The Regulator OmpR in Yersinia enterocolitica Participates in Iron Homeostasis by Modulating Fur Level and Affecting the Expression of Genes Involved in Iron Uptake
Abstract
:1. Introduction
2. Results
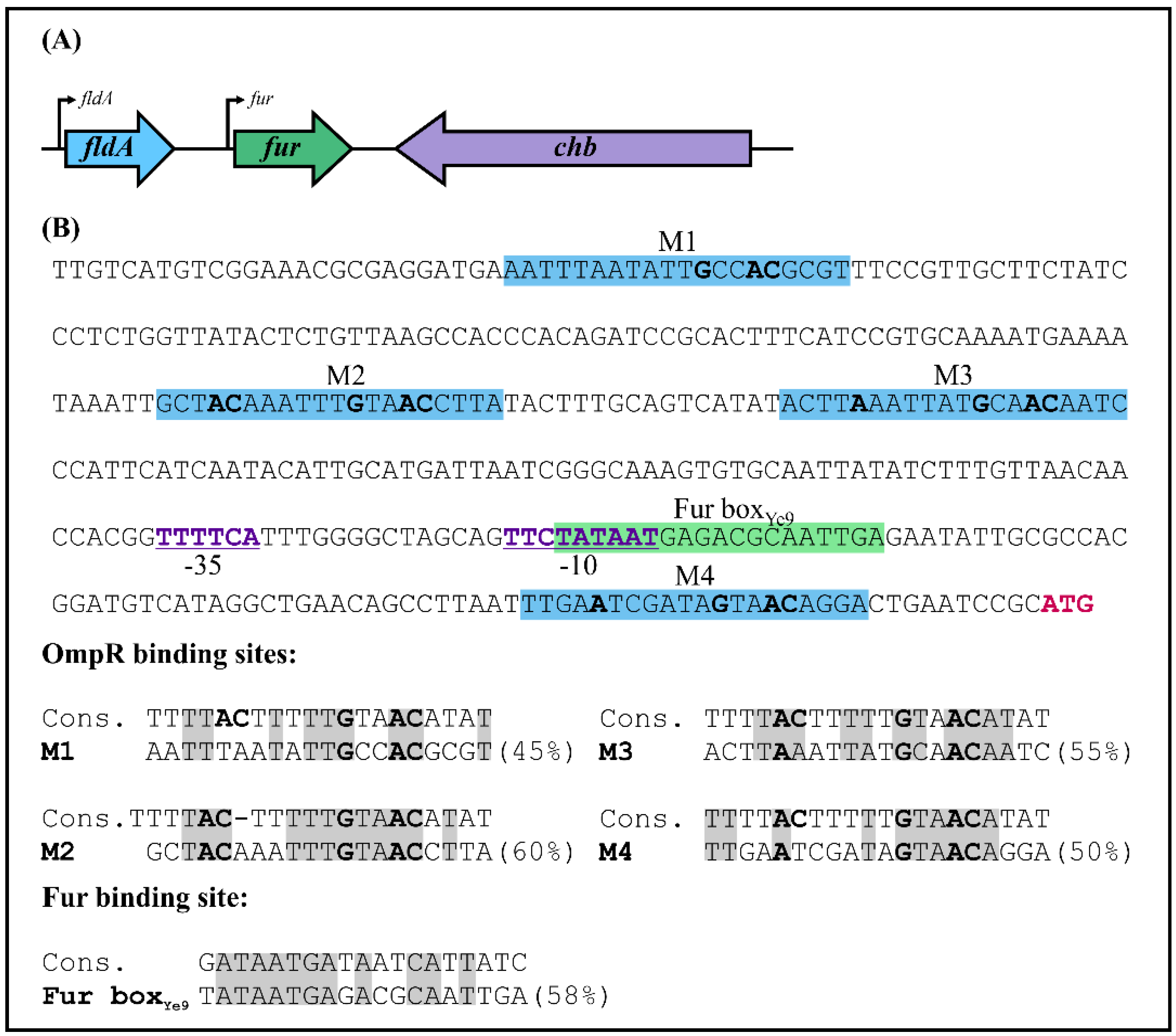
2.1. Characterization of the Fur Promoter Region in Y. enterocolitica Strain Ye9N
2.2. OmpR Inhibits the Activity of the Ye9 Fur Promoter
2.3. Effect of Environmental Signals on OmpR-Dependent Ye9 Fur Expression
2.4. Fur Abundance in Y. enterocolitica Is Modulated by the OmpR Regulator and Certain Environmental Conditions
2.5. The Effect of OmpR and Successive Deletion of the Fur Regulatory Region on the Transcriptional Activity of the YePfur Promoter
2.6. Interaction of OmpR with the Full Length and Truncated Fur Regulatory Regions of Y. enterocolitica
2.7. E. coli Fur Expression Is Negatively Regulated by OmpR
2.8. Effect of OmpR on the Expression of Selected Fur Regulon Members in Y. enterocolitica
3. Discussion
4. Materials and Methods
4.1. Strains, Media and Growth Conditions
4.2. Molecular Biology Techniques
4.3. Construction of a Chromosomal Pfur’::lacZYA Fusion
4.4. Construction of Pfur::lacZ Fusions in Plasmid pCM132Gm
4.5. Complementation of the ΔompR Mutation
4.6. β-Galactosidase Assay
4.7. RT-qPCR
4.8. Construction of Strains Expressing Fur Carrying a 3×FLAG Epitope
4.9. Western Blotting
4.10. Electrophoretic Mobility Shift Assays (EMSAs)
4.11. Bioinformatic and Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fàbrega, A.; Vila, J. Yersinia enterocolitica: Pathogenesis, virulence and antimicrobial resistance. Enferm. Infecc. Microbiol. Clin. 2012, 30, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, T.M.; Brandt, K.; Starke, M.; Rattei, T. Shotgun sequencing of Yersinia enterocolitica strain W22703 (biotype 2, serotype O:9): Genomic evidence for oscillation between invertebrates and mammals. BMC Genom. 2011, 12, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bottone, E.J. Yersinia enterocolitica: Revisitation of an Enduring Human Pathogen. Clin. Microbiol. Newsl. 2015, 37, 1–8. [Google Scholar] [CrossRef]
- Bottone, E.J. Yersinia enterocolitica: A panoramic view of a charismatic microorganism. CRC Crit. Rev. Microbiol. 1977, 5, 211–241. [Google Scholar] [CrossRef]
- Bialas, N.; Kasperkiewicz, K.; Radziejewska-Lebrecht, J.; Skurnik, M. Bacterial cell surface structures in Yersinia enterocolitica. Arch. Immunol. Ther. Exp. 2012, 60, 199–209. [Google Scholar] [CrossRef]
- Zughaier, S.M.; Cornelis, P. Editorial: Role of Iron in Bacterial Pathogenesis. Front. Cell. Infect. Microbiol. 2018, 8, 344. [Google Scholar] [CrossRef] [Green Version]
- Bullen, J.J. The significance of iron in infection. Rev. Infect. Dis. 1981, 3, 1127–1138. [Google Scholar] [CrossRef]
- Stojiljkovic, I.; Hantke, K. Hemin uptake system of Yersinia enterocolitica: Similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1992, 11, 4359–4367. [Google Scholar] [CrossRef]
- Stojiljkovic, I.; BaumLer, A.J.; Hantke, K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 1994, 236, 531–545. [Google Scholar] [CrossRef]
- Straley, S.C.; Perry, R.D. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 1995, 3, 310–317. [Google Scholar] [CrossRef]
- Carniel, E.; Guilvout, I.; Prentice, M. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J. Bacteriol. 1996, 178, 6743–6751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakin, A.; Noelting, C.; Schubert, S.; Heesemann, J. Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect. Immun. 1999, 67, 5265–5274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schubert, S.; Fischer, D.; Heesemann, J. Ferric enterochelin transport in Yersinia enterocolitica: Molecular and evolutionary aspects. J. Bacteriol. 1999, 181, 6387–6395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, R.D.; Fetherston, J.D. Iron and heme uptake systems. In Yersinia Molecular and Cellular Biology; Carniel, E., Hinnebusch, B.J., Eds.; Bioscience Horizons: Wymondham, UK, 2004; pp. 257–283. [Google Scholar]
