
- Serna, G.; Ruiz-Pace, F.; Hernando, J.; Alonso, L.; Fasani, R.; Landolfi, S.; Comas, R.; Jimenez, J.; Elez, E.; Bullman, S.; et al. Fusobacterium nucleatum persistence and risk of recurrence after preoperative treatment in locally advanced rectal cancer. Ann. Oncol. 2020, 31, 1366–1375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yang, Y.; Weng, W.; Guo, B.; Cai, G.; Ma, Y.; Cai, S. Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal cancer. J. Exp. Clin. Cancer Res. 2019, 38, 14. [Google Scholar] [CrossRef] [PubMed]
- Sears, C.L.; Pardoll, D.M. Perspective: Alpha-bugs, their microbial partners, and the link to colon cancer. J. Infect. Dis. 2011, 203, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Rosendahl Huber, A.; van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 2020, 580, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, G.; Zhou, H.; Wu, W.K.K.; Wong, S.H.; Coker, O.O.; Dai, Z.; Li, X.; Szeto, C.H.; Sugimura, N.; Lam, T.Y.; et al. Alterations in Enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology 2018, 155, 529–541.e5. [Google Scholar] [CrossRef]
- Coker, O.O.; Nakatsu, G.; Dai, R.Z.; Wu, W.K.K.; Wong, S.H.; Ng, S.C.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2019, 68, 654–662. [Google Scholar] [CrossRef]
- Coker, O.O.; Wu, W.K.K.; Wong, S.H.; Sung, J.J.Y.; Yu, J. Altered Gut archaea composition and interaction with bacteria are associated with colorectal cancer. Gastroenterology 2020, 159, 1459–1470.e5. [Google Scholar] [CrossRef]
- Bucevic Popovic, V.; Situm, M.; Chow, C.T.; Chan, L.S.; Roje, B.; Terzic, J. The urinary microbiome associated with bladder cancer. Sci. Rep. 2018, 8, 12157. [Google Scholar] [CrossRef]
- Bi, H.; Tian, Y.; Song, C.; Li, J.; Liu, T.; Chen, Z.; Chen, C.; Huang, Y.; Zhang, Y. Urinary microbiota—A potential biomarker and therapeutic target for bladder cancer. J. Med. Microbiol. 2019, 68, 1471–1478. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Wu, X.; Wang, Z.; Zhang, C.; Cao, G.; Liu, K.; Yan, T. Uncovering the microbiota in renal cell carcinoma tissue using 16S rRNA gene sequencing. J. Cancer Res. Clin. Oncol. 2020, 147, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Daisley, B.A.; Chanyi, R.M.; Abdur-Rashid, K.; Al, K.F.; Gibbons, S.; Chmiel, J.A.; Wilcox, H.; Reid, G.; Anderson, A.; Dewar, M.; et al. Abiraterone acetate preferentially enriches for the gut commensal Akkermansia muciniphila in castrate-resistant prostate cancer patients. Nat. Commun. 2020, 11, 4822. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Gnanasekar, A.; Lee, A.; Li, W.T.; Haas, M.; Wang-Rodriguez, J.; Chang, E.Y.; Rajasekaran, M.; Ongkeko, W.M. Influence of Intratumor microbiome on clinical outcome and immune processes in prostate cancer. Cancers 2020, 12, 2524. [Google Scholar] [CrossRef] [PubMed]
- Walther-Antonio, M.R.; Chen, J.; Multinu, F.; Hokenstad, A.; Distad, T.J.; Cheek, E.H.; Keeney, G.L.; Creedon, D.J.; Nelson, H.; Mariani, A.; et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 2016, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.R.; McLaughlin, J.R.; Hung, R.J. Previous lung diseases and lung cancer risk: A systematic review and meta-analysis. PLoS ONE 2011, 6, e17479. [Google Scholar] [CrossRef]
- Tsay, J.J.; Wu, B.G.; Badri, M.H.; Clemente, J.C.; Shen, N.; Meyn, P.; Li, Y.; Yie, T.A.; Lhakhang, T.; Olsen, E.; et al. Airway Microbiota is associated with upregulation of the PI3K Pathway in Lung cancer. Am. J. Respir. Crit. Care Med. 2018, 198, 1188–1198. [Google Scholar] [CrossRef]
- Peters, B.A.; Hayes, R.B.; Goparaju, C.; Reid, C.; Pass, H.I.; Ahn, J. The Microbiome in lung cancer tissue and recurrence-free survival. Cancer Epidemiol. Biomark. Prev. 2019, 28, 731–740. [Google Scholar] [CrossRef]
- Banerjee, S.; Tian, T.; Wei, Z.; Shih, N.; Feldman, M.D.; Peck, K.N.; DeMichele, A.M.; Alwine, J.C.; Robertson, E.S. Distinct Microbial signatures associated with different breast cancer types. Front. Microbiol. 2018, 9, 951. [Google Scholar] [CrossRef]
- Curty, G.; de Carvalho, P.S.; Soares, M.A. The Role of the cervicovaginal microbiome on the genesis and as a biomarker of premalignant cervical intraepithelial neoplasia and invasive cervical cancer. Int J. Mol. Sci. 2019, 21, 222. [Google Scholar] [CrossRef]
- Bruce, J.P.; Yip, K.; Bratman, S.V.; Ito, E.; Liu, F.F. Nasopharyngeal cancer: Molecular landscape. J. Clin. Oncol. 2015, 33, 3346–3355. [Google Scholar] [CrossRef]
- Wang, F.; Wu, J.; Wang, Y.; Jin, Y.; Jiang, X.; Qiu, Z.; Qin, Y.; Liu, Y.; Qi, X.; Ge, X.; et al. Gut Microbiota functional biomolecules with immune-lipid metabolism for a prognostic compound score in Epstein-Barr virus-associated gastric adenocarcinoma: A pilot study. Clin. Transl. Gastroenterol. 2019, 10, e00074. [Google Scholar] [CrossRef]
- Capurso, G.; Lahner, E. The interaction between smoking, alcohol and the gut microbiome. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Peters, B.A.; Dominianni, C.; Zhang, Y.; Pei, Z.; Yang, L.; Ma, Y.; Purdue, M.P.; Jacobs, E.J.; Gapstur, S.M.; et al. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 2016, 10, 2435–2446. [Google Scholar] [CrossRef]
- Taberna, M.; Mena, M.; Pavón, M.A.; Alemany, L.; Gillison, M.L.; Mesía, R. Human papillomavirus-related oropharyngeal cancer. Ann. Oncol. 2017, 28, 2386–2398. [Google Scholar] [CrossRef] [PubMed]
- Salar, A. Gastric MALT lymphoma and Helicobacter pylori. Med. Clin. 2019, 152, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Bornigen, D.; Ren, B.; Pickard, R.; Li, J.; Ozer, E.; Hartmann, E.M.; Xiao, W.; Tickle, T.; Rider, J.; Gevers, D.; et al. Alterations in oral bacterial communities are associated with risk factors for oral and oropharyngeal cancer. Sci. Rep. 2017, 7, 17686. [Google Scholar] [CrossRef]
- Orlandi, E.; Iacovelli, N.A.; Tombolini, V.; Rancati, T.; Polimeni, A.; De Cecco, L.; Valdagni, R.; De Felice, F. Potential role of microbiome in oncogenesis, outcome prediction and therapeutic targeting for head and neck cancer. Oral. Oncol. 2019, 99, 104453. [Google Scholar] [CrossRef]
- Li, N.N.; Bai, C.M.; Zhao, L.; Ge, Y.P. [Gut Microbiome differences between gastrointestinal cancer patients and healthy people]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 2019, 41, 636–645. [Google Scholar]
- Baba, Y.; Iwatsuki, M.; Yoshida, N.; Watanabe, M.; Baba, H. Review of the gut microbiome and esophageal cancer: Pathogenesis and potential clinical implications. Ann. Gastroenterol. Surg. 2017, 1, 99–104. [Google Scholar] [CrossRef]
- Guerrero-Preston, R.; Godoy-Vitorino, F.; Jedlicka, A.; Rodríguez-Hilario, A.; González, H.; Bondy, J.; Lawson, F.; Folawiyo, O.; Michailidi, C.; Dziedzic, A.; et al. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment. Oncotarget 2016, 7, 51320–51334. [Google Scholar] [CrossRef]
- Chen, X.; Winckler, B.; Lu, M.; Cheng, H.; Yuan, Z.; Yang, Y.; Jin, L.; Ye, W. Oral microbiota and Risk for esophageal squamous cell carcinoma in a high-risk area of China. PLoS ONE 2015, 10, e0143603. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.L.; Pang, W.; Huang, Y.; Zhang, Y.; Zhang, C.J. The Gastric microbiome is perturbed in advanced gastric adenocarcinoma identified through shotgun metagenomics. Front. Cell Infect. Microbiol. 2018, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Norder Grusell, E.; Dahlen, G.; Ruth, M.; Ny, L.; Quiding-Jarbrink, M.; Bergquist, H.; Bove, M. Bacterial flora of the human oral cavity, and the upper and lower esophagus. Dis. Esophagus 2013, 26, 84–90. [Google Scholar] [CrossRef]
- Inaba, H.; Sugita, H.; Kuboniwa, M.; Iwai, S.; Hamada, M.; Noda, T.; Morisaki, I.; Lamont, R.J.; Amano, A. Porphyromonas gingivalis promotes invasion of oral squamous cell carcinoma through induction of proMMP9 and its activation. Cell Microbiol. 2014, 16, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, K.; Baba, Y.; Nakagawa, S.; Mima, K.; Miyake, K.; Nakamura, K.; Sawayama, H.; Kinoshita, K.; Ishimoto, T.; Iwatsuki, M.; et al. Human microbiome Fusobacterium nucleatum in Esophageal cancer tissue is associated with prognosis. Clin. Cancer Res. 2016, 22, 5574–5581. [Google Scholar] [CrossRef]
- Yang, H.; Qu, L.; Ma, H.; Chen, L.; Liu, W.; Liu, C.; Meng, L.; Wu, J.; Shou, C. Mycoplasma hyorhinis infection in gastric carcinoma and its effects on the malignant phenotypes of gastric cancer cells. BMC Gastroenterol. 2010, 10, 132. [Google Scholar] [CrossRef]
- Zhu, F.; Willette-Brown, J.; Song, N.Y.; Lomada, D.; Song, Y.; Xue, L.; Gray, Z.; Zhao, Z.; Davis, S.R.; Sun, Z.; et al. Autoreactive T cells and Chronic fungal infection drive esophageal carcinogenesis. Cell Host Microbe 2017, 21, 478–493.e7. [Google Scholar] [CrossRef]
- Ferris, R.; Blumenschein, G.; Harrington, K.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Vokes, E.; Gillison, M.; Even, C.; et al. Evaluation of oral microbiome profiling as a response biomarker in squamous cell carcinoma of the head and neck: Analyses from CheckMate 141. Cancer Res. 2017, 77, CT022. [Google Scholar]
- Xu, Y.; Teng, F.; Huang, S.; Lin, Z.; Yuan, X.; Zeng, X.; Yang, F. Changes of saliva microbiota in nasopharyngeal carcinoma patients under chemoradiation therapy. Arch. Oral Biol. 2014, 59, 176–186. [Google Scholar] [CrossRef]
- Hou, J.; Zheng, H.; Li, P.; Liu, H.; Zhou, H.; Yang, X. Distinct shifts in the oral microbiota are associated with the progression and aggravation of mucositis during radiotherapy. Radiother. Oncol. 2018, 129, 44–51. [Google Scholar] [CrossRef]