- Jaworska, K.; Nieckarz, M.; Ludwiczak, M.; Raczkowska, A.; Brzostek, K. OmpR-Mediated Transcriptional Regulation and Function of Two Heme Receptor Proteins of Yersinia enterocolitica Bio-Serotype 2/O:9. Front. Cell. Infect. Microbiol. 2018, 8, 333. [Google Scholar] [CrossRef] [PubMed]
- Heesemann, J.; Hantke, K.; Vocke, T.; Saken, E.; Rakin, A.; Stojiljkovic, I.; Berner, R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol. Microbiol. 1993, 8, 397–408. [Google Scholar] [CrossRef]
- BäumLer, A.; Koebnik, R.; Stojiljkovic, I.; Heesemann, J.; Braun, V.; Hantke, K. Survey on Newly Characterized Iron Uptake Systems of Yersinia enterocolitica. Zent. Bakteriol. 1993, 278, 416–424. [Google Scholar] [CrossRef]
- KammLer, M.; Schön, C.; Hantke, K. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 1993, 175, 6212–6219. [Google Scholar] [CrossRef] [Green Version]
- Ge, R.; Sun, X. Iron trafficking system in Helicobacter pylori. BioMetals 2012, 25, 247–258. [Google Scholar] [CrossRef]
- O’Connor, L.; Fetherston, J.D.; Perry, R.D. The feoABC Locus of Yersinia pestis Likely Has Two Promoters Causing Unique Iron Regulation. Front. Cell. Infect. Microbiol. 2017, 7, 331. [Google Scholar] [CrossRef]
- Frawley, E.R.; Fang, F.C. The ins and outs of bacterial iron metabolism. Mol. Microbiol. 2014, 93, 609–616. [Google Scholar] [CrossRef] [Green Version]
- Winterbourn, C.C. Toxicity of iron and hydrogen peroxide: The Fenton reaction. Toxicol. Lett. 1995, 82–83, 969–974. [Google Scholar] [CrossRef]
- Masse, E.; Gottesman, S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 4620–4625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McHugh, J.P.; Rodríguez-Quinoñes, F.; Abdul-Tehrani, H.; Svistunenko, D.A.; Poole, R.K.; Cooper, C.E.; Andrews, S.C. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 2003, 278, 29478–29486. [Google Scholar] [CrossRef] [Green Version]
- Escolar, L.; Perez-Martin, J.; de Lorenzo, V. Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 1998, 283, 537–547. [Google Scholar] [CrossRef]
- Hantke, K. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 2001, 4, 172–177. [Google Scholar] [CrossRef]
- Escolar, L.; Perez-Martin, J.; de Lorenzo, V. Opening the iron box: Transcriptional metalloregulation by the Fur protein. J. Bacteriol. 1999, 181, 6223–6229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hantke, K. Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol. Gen. Genet. 1984, 197, 337–341. [Google Scholar] [CrossRef]
- Schaffer, S.; Hantke, K.; Braun, V. Nucleotide sequence of the iron regulatory gene fur. Mol. Gen. Genet. 1985, 200, 110–113. [Google Scholar] [CrossRef]
- Tardat, B.; Touati, D. Iron and oxygen regulation of Escherichia coli MnSOD expression: Competition between the global regulators Fur and ArcA for binding to DNA. Mol. Microbiol. 1993, 9, 53–63. [Google Scholar] [CrossRef]
- Lee, H.S.; Lee, Y.S.; Kim, H.S.; Choi, J.Y.; Hassan, H.M.; Chung, M.H. Mechanism of regulation of 8-hydroxyguanine endonuclease by oxidative stress: Roles of FNR, ArcA, and Fur. Free Radic. Biol. Med. 1998, 24, 1193–1201. [Google Scholar] [CrossRef]
- Hantke, K. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K-12: Fur not only affects iron metabolism. Mol. Gen. Genet. 1987, 210, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Hall, H.K.; Foster, J.W. The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J. Bacteriol. 1996, 178, 5683–5691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fillat, M.F. The FUR (ferric uptake regulator) superfamily: Diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 2014, 546, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Ernst, F.D.; Bereswill, S.; Waidner, B.; Stoof, J.; Mäder, U.; Kusters, J.G.; Kuipers, E.J.; Kist, M.; van Vliet, A.H.M.; Homuth, G. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 2005, 151, 533–546. [Google Scholar] [CrossRef] [Green Version]
- Yuhara, S.; Komatsu, H.; Goto, H.; Ohtsubo, Y.; Nagata, Y.; Tsuda, M. Pleiotropic roles of iron-responsive transcriptional regulator Fur in Burkholderia multivorans. Microbiology 2008, 154, 1763–1774. [Google Scholar] [CrossRef] [Green Version]