- Yu, L.X.; Schwabe, R.F. The gut microbiome and liver cancer: Mechanisms and clinical translation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Cani, P.D.; Mayer, E.A. Gut microbiome and liver diseases. Gut 2016, 65, 2035–2044. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ma, C.; Duan, Y.; Heinrich, B.; Rosato, U.; Diggs, L.P.; Ma, L.; Roy, S.; Fu, Q.; Brown, Z.J.; et al. Gut microbiome directs hepatocytes to recruit MDSC and promote cholangiocarcinoma. Cancer Discov. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chakladar, J.; Wong, L.M.; Kuo, S.Z.; Li, W.T.; Yu, M.A.; Chang, E.Y.; Wang, X.Q.; Ongkeko, W.M. The Liver microbiome is implicated in cancer prognosis and modulated by alcohol and hepatitis B. Cancers 2020, 12, 1642. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018, 8, 403. [Google Scholar] [CrossRef]
- Chakladar, J.; Kuo, S.Z.; Castaneda, G.; Li, W.T.; Gnanasekar, A.; Yu, M.A.; Chang, E.Y.; Wang, X.Q.; Ongkeko, W.M. The Pancreatic microbiome is associated with carcinogenesis and worse prognosis in males and smokers. Cancers 2020, 12, 2672. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, T.; Tu, X.; Huang, Y.; Zhang, H.; Tan, D.; Jiang, W.; Cai, S.; Zhao, P.; Song, R.; et al. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J. Immunother. Cancer 2019, 7, 193. [Google Scholar] [CrossRef]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A bacterial driver-passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef]
- Amitay, E.L.; Werner, S.; Vital, M.; Pieper, D.H.; Hofler, D.; Gierse, I.J.; Butt, J.; Balavarca, Y.; Cuk, K.; Brenner, H. Fusobacterium and colorectal cancer: Causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis 2017, 38, 781–788. [Google Scholar] [CrossRef]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum Increases Proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kB, and up-regulating expression of microRNA-21. Gastroenterology 2017, 152, 851–866.e24. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 protein of Fusobacterium nucleatum to Human inhibitory receptor TIGIT Protects tumors from immune cell attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef]
- Gur, C.; Maalouf, N.; Shhadeh, A.; Berhani, O.; Singer, B.B.; Bachrach, G.; Mandelboim, O. Fusobacterium nucleatum supresses anti-tumor immunity by activating CEACAM1. Oncoimmunology 2019, 8, e1581531. [Google Scholar] [CrossRef] [PubMed]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Mima, K.; Cao, Y.; Chan, A.T.; Qian, Z.R.; Nowak, J.A.; Masugi, Y.; Shi, Y.; Song, M.; da Silva, A.; Gu, M.; et al. Fusobacterium nucleatum in Colorectal carcinoma tissue according to tumor location. Clin. Transl Gastroenterol. 2016, 7, e200. [Google Scholar] [CrossRef]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef]
- Lewis, D.A.; Brown, R.; Williams, J.; White, P.; Jacobson, S.K.; Marchesi, J.R.; Drake, M.J. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front. Cell Infect. Microbiol. 2013, 3, 41. [Google Scholar] [CrossRef]
- Burger, M.; Catto, J.W.; Dalbagni, G.; Grossman, H.B.; Herr, H.; Karakiewicz, P.; Kassouf, W.; Kiemeney, L.A.; La Vecchia, C.; Shariat, S.; et al. Epidemiology and risk factors of urothelial bladder cancer. Eur. Urol. 2013, 63, 234–241. [Google Scholar] [CrossRef]
- Banerjee, S.; Tian, T.; Wei, Z.; Shih, N.; Feldman, M.D.; Alwine, J.C.; Coukos, G.; Robertson, E.S. The ovarian cancer oncobiome. Oncotarget 2017, 8, 36225–36245. [Google Scholar] [CrossRef]
- Morales-Sanchez, A.; Fuentes-Panana, E.M. Epstein-Barr virus-associated gastric cancer and potential mechanisms of oncogenesis. Curr. Cancer Drug Targets 2017, 1, 534–554. [Google Scholar] [CrossRef] [PubMed]
- Brianti, P.; De Flammineis, E.; Mercuri, S.R. Review of HPV-related diseases and cancers. New Microbiol. 2017, 40, 80–85. [Google Scholar] [PubMed]
- Carbone, A.; Gloghini, A. Epstein Barr Virus-associated Hodgkin lymphoma. Cancers 2018, 10, 163. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.U.; Jung, D.R.; Lee, Y.H.; Jeon, S.Y.; Han, H.S.; Chong, G.O.; Shin, J.H. Dynamics of Fecal microbiota with and without invasive cervical cancer and its application in early diagnosis. Cancers 2020, 12, 3800. [Google Scholar] [CrossRef]
- Picardo, S.L.; Coburn, B.; Hansen, A.R. The microbiome and cancer for clinicians. Crit. Rev. Oncol. Hematol. 2019, 141, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Maddi, A.; Sabharwal, A.; Violante, T.; Manuballa, S.; Genco, R.; Patnaik, S.; Yendamuri, S. The microbiome and lung cancer. J. Thorac Dis. 2019, 11, 280–291. [Google Scholar] [CrossRef]
- Garrido-Martin, E.M.; Paz-Ares, L. Lung cancer and microbiome. Arch. Bronconeumol. 2020, 56, 3–4. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the gut microbiome on cancer, Immunity, and cancer immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef]