- Porcheron, G.; Dozois, C.M. Interplay between iron homeostasis and virulence: Fur and RyhB as major regulators of bacterial pathogenicity. Vet. Microbiol. 2015, 179, 2–14. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, S.; Cissé, C.; Vitale, S.; Ahmadova, A.; Degardin, M.; Pérard, J.; Colas, P.; Miras, R.; Boturyn, D.; Covès, J.; et al. From Peptide Aptamers to Inhibitors of FUR, Bacterial Transcriptional Regulator of Iron Homeostasis and Virulence. ACS Chem. Biol. 2016, 11, 2519–2528. [Google Scholar] [CrossRef]
- Staggs, T.M.; Perry, R.D. Identification and cloning of a fur regulatory gene in Yersinia pestis. J. Bacteriol. 1991, 173, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Schwiesow, L.; Mettert, E.; Wei, Y.; Miller, H.K.; Herrera, N.G.; Balderas, D.; Kiley, P.J.; Auerbuch, V. Control of hmu Heme Uptake Genes in Yersinia pseudotuberculosis in Response to Iron Sources. Front. Cell. Infect. Microbiol. 2018, 8, 47. [Google Scholar] [CrossRef]
- Gao, H.; Zhou, D.; Li, Y.; Guo, Z.; Han, Y.; Song, Y.; Zhai, J.; Du, Z.; Wang, X.; Lu, J.; et al. The iron-responsive Fur regulon in Yersinia pestis. J. Bacteriol. 2008, 190, 3063–3075. [Google Scholar] [CrossRef] [Green Version]
- Zhou, D.; Qin, L.; Han, Y.; Qiu, J.; Chen, Z.; Li, B.; Song, Y.; Wang, J.; Guo, Z.; Zhai, J.; et al. Global analysis of iron assimilation and fur regulation in Yersinia pestis. FEMS Microbiol. Lett. 2006, 258, 9–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lesic, B.; Carniel, E. The high pathogenicity island: A broad-host-range pathogenicity island. In Yersinia: Molecular and Cellular Biology; Bioscience Horizons: Norfolk, UK, 2004; pp. 285–306. [Google Scholar]
- Beauchene, N.A.; Mettert, E.L.; Moore, L.J.; Keles, S.; Willey, E.R.; Kiley, P.J. O2 availability impacts iron homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2017, 114, 12261–12266. [Google Scholar] [CrossRef] [Green Version]
- van Vliet, A.H.; Rock, J.D.; Madeleine, L.N.; Ketley, J.M. The iron-responsive regulator Fur of Campylobacter jejuni is expressed from two separate promoters. FEMS Microbiol. Lett. 2000, 188, 115–118. [Google Scholar] [CrossRef]
- De Lorenzo, V.; Herrero, M.; Giovannini, F.; Neilands, J.B. Fur (ferric uptake regulation) protein and CAP (catabolite-activator protein) modulate transcription of fur gene in Escherichia coli. Eur. J. Biochem. 1988, 173, 537–546. [Google Scholar] [CrossRef]
- Zheng, M.; Doan, B.; Schneider, T.D.; Storz, G. OxyR and SoxRS regulation of fur. J. Bacteriol. 1999, 181, 4639–4643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bijlsma, J.J.E.; Waidner, B.; van Vliet, A.H.M.; Hughes, N.J.; Hag, S.; Bereswill, S.; Kelly, D.J.; Vandenbroucke-Grauls, C.M.J.E.; Kist, M.; Kusters, J.G. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect. Immun. 2002, 70, 606–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delany, I.; Spohn, G.; Pacheco, A.-B.F.; Ieva, R.; Alaimo, C.; Rappuoli, R.; Scarlato, V. Autoregulation of Helicobacter pylori Fur revealed by functional analysis of the iron-binding site. Mol. Microbiol. 2002, 46, 1107–1122. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-J.; Bang, S.H.; Lee, K.-H.; Park, S.-J. Positive regulation of fur gene expression via direct interaction of fur in a pathogenic bacterium, Vibrio vulnificus. J. Bacteriol. 2007, 189, 2629–2636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vecerek, B.; Moll, I.; Blasi, U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J. 2007, 26, 965–975. [Google Scholar] [CrossRef] [Green Version]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-Component Signal Transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef] [Green Version]
- Soncini, F.C.; Groisman, E.A. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 1996, 178, 6796–6801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pratt, L.A.; Hsing, W.; Gibson, K.E.; Silhavy, T.J. From acids to osmZ: Multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 1996, 20, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.J.; Inouye, M. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 2002, 277, 24155–24161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenney, L.J. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 2002, 5, 135–141. [Google Scholar] [CrossRef]
- Stincone, A.; Daudi, N.; Rahman, A.S.; Antczak, P.; Henderson, I.; Cole, J.; Johnson, M.D.; Lund, P.; Falciani, F. A systems biology approach sheds new light on Escherichia coli acid resistance. Nucleic Acids Res. 2011, 39, 7512–7528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, H.J.; Cameron, A.D.S.; Dorman, C.J. Bacterial regulon evolution: Distinct responses and roles for the identical OmpR proteins of Salmonella Typhimurium and Escherichia coli in the acid stress response. PLoS Genet. 2014, 10, e1004215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, S.; Mizusaki, H.; Kenney, L.J. A FRET-based DNA biosensor tracks OmpR-dependent acidification of Salmonella during macrophage infection. PLoS Biol. 2015, 13, e1002116. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, S.; Winardhi, R.S.; Morgan, L.K.; Yan, J.; Kenney, L.J. Non-canonical activation of OmpR drives acid and osmotic stress responses in single bacterial cells. Nat. Commun. 2017, 8, 1587. [Google Scholar] [CrossRef]
- Chakraborty, S.; Kenney, L.J. A New Role of OmpR in Acid and Osmotic Stress in Salmonella and E. coli. Front. Microbiol. 2018, 9, 2656. [Google Scholar] [CrossRef] [Green Version]
- Bontemps-Gallo, S.; Fernandez, M.; Dewitte, A.; Raphaël, E.; Gherardini, F.C.; Elizabeth, P.; Koch, L.; Biot, F.; Reboul, A.; Sebbane, F. Nutrient depletion may trigger the Yersinia pestis OmpR-EnvZ regulatory system to promote flea-borne plague transmission. Mol. Microbiol. 2019, 112, 1471–1482. [Google Scholar] [CrossRef]
- Slauch, J.M.; Silhavy, T.J. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J. Mol. Biol. 1989, 210, 281–292. [Google Scholar] [CrossRef]
- Shin, S.; Park, C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 1995, 177, 4696–4702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidal, O.; Longin, R.; Prigent-Combaret, C.; Dorel, C.; Hooreman, M.; Lejeune, P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: Involvement of a new ompR allele that increases curli expression. J. Bacteriol. 1998, 180, 2442–2449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, K.; Nagura, R.; Tanabe, H.; Fujita, N.; Ishihama, A.; Utsumi, R. Negative regulation of the bolA1p of Escherichia coli K-12 by the transcription factor OmpR for osmolarity response genes. FEMS Microbiol. Lett. 2000, 186, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, M.L.; Fontaine, A.; Sansonetti, P.J. The two-component regulatory system ompR-envZ controls the virulence of Shigella flexneri. J. Bacteriol. 1990, 172, 6274–6281. [Google Scholar] [CrossRef] [Green Version]
- Perkins, T.T.; Davies, M.R.; Klemm, E.J.; Rowley, G.; Wileman, T.; James, K.; Keane, T.; Maskell, D.; Hinton, J.C.D.; Dougan, G.; et al. ChIP-seq and transcriptome analysis of the OmpR regulon of Salmonella enterica serovars Typhi and Typhimurium reveals accessory genes implicated in host colonization. Mol. Microbiol. 2013, 87, 526–538. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Zhang, Y.; Han, Y.; Yang, L.; Liu, X.; Guo, Z.; Tan, Y.; Huang, X.; Zhou, D.; Yang, R. Phenotypic and transcriptional analysis of the osmotic regulator OmpR in Yersinia pestis. BMC Microbiol. 2011, 11, 39. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Wang, Y.; Ding, L.; Lu, P.; Atkinson, S.; Chen, S. Positive regulation of flhDC expression by OmpR in Yersinia pseudotuberculosis. Microbiology 2009, 155, 3622–3631. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Lu, P.; Wang, Y.; Ding, L.; Atkinson, S.; Chen, S. OmpR positively regulates urease expression to enhance acid survival of Yersinia pseudotuberculosis. Microbiology 2009, 155, 2522–2531. [Google Scholar] [CrossRef] [Green Version]