- Lazar, V.; Ditu, L.M.; Pircalabioru, G.G.; Gheorghe, I.; Curutiu, C.; Holban, A.M.; Picu, A.; Petcu, L.; Chifiriuc, M.C. Aspects of Gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front. Immunol. 2018, 9, 1830. [Google Scholar] [CrossRef]
- Khosravi, A.; Yáñez, A.; Price, J.G.; Chow, A.; Merad, M.; Goodridge, H.S.; Mazmanian, S.K. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 2014, 15, 374–381. [Google Scholar] [CrossRef]
- Sanos, S.L.; Bui, V.L.; Mortha, A.; Oberle, K.; Heners, C.; Johner, C.; Diefenbach, A. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 2009, 10, 83–91. [Google Scholar] [CrossRef]
- Honda, K.; Littman, D.R. The microbiota in adaptive immune homeostasis and disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 453, 620–625. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S.; Kiguchi, Y.; Yasuma, K.; Watanabe, E.; Tanoue, T.; et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature 2012, 487, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef]
- Sano, T.; Huang, W.; Hall, J.A.; Yang, Y.; Chen, A.; Gavzy, S.J.; Lee, J.Y.; Ziel, J.W.; Miraldi, E.R.; Domingos, A.I.; et al. An IL-23R/IL-22 Circuit regulates epithelial serum amyloid a to promote local effector Th17 responses. Cell 2015, 163, 381–393. [Google Scholar] [CrossRef]
- Tanoue, T.; Morita, S.; Plichta, D.R.; Skelly, A.N.; Suda, W.; Sugiura, Y.; Narushima, S.; Vlamakis, H.; Motoo, I.; Sugita, K.; et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 2019, 565, 600–605. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Mazmanian, S.K. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol 2013, 14, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Yuan, Y.; Yu, H.; Dai, X.; Wang, S.; Sun, Z.; Wang, F.; Fei, H.; Lin, Q.; Jiang, H.; et al. The gut microbial metabolite trimethylamine N-oxide aggravates GVHD by inducing M1 macrophage polarization in mice. Blood 2020, 136, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Schulthess, J.; Pandey, S.; Capitani, M.; Rue-Albrecht, K.C.; Arnold, I.; Franchini, F.; Chomka, A.; Ilott, N.E.; Johnston, D.G.W.; Pires, E.; et al. The Short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 2019, 50, 432–445.e7. [Google Scholar] [CrossRef] [PubMed]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Wind, T.T.; Gacesa, R.; Vich Vila, A.; de Haan, J.J.; Jalving, M.; Weersma, R.K.; Hospers, G.A.P. Gut microbial species and metabolic pathways associated with response to treatment with immune checkpoint inhibitors in metastatic melanoma. Melanoma Res. 2020, 30, 235–246. [Google Scholar] [CrossRef]
- Hakozaki, T.; Richard, C.; Elkrief, A.; Hosomi, Y.; Benlaïfaoui, M.; Mimpen, I.; Terrisse, S.; Derosa, L.; Zitvogel, L.; Routy, B.; et al. The Gut microbiome associates with immune checkpoint inhibition outcomes in patients with advanced non-small cell lung cancer. Cancer Immunol. Res. 2020. [Google Scholar] [CrossRef]
- Fukuoka, S.; Daisuke, M.; Togashi, Y.; Sugiyama, E.; Udagawa, H.; Kirita, K.; Kamada, T.; Kawazoe, A.; Goto, K.; Doi, T.; et al. Association of gut microbiome with immune status and clinical response in solid tumor patients who received on anti-PD-1 therapies. J. Clin. Oncol. 2018, 36, 3011. [Google Scholar] [CrossRef]
- Baruch, E.N.; Youngster, I.; Ben-Betzalel, G.; Ortenberg, R.; Lahat, A.; Katz, L.; Adler, K.; Dick-Necula, D.; Raskin, S.; Bloch, N.; et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2020. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, V.P.; Łuksza, M.; Zhao, J.N.; Makarov, V.; Moral, J.A.; Remark, R.; Herbst, B.; Askan, G.; Bhanot, U.; Senbabaoglu, Y.; et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 2017, 551, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Fessler, J.; Matson, V.; Gajewski, T.F. Exploring the emerging role of the microbiome in cancer immunotherapy. J. ImmunoTher. Cancer 2019, 7, 108. [Google Scholar] [CrossRef] [PubMed]
- Fluckiger, A.; Daillère, R.; Sassi, M.; Sixt, B.S.; Liu, P.; Loos, F.; Richard, C.; Rabu, C.; Alou, M.T.; Goubet, A.G.; et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science 2020, 369, 936–942. [Google Scholar] [CrossRef] [PubMed]
- Paulos, C.M.; Wrzesinski, C.; Kaiser, A.; Hinrichs, C.S.; Chieppa, M.; Cassard, L.; Palmer, D.C.; Boni, A.; Muranski, P.; Yu, Z.; et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J. Clin. Invest. 2007, 117, 2197–2204. [Google Scholar] [CrossRef]
- Hall, J.A.; Bouladoux, N.; Sun, C.M.; Wohlfert, E.A.; Blank, R.B.; Zhu, Q.; Grigg, M.E.; Berzofsky, J.A.; Belkaid, Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 2008, 29, 637–649. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.A.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M.; et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489. [Google Scholar] [CrossRef]