- Dorrell, N.; Li, S.R.; Everest, P.H.; Dougan, G.; Wren, B.W. Construction and characterisation of a Yersinia enterocolitica O:8 ompR mutant. FEMS Microbiol. Lett. 1998, 165, 145–151. [Google Scholar] [CrossRef] [Green Version]
- Brzostek, K.; Raczkowska, A.; Zasada, A. The osmotic regulator OmpR is involved in the response of Yersinia enterocolitica O:9 to environmental stresses and survival within macrophages. FEMS Microbiol. Lett. 2003, 228, 265–271. [Google Scholar] [CrossRef] [Green Version]
- Brzostek, K.; Brzostkowska, M.; Bukowska, I.; Karwicka, E.; Raczkowska, A. OmpR negatively regulates expression of invasin in Yersinia enterocolitica. Microbiology 2007, 153, 2416–2425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brzostkowska, M.; Raczkowska, A.; Brzostek, K. OmpR, a response regulator of the two-component signal transduction pathway, influences inv gene expression in Yersinia enterocolitica O9. Front. Cell. Infect. Microbiol. 2012, 2, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skorek, K.; Raczkowska, A.; Dudek, B.; Miętka, K.; Guz-Regner, K.; Pawlak, A.; Klausa, E.; Bugla-Płoskońska, G.; Brzostek, K. Regulatory protein OmpR influences the serum resistance of Yersinia enterocolitica O:9 by modifying the structure of the outer membrane. PLoS ONE 2013, 8, e79525. [Google Scholar] [CrossRef] [PubMed]
- Raczkowska, A.; Trzos, J.; Lewandowska, O.; Nieckarz, M.; Brzostek, K. Expression of the AcrAB Components of the AcrAB-TolC Multidrug Efflux Pump of Yersinia enterocolitica Is Subject to Dual Regulation by OmpR. PLoS ONE 2015, 10, e0124248. [Google Scholar] [CrossRef] [PubMed]
- Nieckarz, M.; Raczkowska, A.; Jaworska, K.; Stefańska, E.; Skorek, K.; Stosio, D.; Brzostek, K. The Role of OmpR in the Expression of Genes of the KdgR Regulon Involved in the Uptake and Depolymerization of Oligogalacturonides in Yersinia enterocolitica. Front. Cell. Infect. Microbiol. 2017, 7, 366. [Google Scholar] [CrossRef]
- Raczkowska, A.; Skorek, K.; Bielecki, J.; Brzostek, K. OmpR controls Yersinia enterocolitica motility by positive regulation of flhDC expression. Antonie Van Leeuwenhoek 2011, 99, 381–394. [Google Scholar] [CrossRef] [Green Version]
- Nieckarz, M.; Raczkowska, A.; Debski, J.; Kistowski, M.; Dadlez, M.; Heesemann, J.; Rossier, O.; Brzostek, K. Impact of OmpR on the membrane proteome of Yersinia enterocolitica in different environments: Repression of major adhesin YadA and heme receptor HemR. Environ. Microbiol. 2016, 18, 997–1021. [Google Scholar] [CrossRef]
- Solovyev, V.; Salamov, A. Automatic Annotation of Microbial Genomes and Metagenomic Sequences. In Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies; Nova Science Publishers, Inc.: New York, NY, USA, 2011; pp. 61–78. [Google Scholar]
- Maeda, S.; Takayanagi, K.; Nishimura, Y.; Maruyama, T.; Sato, K.; Mizuno, T. Activation of the osmoregulated ompC gene by the OmpR protein in Escherichia coli: A study involving synthetic OmpR-binding sequences. J. Biochem. 1991, 110, 324–327. [Google Scholar] [CrossRef] [Green Version]
- Lucas, R.L.; Lee, C.A. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2001, 183, 2733–2745. [Google Scholar] [CrossRef] [Green Version]
- Cameron, A.D.S.; Dorman, C.J. A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet. 2012, 8, e1002615. [Google Scholar] [CrossRef] [PubMed]
- de Lorenzo, V.; Wee, S.; Herrero, M.; Neilands, J.B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 1987, 169, 2624–2630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keseler, I.M.; Mackie, A.; Santos-Zavaleta, A.; Billington, R.; Bonavides-Martínez, C.; Caspi, R.; Fulcher, C.; Gama-Castro, S.; Kothari, A.; Krummenacker, M.; et al. The EcoCyc database: Reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2016, 45, D543–D550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oshima, T.; Aiba, H.; Masuda, Y.; Kanaya, S.; Sugiura, M.; Wanner, B.L.; Mori, H.; Mizuno, T. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 2002, 46, 281–291. [Google Scholar] [CrossRef]