- Ma, W.; Mao, Q.; Xia, W.; Dong, G.; Yu, C.; Jiang, F. Gut Microbiota shapes the efficiency of cancer therapy. Front. Microbiol. 2019, 10, 1050. [Google Scholar] [CrossRef]
- Soldati, L.; Di Renzo, L.; Jirillo, E.; Ascierto, P.A.; Marincola, F.M.; De Lorenzo, A. The influence of diet on anti-cancer immune responsiveness. J. Transl. Med. 2018, 16, 75. [Google Scholar] [CrossRef]
- Peng, W.; Chen, J.Q.; Liu, C.; Malu, S.; Creasy, C.; Tetzlaff, M.T.; Xu, C.; McKenzie, J.A.; Zhang, C.; Liang, X.; et al. Loss of PTEN Promotes resistance to T Cell-Mediated immunotherapy. Cancer Discov. 2016, 6, 202–216. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, J.; Kumar, A.; Parikh, K.; Anwar, A.; Knoll, B.M.; Puccio, C.; Chun, H.; Fanucchi, M.; Lim, S.H. Use of broad-spectrum antibiotics impacts outcome in patients treated with immune checkpoint inhibitors. Oncoimmunology 2018, 7, e1507670. [Google Scholar] [CrossRef] [PubMed]
- Derosa, L.; Hellmann, M.D.; Spaziano, M.; Halpenny, D.; Fidelle, M.; Rizvi, H.; Long, N.; Plodkowski, A.J.; Arbour, K.C.; Chaft, J.E.; et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann. Oncol. 2018, 29, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Elkrief, A.; El Raichani, L.; Richard, C.; Messaoudene, M.; Belkaid, W.; Malo, J.; Belanger, K.; Miller, W.; Jamal, R.; Letarte, N.; et al. Antibiotics are associated with decreased progression-free survival of advanced melanoma patients treated with immune checkpoint inhibitors. Oncoimmunology 2019, 8, e1568812. [Google Scholar] [CrossRef] [PubMed]
- Pinato, D.J.; Howlett, S.; Ottaviani, D.; Urus, H.; Patel, A.; Mineo, T.; Brock, C.; Power, D.; Hatcher, O.; Falconer, A.; et al. Association of Prior antibiotic treatment with survival and response to immune checkpoint inhibitor therapy in patients with cancer. JAMA Oncol. 2019, 5, 1774–1778. [Google Scholar] [CrossRef]
- Wilson, B.E.; Routy, B.; Nagrial, A.; Chin, V.T. The effect of antibiotics on clinical outcomes in immune-checkpoint blockade: A systematic review and meta-analysis of observational studies. Cancer Immunol. Immunother. 2020, 69, 343–354. [Google Scholar] [CrossRef]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019, 25, 377–388. [Google Scholar] [CrossRef]
- Chen, D.; Wu, J.; Jin, D.; Wang, B.; Cao, H. Fecal microbiota transplantation in cancer management: Current status and perspectives. Int. J. Cancer 2019, 145, 2021–2031. [Google Scholar] [CrossRef]
- Shui, L.; Yang, X.; Li, J.; Yi, C.; Sun, Q.; Zhu, H. Gut Microbiome as a potential factor for modulating resistance to cancer immunotherapy. Front. Immunol. 2019, 10, 2989. [Google Scholar] [CrossRef]
- Villeger, R.; Lopes, A.; Carrier, G.; Veziant, J.; Billard, E.; Barnich, N.; Gagniere, J.; Vazeille, E.; Bonnet, M. Intestinal microbiota: A Novel target to improve anti-tumor treatment? Int. J. Mol. Sci. 2019, 20, 4584. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Kolida, S.; Gibson, G.R. Synbiotics in health and disease. Annu. Rev. Food Sci. Technol. 2011, 2, 373–393. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Kathera, C.; Janardhan, A.; Praveen Kumar, A.; Viswanath, B. Surfacing role of probiotics in cancer prophylaxis and therapy: A systematic review. Clin. Nutr. 2017, 36, 1465–1472. [Google Scholar] [CrossRef] [PubMed]
- Rafter, J.; Bennett, M.; Caderni, G.; Clune, Y.; Hughes, R.; Karlsson, P.C.; Klinder, A.; O’Riordan, M.; O’Sullivan, G.C.; Pool-Zobel, B.; et al. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am. J. Clin. Nutr. 2007, 85, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Cammarota, G.; Ianiro, G.; Tilg, H.; Rajilic-Stojanovic, M.; Kump, P.; Satokari, R.; Sokol, H.; Arkkila, P.; Pintus, C.; Hart, A.; et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut 2017, 66, 569–580. [Google Scholar] [CrossRef]
- Wonderlick, J.S.; D’Agostino, R. Fecal microbiota transplantation via fluoroscopy-guided nasojejunal catheter placement: Indications, technique, and the role of radiology. Abdom. Radiol. 2016, 41, 2020–2025. [Google Scholar] [CrossRef]
- Halkjaer, S.I.; Christensen, A.H.; Lo, B.Z.S.; Browne, P.D.; Gunther, S.; Hansen, L.H.; Petersen, A.M. Faecal microbiota transplantation alters gut microbiota in patients with irritable bowel syndrome: Results from a randomised, double-blind placebo-controlled study. Gut 2018, 67, 2107–2115. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; von Wright, A.; Vilpponen-Salmela, T.; Ben-Amor, K.; Akkermans, A.D.; de Vos, W.M. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 2002, 68, 3401–3407. [Google Scholar] [CrossRef]
- Tang, Q.; Jin, G.; Wang, G.; Liu, T.; Liu, X.; Wang, B.; Cao, H. Current Sampling methods for gut microbiota: A Call for more precise devices. Front. Cell Infect. Microbiol. 2020, 10, 151. [Google Scholar] [CrossRef]