- Nieckarz, M.; Kaczor, P.; Jaworska, K.; Raczkowska, A.; Brzostek, K. Urease Expression in Pathogenic Yersinia enterocolitica Strains of Bio-Serotypes 2/O:9 and 1B/O:8 Is Differentially Regulated by the OmpR Regulator. Front. Microbiol. 2020, 11, 607. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S.C.; Robinson, A.K.; Rodriguez-Quinones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Perry, R.D.; Fetherston, J.D. Yersiniabactin iron uptake: Mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect. 2011, 13, 808–817. [Google Scholar] [CrossRef] [Green Version]
- Achenbach, L.A.; Yang, W. The fur gene from Klebsiella pneumoniae: Characterization, genomic organization and phylogenetic analysis. Gene 1997, 185, 201–207. [Google Scholar] [CrossRef]
- Delany, I.; Ieva, R.; Alaimo, C.; Rappuoli, R.; Scarlato, V. The iron-responsive regulator fur is transcriptionally autoregulated and not essential in Neisseria meningitidis. J. Bacteriol. 2003, 185, 6032–6041. [Google Scholar] [CrossRef] [Green Version]
- Lowe, C.A.; Asghar, A.H.; Shalom, G.; Shaw, J.G.; Thomas, M.S. The Burkholderia cepacia fur gene: Co-localization with omLA and absence of regulation by iron. Microbiology 2001, 147, 1303–1314. [Google Scholar] [CrossRef] [Green Version]
- Russo, F.D.; Silhavy, T.J. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 1991, 222, 567–580. [Google Scholar] [CrossRef]
- Head, C.G.; Tardy, A.; Kenney, L.J. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J. Mol. Biol. 1998, 281, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Owen-Hughes, T.A.; Pavitt, G.D.; Santos, D.S.; Sidebotham, J.M.; Hulton, C.S.; Hinton, J.C.; Higgins, C.F. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell 1992, 71, 255–265. [Google Scholar] [CrossRef]
- Lucht, J.M.; Dersch, P.; Kempf, B.; Bremer, E. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J. Biol. Chem. 1994, 269, 6578. [Google Scholar] [CrossRef]
- Puente, J.L.; Verdugo-Rodríguez, A.; Calva, E. Expression of Salmonella typhi and Escherichia coli OmpC is influenced differently by medium osmolarity; dependence on Escherichia coli OmpR. Mol. Microbiol. 1991, 5, 1205–1210. [Google Scholar] [CrossRef]
- Martínez-Flores, I.; Cano, R.; Bustamante, V.H.; Calva, E.; Puente, J.L. The ompB operon partially determines differential expression of OmpC in Salmonella typhi and Escherichia coli. J. Bacteriol. 1999, 181, 556–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goh, E.-B.; Siino, D.F.; Igo, M.M. The Escherichia coli tppB (ydgR) gene represents a new class of OmpR-regulated genes. J. Bacteriol. 2004, 186, 4019–4024. [Google Scholar] [CrossRef] [Green Version]
- Argandoña, M.; Nieto, J.J.; Iglesias-Guerra, F.; Calderón, M.I.; García-Estepa, R.; Vargas, C. Interplay between iron homeostasis and the osmotic stress response in the halophilic bacterium Chromohalobacter salexigens. Appl. Environ. Microbiol. 2010, 76, 3575–3589. [Google Scholar] [CrossRef] [Green Version]
- Higashitani, A.; Nishimura, Y.; Hara, H.; Aiba, H.; Mizuno, T.; Horiuchi, K. Osmoregulation of the fatty acid receptor gene fadL in Escherichia coli. Mol. Gen. Genet. 1993, 240, 339–347. [Google Scholar] [CrossRef]
- Bang, I.S.; Audia, J.P.; Park, Y.K.; Foster, J.W. Autoinduction of the ompR response regulator by acid shock and control of the Salmonella enterica acid tolerance response. Mol. Microbiol. 2002, 44, 1235–1250. [Google Scholar] [CrossRef]
- Rentschler, A.E.; Lovrich, S.D.; Fitton, R.; Enos-Berlage, J.; Schwan, W.R. OmpR regulation of the uropathogenic Escherichia coli fimB gene in an acidic/high osmolality environment. Microbiology 2013, 159, 316–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, S.W.; Kim, D.; Szubin, R.; Palsson, B.O. Genome-wide Reconstruction of OxyR and SoxRS Transcriptional Regulatory Networks under Oxidative Stress in Escherichia coli K-12 MG1655. Cell Rep. 2015, 12, 1289–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loprasert, S.; Sallabhan, R.; Whangsuk, W.; Mongkolsuk, S. Characterization and mutagenesis of fur gene from Burkholderia pseudomallei. Gene 2000, 254, 129–137. [Google Scholar] [CrossRef]