- Mansour, B.; Monyok, A.; Makra, N.; Gajdacs, M.; Vadnay, I.; Ligeti, B.; Juhasz, J.; Szabo, D.; Ostorhazi, E. Bladder cancer-related microbiota: Examining differences in urine and tissue samples. Sci. Rep. 2020, 10, 11042. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Miller, S.A. Clinical metagenomics. Nat. Rev. Genet. 2019, 20, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Byrd, D.A.; Chen, J.; Vogtmann, E.; Hullings, A.; Song, S.J.; Amir, A.; Kibriya, M.G.; Ahsan, H.; Chen, Y.; Nelson, H.; et al. Reproducibility, stability, and accuracy of microbial profiles by fecal sample collection method in three distinct populations. PLoS ONE 2019, 14, e0224757. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Tomasch, J.; Deng, Z.; Jarek, M.; Husemann, P.; Wagner-Dobler, I.; COMBACTE Consortium. Stool metatranscriptomics: A technical guideline for mRNA stabilisation and isolation. BMC Genom. 2015, 16, 494. [Google Scholar] [CrossRef] [PubMed]
- Angebault, C.; Ghozlane, A.; Volant, S.; Botterel, F.; d’Enfert, C.; Bougnoux, M.E. Combined bacterial and fungal intestinal microbiota analyses: Impact of storage conditions and DNA extraction protocols. PLoS ONE 2018, 13, e0201174. [Google Scholar] [CrossRef] [PubMed]
- Gudra, D.; Shoaie, S.; Fridmanis, D.; Klovins, J.; Wefer, H.; Silamikelis, I.; Peculis, R.; Kalnina, I.; Elbere, I.; Radovica-Spalvina, I.; et al. A widely used sampling device in colorectal cancer screening programmes allows for large-scale microbiome studies. Gut 2019, 68, 1723–1725. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.M.; Segata, N. Multiple levels of the unknown in microbiome research. BMC Biol. 2019, 17, 48. [Google Scholar] [CrossRef] [PubMed]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glockner, F.O.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B.; Euzeby, J.; Amann, R.; Rossello-Mora, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Franzen, O.; Hu, J.; Bao, X.; Itzkowitz, S.H.; Peter, I.; Bashir, A. Improved OTU-picking using long-read 16S rRNA gene amplicon sequencing and generic hierarchical clustering. Microbiome 2015, 3, 43. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.H.; Siddle, K.J.; Park, D.J.; Sabeti, P.C. Benchmarking Metagenomics Tools for taxonomic classification. Cell 2019, 178, 779–794. [Google Scholar] [CrossRef] [PubMed]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A review of methods and databases for metagenomic classification and assembly. Brief. Bioinform. 2019, 20, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome datasets are compositional: And this is not optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, D.; Kathagen, G.; D’Hoe, K.; Vieira-Silva, S.; Valles-Colomer, M.; Sabino, J.; Wang, J.; Tito, R.Y.; De Commer, L.; Darzi, Y.; et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 2017, 551, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Boernigen, D.; Tickle, T.L.; Morgan, X.C.; Garrett, W.S.; Huttenhower, C. Computational meta’omics for microbial community studies. Mol. Syst. Biol. 2013, 9, 666. [Google Scholar] [CrossRef]
- Mark Welch, J.L.; Rossetti, B.J.; Rieken, C.W.; Dewhirst, F.E.; Borisy, G.G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. USA 2016, 113, E791–E800. [Google Scholar] [CrossRef]
- Amann, R.I.; Krumholz, L.; Stahl, D.A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 1990, 172, 762–770. [Google Scholar] [CrossRef]
- Valm, A.M.; Mark Welch, J.L.; Rieken, C.W.; Hasegawa, Y.; Sogin, M.L.; Oldenbourg, R.; Dewhirst, F.E.; Borisy, G.G. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl. Acad. Sci. USA 2011, 108, 4152–4157. [Google Scholar] [CrossRef]
- Wang, H.; Wang, M.X.; Su, N.; Wang, L.C.; Wu, X.; Bui, S.; Nielsen, A.; Vo, H.T.; Nguyen, N.; Luo, Y.; et al. RNAscope for in situ detection of transcriptionally active human papillomavirus in head and neck squamous cell carcinoma. J. Vis. Exp. 2014. [Google Scholar] [CrossRef]
- Baker, K. Organoids provide an important window on inflammation in cancer. Cancers 2018, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Al-Asmakh, M.; Zadjali, F. Use of germ-free animal models in microbiota-related research. J. Microbiol. Biotechnol. 2015, 25, 1583–1588. [Google Scholar] [CrossRef] [PubMed]
- Guzman-Rodriguez, M.; McDonald, J.A.K.; Hyde, R.; Allen-Vercoe, E.; Claud, E.C.; Sheth, P.M.; Petrof, E.O. Using bioreactors to study the effects of drugs on the human microbiota. Methods 2018, 149, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Fair, K.L.; Colquhoun, J.; Hannan, N.R.F. Intestinal organoids for modelling intestinal development and disease. Philos. Trans. R. Soc. Lond B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [PubMed]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal Bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967. [Google Scholar] [CrossRef]