- Wei, Q.; Le Minh, P.N.; Dötsch, A.; Hildebrand, F.; Panmanee, W.; Elfarash, A.; Schulz, S.; Plaisance, S.; Charlier, D.; Hassett, D.; et al. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 2012, 40, 4320–4333. [Google Scholar] [CrossRef] [PubMed]
- Nouaille, S.; Mondeil, S.; Finoux, A.-L.; Moulis, C.; Girbal, L.; Cocaign-Bousquet, M. The stability of an mRNA is influenced by its concentration: A potential physical mechanism to regulate gene expression. Nucleic Acids Res. 2017, 45, 11711–11724. [Google Scholar] [CrossRef] [Green Version]
- Bernhardt, H.S.; Tate, W.P. Primordial soup or vinaigrette: Did the RNA world evolve at acidic pH? Biol. Direct 2012, 7, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marinho, H.S.; Real, C.; Cyrne, L.; Soares, H.; Antunes, F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014, 2, 535–562. [Google Scholar] [CrossRef] [Green Version]
- Pratt, L.A.; Silhavy, T.J. Identification of base pairs important for OmpR-DNA interaction. Mol. Microbiol. 1995, 17, 565–573. [Google Scholar] [CrossRef]
- Shimada, T.; Takada, H.; Yamamoto, K.; Ishihama, A. Expanded roles of two-component response regulator OmpR in Escherichia coli: Genomic SELEX search for novel regulation targets. Genes Cells 2015, 20, 915–931. [Google Scholar] [CrossRef]
- Griffith, K.L.; Wolf, R.E.J. Systematic mutagenesis of the DNA binding sites for SoxS in the Escherichia coli zwf and fpr promoters: Identifying nucleotides required for DNA binding and transcription activation. Mol. Microbiol. 2001, 40, 1141–1154. [Google Scholar] [CrossRef]
- Dunn, T.M.; Hahn, S.; Ogden, S.; Schleif, R.F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: Addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc. Natl. Acad. Sci. USA 1984, 81, 5017–5020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortuno, M.J.; Lawther, R.P. Effect of the deletion of upstream DNA sequences on expression from the ilvGp2 promoter of the ilvGMEDA operon of Escherichia coli K-12. Nucleic Acids Res. 1987, 15, 1521–1542. [Google Scholar] [CrossRef] [Green Version]
- Rampersaud, A.; Norioka, S.; Inouye, M. Characterization of OmpR binding sequences in the upstream region of the ompF promoter essential for transcriptional activation. J. Biol. Chem. 1989, 264, 18693–18700. [Google Scholar] [CrossRef]
- Rampersaud, A.; Harlocker, S.L.; Inouye, M. The OmpR protein of Escherichia coli binds to sites in the ompF promoter region in a hierarchical manner determined by its degree of phosphorylation. J. Biol. Chem. 1994, 269, 12559–12566. [Google Scholar] [CrossRef]
- Harlocker, S.L.; Bergstrom, L.; Inouye, M. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J. Biol. Chem. 1995, 270, 26849–26856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.J.; Lan, C.Y.; Igo, M.M. Phosphorylation stimulates the cooperative DNA-binding properties of the transcription factor OmpR. Proc. Natl. Acad. Sci. USA 1997, 94, 2828–2832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergstrom, L.C.; Qin, L.; Harlocker, S.L.; Egger, L.A.; Inouye, M. Hierarchical and co-operative binding of OmpR to a fusion construct containing the ompC and ompF upstream regulatory sequences of Escherichia coli. Genes Cells 1998, 3, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Gerstel, U.; Park, C.; RömLing, U. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 2003, 49, 639–654. [Google Scholar] [CrossRef] [Green Version]
- Jubelin, G.; Vianney, A.; Beloin, C.; Ghigo, J.-M.; Lazzaroni, J.-C.; Lejeune, P.; Dorel, C. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 2005, 187, 2038–2049. [Google Scholar] [CrossRef] [Green Version]
- Perini, L.T.; Doherty, E.A.; Werner, E.; Senear, D.F. Multiple specific CytR binding sites at the Escherichia coli deoP2 promoter mediate both cooperative and competitive interactions between CytR and cAMP receptor protein. J. Biol. Chem. 1996, 271, 33242–33255. [Google Scholar] [CrossRef] [Green Version]
- Hantke, K. Is the bacterial ferrous iron transporter FeoB a living fossil? Trends Microbiol. 2003, 11, 192–195. [Google Scholar] [CrossRef]
- Perry, R.D.; Mier, I.J.; Fetherston, J.D. Roles of the Yfe and Feo transporters of Yersinia pestis in iron uptake and intracellular growth. BioMetals 2007, 20, 699–703. [Google Scholar] [CrossRef] [PubMed]
- Kanaujia, P.K.; Bajaj, P.; Virdi, J.S. Analysis of iron acquisition and storage-related genes in clinical and non-clinical strains of Yersinia enterocolitica biovar 1A. APMIS 2015, 123, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.D.; Pettis, G.S.; McIntosh, M.A. Promoter and operator determinants for fur-mediated iron regulation in the bidirectional fepA-fes control region of the Escherichia coli enterobactin gene system. J. Bacteriol. 1994, 176, 3944–3955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angerer, A.; Braun, V. Iron regulates transcription of the Escherichia coli ferric citrate transport genes directly and through the transcription initiation proteins. Arch. Microbiol. 1998, 169, 483–490. [Google Scholar] [CrossRef]
- Noinaj, N.; Guillier, M.; Barnard, T.J.; Buchanan, S.K. TonB-dependent transporters: Regulation, structure, and function. Annu. Rev. Microbiol. 2010, 64, 43–60. [Google Scholar] [CrossRef] [Green Version]
- Miethke, M.; Marahiel, M.A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef] [Green Version]
- Guillier, M.; Gottesman, S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006, 59, 231–247. [Google Scholar] [CrossRef]
- Fernandez-Beros, M.E.; Gonzalez, C.; McIntosh, M.A.; Cabello, F.C. Immune response to the iron-deprivation-induced proteins of Salmonella typhi in typhoid fever. Infect. Immun. 1989, 57, 1271–1275. [Google Scholar] [CrossRef] [Green Version]
- Gerken, H.; Vuong, P.; Soparkar, K.; Misra, R. Roles of the EnvZ/OmpR Two-Component System and Porins in Iron Acquisition in Escherichia coli. mBio 2020, 11. [Google Scholar] [CrossRef]
- Troxell, B.; Hassan, H.M. Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 2013, 3, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambrook, J.; Russel, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Marx, C.J.; Lidstrom, M.E. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 2001, 147, 2065–2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory: Cold Spring Harbour, NY, USA, 1992; ISBN 0-87969-106-9. [Google Scholar]
- Philippe, N.; Alcaraz, J.-P.; Coursange, E.; Geiselmann, J.; Schneider, D. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 2004, 51, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Kędzierska, S.; Chesnokova, L.S.; Witt, S.N.; Zolkiewski, M. Interactions within the ClpB/DnaK bi-chaperone system from Escherichia coli. Arch. Biochem. Biophys. 2005, 444, 61–65. [Google Scholar] [CrossRef]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaworska, K.; Ludwiczak, M.; Murawska, E.; Raczkowska, A.; Brzostek, K. The Regulator OmpR in Yersinia enterocolitica Participates in Iron Homeostasis by Modulating Fur Level and Affecting the Expression of Genes Involved in Iron Uptake. Int. J. Mol. Sci. 2021, 22, 1475. https://doi.org/10.3390/ijms22031475
Jaworska K, Ludwiczak M, Murawska E, Raczkowska A, Brzostek K. The Regulator OmpR in Yersinia enterocolitica Participates in Iron Homeostasis by Modulating Fur Level and Affecting the Expression of Genes Involved in Iron Uptake. International Journal of Molecular Sciences. 2021; 22(3):1475. https://doi.org/10.3390/ijms22031475
Chicago/Turabian StyleJaworska, Karolina, Marta Ludwiczak, Emilia Murawska, Adrianna Raczkowska, and Katarzyna Brzostek. 2021. "The Regulator OmpR in Yersinia enterocolitica Participates in Iron Homeostasis by Modulating Fur Level and Affecting the Expression of Genes Involved in Iron Uptake" International Journal of Molecular Sciences 22, no. 3: 1475. https://doi.org/10.3390/ijms22031475
APA StyleJaworska, K., Ludwiczak, M., Murawska, E., Raczkowska, A., & Brzostek, K. (2021). The Regulator OmpR in Yersinia enterocolitica Participates in Iron Homeostasis by Modulating Fur Level and Affecting the Expression of Genes Involved in Iron Uptake. International Journal of Molecular Sciences, 22(3), 1475. https://doi.org/10.3390/ijms22031475