- McCracken, K.W.; Cata, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rockich, B.E.; Tsai, Y.H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Allen-Vercoe, E. Bringing the gut microbiota into focus through microbial culture: Recent progress and future perspective. Curr. Opin. Microbiol. 2013, 16, 625–629. [Google Scholar] [CrossRef]
- McDonald, J.A.; Schroeter, K.; Fuentes, S.; Heikamp-Dejong, I.; Khursigara, C.M.; de Vos, W.M.; Allen-Vercoe, E. Evaluation of microbial community reproducibility, stability and composition in a human distal gut chemostat model. J. Microbiol. Methods. 2013, 95, 167–174. [Google Scholar] [CrossRef]
- Oliphant, K.; Cochrane, K.; Schroeter, K.; Daigneault, M.C.; Yen, S.; Verdu, E.F.; Allen-Vercoe, E. Effects of Antibiotic pretreatment of an ulcerative colitis-derived fecal microbial community on the integration of therapeutic bacteria in vitro. mSystems 2020, 5. [Google Scholar] [CrossRef]
- Jumas-Bilak, E.; Bouvet, P.; Allen-Vercoe, E.; Aujoulat, F.; Lawson, P.A.; Jean-Pierre, H.; Marchandin, H. Rarimicrobium hominis gen. nov., sp. nov., representing the fifth genus in the phylum Synergistetes that includes human clinical isolates. Int. J. Syst. Evol. Microbiol. 2015, 65, 3965–3970. [Google Scholar] [CrossRef]
- Schmidt, T.S.B.; Raes, J.; Bork, P. The Human gut microbiome: From Association to modulation. Cell 2018, 172, 1198–1215. [Google Scholar] [CrossRef] [PubMed]
- Panek, M.; Cipcic Paljetak, H.; Baresic, A.; Peric, M.; Matijasic, M.; Lojkic, I.; Vranesic Bender, D.; Krznaric, Z.; Verbanac, D. Methodology challenges in studying human gut microbiota—Effects of collection, storage, DNA extraction and next generation sequencing technologies. Sci. Rep. 2018, 8, 5143. [Google Scholar] [CrossRef]
- Malagon, M.; Ramio-Pujol, S.; Serrano, M.; Amoedo, J.; Oliver, L.; Bahi, A.; Miquel-Cusachs, J.O.; Ramirez, M.; Queralt-Moles, X.; Gilabert, P.; et al. New fecal bacterial signature for colorectal cancer screening reduces the fecal immunochemical test false-positive rate in a screening population. PLoS ONE 2020, 15, e0243158. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.; Abu-Ali, G.; Vogtmann, E.; Fodor, A.A.; Ren, B.; Amir, A.; Schwager, E.; Crabtree, J.; Ma, S.; Microbiome Quality Control Project, C.; et al. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat. Biotechnol. 2017, 35, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Integrative HMP (iHMP) Research Network Consortium. The Integrative human microbiome project: Dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe 2014, 16, 276–289. [Google Scholar] [CrossRef]
- Schloissnig, S.; Arumugam, M.; Sunagawa, S.; Mitreva, M.; Tap, J.; Zhu, A.; Waller, A.; Mende, D.R.; Kultima, J.R.; Martin, J.; et al. Genomic variation landscape of the human gut microbiome. Nature 2013, 493, 45–50. [Google Scholar] [CrossRef]
- Minot, S.; Bryson, A.; Chehoud, C.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. USA 2013, 110, 12450–12455. [Google Scholar] [CrossRef]
- Bagel, S.; Hullen, V.; Wiedemann, B.; Heisig, P. Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob. Agents Chemother. 1999, 43, 868–875. [Google Scholar] [CrossRef]
- Harrison, K.L.; Farrell, R.M.; Brinich, M.A.; Highland, J.; Mercer, M.; McCormick, J.B.; Tilburt, J.; Geller, G.; Marshall, P.; Sharp, R.R. ‘Someone should oversee it’: Patient perspectives on the ethical issues arising with the regulation of probiotics. Health Expect. 2015, 18, 250–261. [Google Scholar] [CrossRef]

| Disease Site | Tumor Type | Sample Type | Tumor-Associated Taxa | Potential Clinical Utility Based on Recent Evidence |
|---|---|---|---|---|
| Head and Neck and Upper Gastrointestinal Tumors | Head and neck SCC | Saliva tumor tissue | Kingella and Corynebacterium [20,30] | Cancer prevention |
| Porphyromonas gingivalis [31] | Diagnostic | |||
| Fusobacterium nucleatum [32,33] | Prognostic | |||
| Esophageal ADC | Tumor tissue | Campylobacter species [34,35,36] | Diagnostic | |
| Esophageal SCC | Tumor tissue | Fusobacterium nucleatum [13,33] | Prognostic | |
| Gastric carcinoma | Tumor tissue | Helicobacter pylori [6] | Diagnostic | |
| Hepatocarcinoma and Pancreatic Cancer | Ductal ADC | Tumor tissue | VHB, VHC [5] | Screening Diagnostic |
| Normal tissue | H. pylori, P gingivalis, Fusobacterium sp., Aggregatibacter sp., Prevotella sp., or Capnocytophaga sp. [37] | Diagnostic | ||
| Pancreatic ductal ADC | Tumor tissue | Pseudoxanthomonas sp., Streptomyces sp., Saccharopolyspora sp., Bacillus clausii, Proteobacteri sp. [26] | Prognostic | |
| Gammaproteobacteria [38] | Predictive | |||
| Colorectal Cancer | Colorectal ADC | Tumor tissue Stool Saliva | Fusobacterium nucleatum [39,40,41,42,43] | Diagnostic Prognostic Predictive Therapeutic |
| Tumor tissue | Enterotoxigenic Bacteroides fragilis [44] | Cancer Prevention | ||
| Escherichia coli (pk+) [45] | ||||
| Stool | Peptostreptococcus stomatis, Parvimonas, Porphyromonas [39,40] Ascomycota, Basidiomycota, Orthobunyavirus [46,47,48] | Diagnostic Screening tool | ||
| Genitourinary tumors | Urothelial carcinoma | Urine | Fusobacterium, Firmicute [49,50] | Diagnostic |
| Renal cell carcinoma | Tumor tissue | Chloroplast, Streptophyta [51] | Diagnostic | |
| Prostate SDC | Tumor tissue | Akkermansia muciniphila [52] | Predictive of response | |
| Listeria monocytogenes [53] | Prognostic | |||
| Endometrial cancer | Tumor tissue | Porphyromonas sp., Atopobium vaginae [54] | Diagnostic | |
| Lung cancer | Lung ADC and SCC | Normal site | Chlamydia pneumonia, Mycobacterium tuberculosi [55] | Cancer prevention |
| Saliva Tumor tissue | Veillonella, Capnocytophaga, Selenomonas Megasphaera, Neisseria [56] | Diagnostic | ||
| Family Lachnospiraceae, genera Faecalibacterium and Ruminococcus [57] | Prognostic | |||
| Faeces | Akkermansia muciniphila [14] | Predictive of response | ||
| Breast cancer | Triple-positive ductal ADC (HR/HER-2+) | Tumor tissue | Bordetella, Campylobacter, Chlamydia, Chlamydophila, Legionella, Pasteurella [58] | Diagnostic |
| Triple-negative ductal ADC (HR/HER-2 -) | Tumor tissue | Aerococcus, Arcobacter, Geobacillus, Orientia, Rothia [58] | Diagnostic | |
| HPV-related cancers | Oropharyngeal SCC | Saliva | Lactobacillus-enriched [32] | Diagnostic |
| Cervical SCC | Tumor tissue | HPV16 [5] | Prognostic | |
| Vaginal fluid | Lactobacillus, Gardnerella, Atopobium, Fusobacterium, Sneathia [59] | Diagnostic Prognostic | ||
| EBV-related cancers | Nasopharyngeal carcinoma | Tumor tissue | EBV [60] | Diagnostic Prognostic |
| Gut | Functional metabolic signature [61] | Prognostic |
| Type | Technique | Problem | Solution/Alternative |
|---|---|---|---|
| Sequencing technique | 16S rRNA-seq | Low taxonomic resolution Limited functional analysis | Full-length 16S sequencing, shotgun sequencing |
| Whole shotgun sequencing | More expensive Human DNA also gets sequenced | Sequencing at low coverage Adequate source material, enrichment of microbial material before sequencing | |
| Long read sequencing | Sequencing errors are difficult to detect | Combining long read sequencing with short read shotgun | |
| 16S bioinformatics | OTU-based methods | Loss of information in clustering | ASV-based methods |
| ASV-based methods | Reliance on the algorithm to detect sequencing errors | ||
| Shotgun bioinformatics | Taxonomic profiling | Reliance on incomplete databases | New assemblies will provide more complete databases |
| Functional profiling | Reliance on incomplete databases, proteins of unknown function | Further characterization of microbial proteins is still needed | |
| De novo assembly | Incomplete assemblies, chimeric genomes, strain heterogeneity | Strict quality control Long-read sequencing will provide better assemblies | |
| Biostatistics | Traditional statistics | Datasets are compositional | Compositional methods, estimation of total microbial presence to avoid compositionality |
| Compositional analysis | Presence of zeroes Difficult to interpret | Zero-replacement | |
| Spatial in situ resolution | RNA in situ hybridization | Low-throughput (only 2-3 bacterium can be detected) | Use it when information about spatial resolution is needed |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliva, M.; Mulet-Margalef, N.; Ochoa-De-Olza, M.; Napoli, S.; Mas, J.; Laquente, B.; Alemany, L.; Duell, E.J.; Nuciforo, P.; Moreno, V. Tumor-Associated Microbiome: Where Do We Stand? Int. J. Mol. Sci. 2021, 22, 1446. https://doi.org/10.3390/ijms22031446
Oliva M, Mulet-Margalef N, Ochoa-De-Olza M, Napoli S, Mas J, Laquente B, Alemany L, Duell EJ, Nuciforo P, Moreno V. Tumor-Associated Microbiome: Where Do We Stand? International Journal of Molecular Sciences. 2021; 22(3):1446. https://doi.org/10.3390/ijms22031446
Chicago/Turabian StyleOliva, Marc, Nuria Mulet-Margalef, Maria Ochoa-De-Olza, Stefania Napoli, Joan Mas, Berta Laquente, Laia Alemany, Eric J. Duell, Paolo Nuciforo, and Victor Moreno. 2021. "Tumor-Associated Microbiome: Where Do We Stand?" International Journal of Molecular Sciences 22, no. 3: 1446. https://doi.org/10.3390/ijms22031446
APA StyleOliva, M., Mulet-Margalef, N., Ochoa-De-Olza, M., Napoli, S., Mas, J., Laquente, B., Alemany, L., Duell, E. J., Nuciforo, P., & Moreno, V. (2021). Tumor-Associated Microbiome: Where Do We Stand? International Journal of Molecular Sciences, 22(3), 1446. https://doi.org/10.3390/ijms22031446








